Long COVID and Biomarker Dysregulation—A Shift Toward Immune Exhaustion?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. The Follow-Up at Three and Six Months After Acute COV Infection
2.3. Profiling of Plasma Inflammation-Related Biomarkers by Olink® Extension Proximity Assay
2.4. Data Management and Ethics Approval
2.5. Statistical Analysis
3. Results
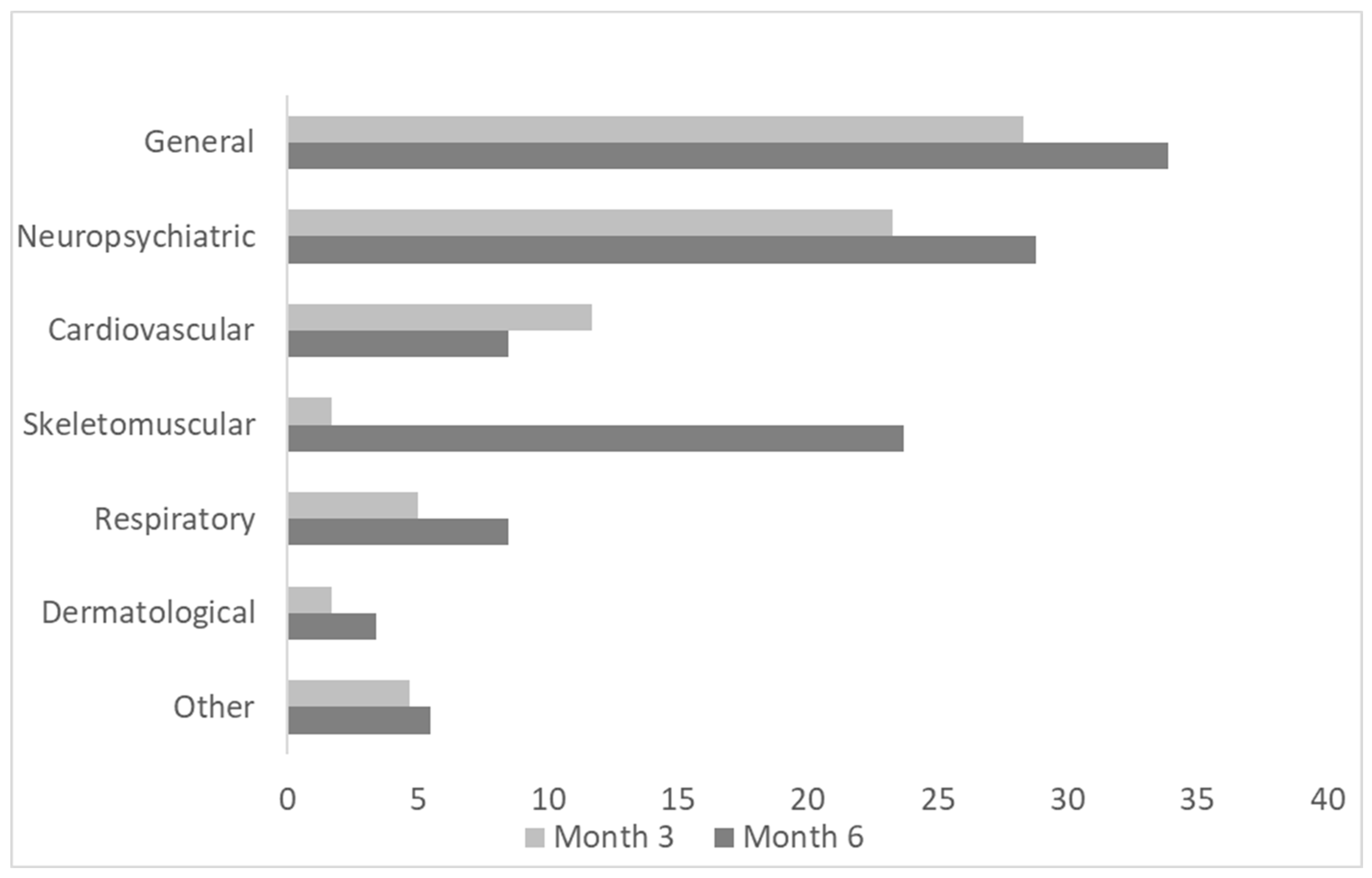
3.1. Demographics, Clinical Features, and Long-Term Symptoms at Follow-Ups
3.2. Routine Laboratory Biomarkers at Follow-Ups of Non-COV, LC, and R Patients
3.3. Univariant Analysis of Inflammatory-Related Biomarkers
3.3.1. Post-Infection Plasma Proteomic Profiles of COV Patients Compared to Non-COV Patients
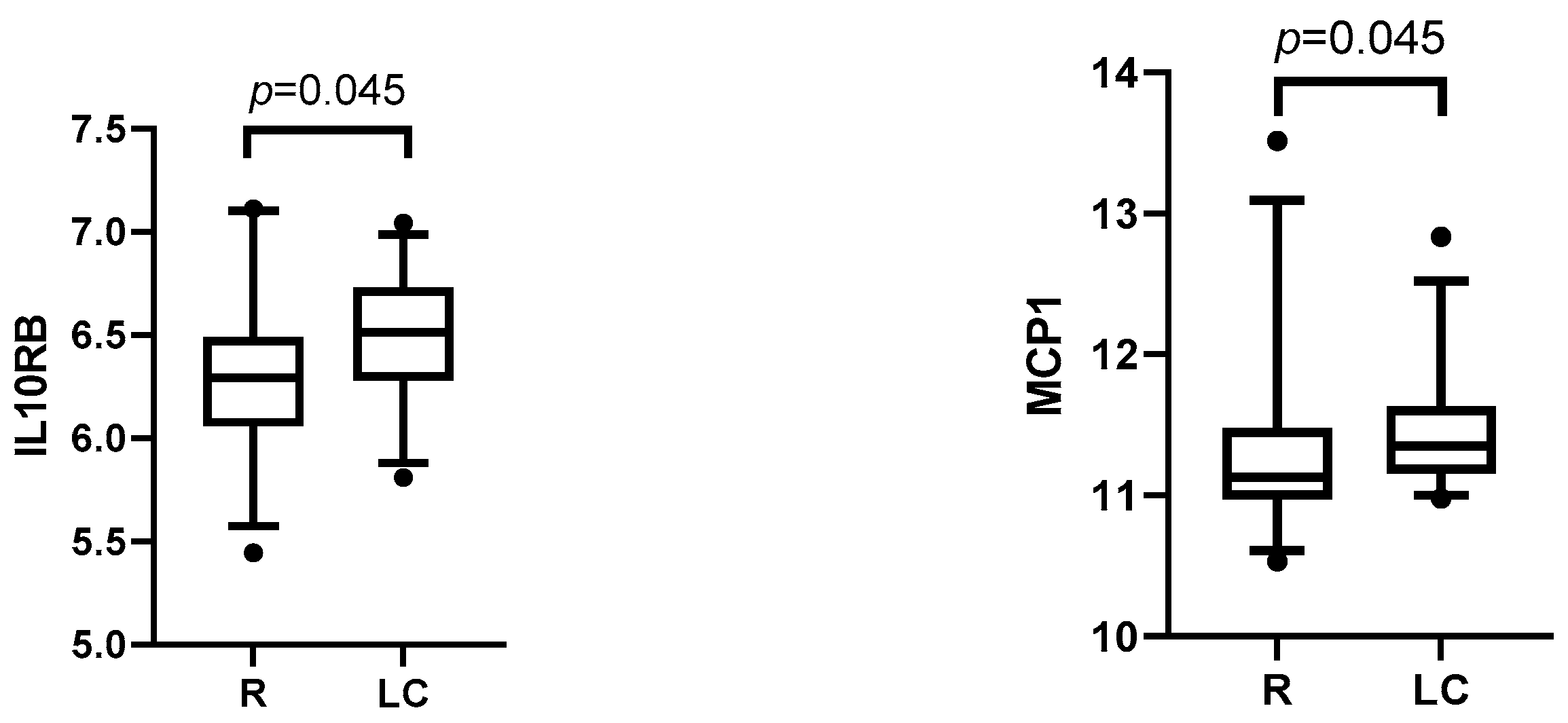
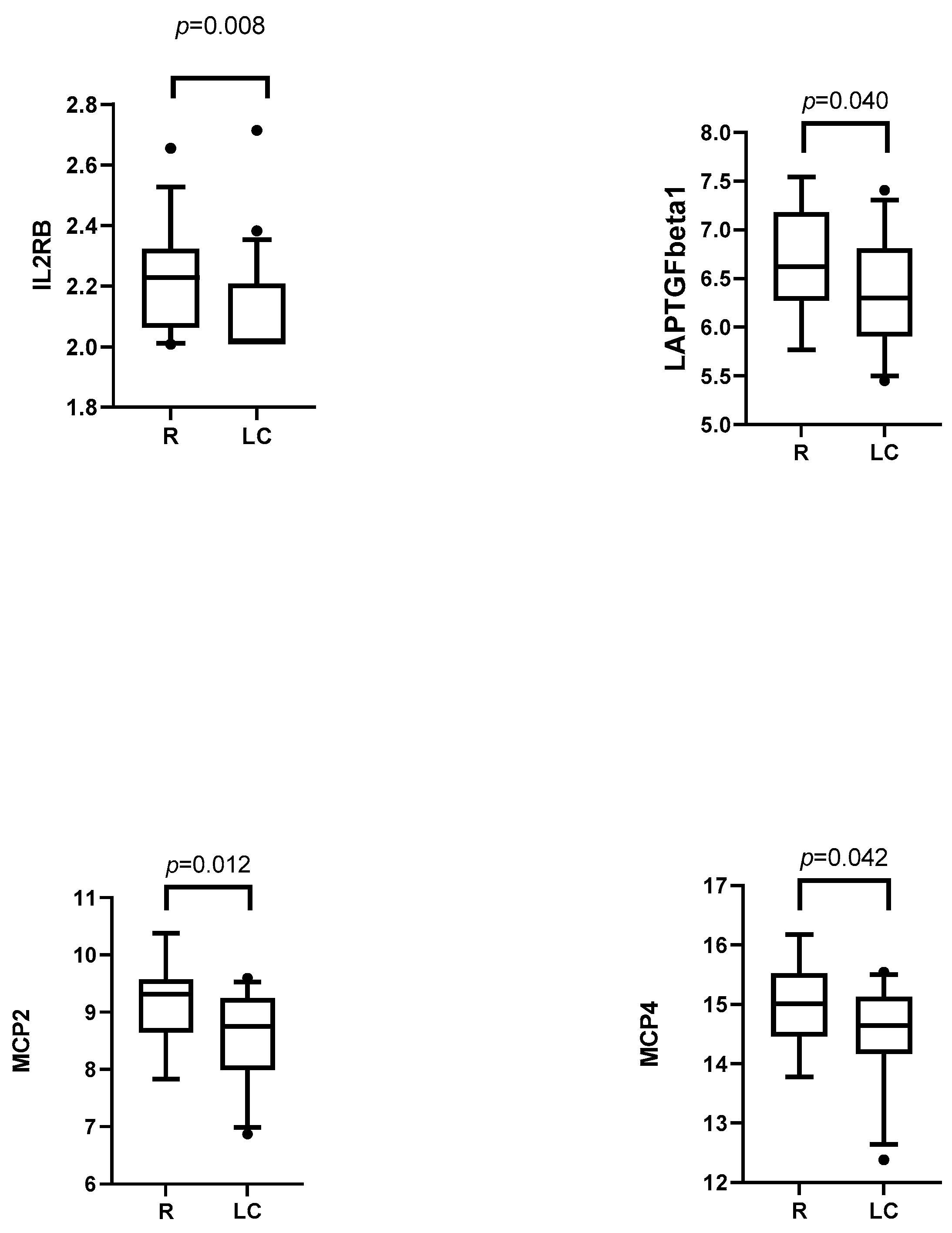
3.3.2. Post-Infection Plasma Proteomic Profiles of LC Compared to R Patients
3.4. Multivariant Analysis of Inflammatory-Related Biomarkers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. A Clinical Casedefinition of Post COVID-19 Condition by a Delphiconsensus; WHO: Geneva, Switzerland, 2021. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 (accessed on 25 April 2025).
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Shetty, A.; Delanerolle, G.; Zeng, Y.; Zhang, Y.; Raymont, V.; Rathod, S.; Halabi, S.; Elliot, K.; Shi, J.Q.; et al. A systematic review and meta-analysis of long COVID symptoms. Syst. Rev. 2023, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Perumal, R.; Shunmugam, L.; Naidoo, K.; Karim, S.S.A.; Wilkins, D.; Garzino-Demo, A.; Brechot, C.; Parthasarathy, S.; Vahlne, A.; Nikolich, J.Ž. Long COVID: A review and proposed visualization of the complexity of long COVID. Front. Immunol. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Zhao, Y.; Zeng, N.; Liu, X.; Zheng, Y.; Sun, J.; Zhong, Y.; Wu, S.; Ni, S.; Gong, Y.; et al. Epidemiology, clinical presentation, pathophysiology, and management of long COVID: An update. Mol. Psychiatry 2023, 28, 4056–4069. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 698169. [Google Scholar] [CrossRef]
- Tsilingiris, D.; Vallianou, N.G.; Karampela, I.; Christodoulatos, G.S.; Papavasileiou, G.; Petropoulou, D.; Magkos, F.; Dalamaga, M. Laboratory Findings and Biomarkers in Long COVID: What Do We Know So Far? Insights into Epidemiology, Pathogenesis, Therapeutic Perspectives and Challenges. Int. J. Mol. Sci. 2023, 24, 10458. [Google Scholar] [CrossRef]
- Lai, Y.-J.; Liu, S.-H.; Manachevakul, S.; Lee, T.-A.; Kuo, C.-T.; Bello, D. Biomarkers in long COVID-19: A systematic review. Front. Med. 2023, 10, 1085988. [Google Scholar] [CrossRef]
- Peluso, M.J.; Lu, S.; Tang, A.F.; Durstenfeld, M.S.; Ho, H.-E.; Goldberg, S.A.; Forman, C.A.; Munter, S.E.; Hoh, R.; Tai, V.; et al. Markers of Immune Activation and Inflammation in Individuals with Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2021, 224, 1839–1848. [Google Scholar] [CrossRef]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef]
- Zhao, J.; Schank, M.; Wang, L.; Dang, X.; Cao, D.; Khanal, S.; Nguyen, L.N.; Zhang, Y.; Wu, X.Y.; Adkins, J.L.; et al. Plasma biomarkers for systemic inflammation in COVID-19 survivors. Proteom.–Clin. Appl. 2022, 16, e2200031. [Google Scholar] [CrossRef]
- Williams, E.S.; Martins, T.B.; Shah, K.S.; Hill, H.R.; Coiras, M.; Spivak, A.M.; Planelles, V. Cytokine Deficiencies in Patients with Long-COVID. J. Clin. Cell. Immunol. 2022, 13, 672. [Google Scholar] [PubMed]
- Kallaste, A.; Kisand, K.; Aart, A.; Peterson, P.; Lember, M. Antibody levels remain high to one-year’s follow-up after moderate and severe COVID-19, but not after mild cases. Infect. Dis. 2021, 54, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Choi, T.; Al-Aly, Z. Long-term outcomes following hospital admission for COVID-19 versus seasonal influenza: A cohort study. Lancet Infect. Dis. 2023, 24, 239–255. [Google Scholar] [CrossRef]
- Pacheco-Hernández, L.M.; Ramírez-Noyola, J.A.; Gómez-García, I.A.; Ignacio-Cortés, S.; Zúñiga, J.; Choreño-Parra, J.A. Comparing the Cytokine Storms of COVID-19 and Pandemic Influenza. J. Interf. Cytokine Res. 2022, 42, 369–392. [Google Scholar] [CrossRef]
- Espín, E.; Yang, C.; Shannon, C.P.; Assadian, S.; He, D.; Tebbutt, S.J. Cellular and molecular biomarkers of long COVID: A scoping review. eBioMedicine 2023, 91, 104552. [Google Scholar] [CrossRef]
- Ahn, D.; Prince, A. Participation of the IL-10RB Related Cytokines, IL-22 and IFN-λ in Defense of the Airway Mucosal Barrier. Front. Cell. Infect. Microbiol. 2020, 10, 300. [Google Scholar] [CrossRef]
- Islam, H.; Chamberlain, T.C.; Mui, A.L.; Little, J.P. Elevated Interleukin-10 Levels in COVID-19: Potentiation of Pro-Inflammatory Responses or Impaired Anti-Inflammatory Action? Front. Immunol. 2021, 12, 677008. [Google Scholar] [CrossRef]
- Voloudakis, G.; Vicari, J.M.; Venkatesh, S.; Hoffman, G.E.; Dobrindt, K.; Zhang, W.; Beckmann, N.D.; Higgins, C.A.; Argyriou, S.; Jiang, S.; et al. A translational genomics approach identifies IL10RB as the top candidate gene target for COVID-19 susceptibility. npj Genom. Med. 2022, 7, 52. [Google Scholar] [CrossRef]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101, 107598. [Google Scholar] [CrossRef]
- Ma, A.; Zhang, L.; Ye, X.; Chen, J.; Yu, J.; Zhuang, L.; Weng, C.; Petersen, F.; Wang, Z.; Yu, X. High Levels of Circulating IL-8 and Soluble IL-2R Are Associated with Prolonged Illness in Patients with Severe COVID-19. Front. Immunol. 2021, 12, 626235. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Liu, C.; Su, L.; Zhang, D.; Fan, J.; Yang, Y.; Xiao, M.; Xie, J.; Xu, Y.; et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020, 26, 97. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Shahbaz, S.; Osman, M.; Redmond, D.; Bozorgmehr, N.; Rosychuk, R.J.; Lam, G.; Sligl, W.; Tervaert, J.W.C.; Elahi, S. Diverse immunological dysregulation, chronic inflammation, and impaired erythropoiesis in long COVID patients with chronic fatigue syndrome. J. Autoimmun. 2024, 147, 103267. [Google Scholar] [CrossRef] [PubMed]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dai, F.; Wang, L.; Sun, Y.; Mei, L.; Ran, Y.; Ye, F. CCL13 and human diseases. Front. Immunol. 2023, 14, 1176639. [Google Scholar] [CrossRef]
- Abbas, A.K.; Trotta, E.; Simeonov, D.R.; Marson, A.; Bluestone, J.A. Revisiting IL-2: Biology and therapeutic prospects. Sci. Immunol. 2018, 3, eaat1482. [Google Scholar] [CrossRef]
- Chang, Y.; Bai, M.; You, Q. Associations between Serum Interleukins (IL-1β, IL-2, IL-4, IL-6, IL-8, and IL-10) and Disease Severity of COVID-19: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2022, 2022, 2755246. [Google Scholar] [CrossRef]
- Arguinchona, L.M.; Zagona-Prizio, C.; Joyce, M.E.; Chan, E.D.; Maloney, J.P. Microvascular significance of TGF-β axis activation in COVID-19. Front. Cardiovasc. Med. 2023, 9, 1054690. [Google Scholar] [CrossRef]
- Patterson, B.K.; Guevara-Coto, J.; Yogendra, R.; Francisco, E.B.; Long, E.; Pise, A.; Rodrigues, H.; Parikh, P.; Mora, J.; Mora-Rodríguez, R.A. Immune-Based Prediction of COVID-19 Severity and Chronicity Decoded Using Machine Learning. Front. Immunol. 2021, 12, 700782. [Google Scholar] [CrossRef]
- Colarusso, C.; Maglio, A.; Terlizzi, M.; Vitale, C.; Molino, A.; Pinto, A.; Vatrella, A.; Sorrentino, R. Post-COVID-19 Patients Who Develop Lung Fibrotic-like Changes Have Lower Circulating Levels of IFN-β but Higher Levels of IL-1α and TGF-β. Biomedicines 2021, 9, 1931. [Google Scholar] [CrossRef]
- Dulfer, E.A.; Joosten, L.A.; Netea, M.G. Enduring echoes: Post-infectious long-term changes in innate immunity. Eur. J. Intern. Med. 2023, 123, 15–22. [Google Scholar] [CrossRef]
- Rigo, S.; Barbic, F.; Khalaf, K.; Bisoglio, A.; Pani, M.; Minonzio, M.; Rinaldi, L.; Ciccarelli, M.; Bordoni, M.G.; Verzeletti, P.; et al. The Long-COVID autonomic syndrome in hospitalized patients: A one-year prospective cohort study. Eur. J. Intern. Med. 2023, 120, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y. The uPA/uPAR System Orchestrates the Inflammatory Response, Vascular Homeostasis, and Immune System in Fibrosis Progression. Int. J. Mol. Sci. 2023, 24, 1796. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, E.; Vlok, M.; Venter, C.; Bezuidenhout, J.A.; Laubscher, G.J.; Steenkamp, J.; Kell, D.B. Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 2021, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Appelman, B.; Charlton, B.T.; Goulding, R.P.; Kerkhoff, T.J.; Breedveld, E.A.; Noort, W.; Offringa, C.; Bloemers, F.W.; van Weeghel, M.; Schomakers, B.V.; et al. Muscle abnormalities worsen after post-exertional malaise in long COVID. Nat. Commun. 2024, 15, 17. [Google Scholar] [CrossRef]



| COV (n = 60) | Non-COV (n = 45) | p-Value * | |
|---|---|---|---|
| Age, mean (SD) (years) | 56.9 (16.4) | 53.8 (13.9) | 0.403 |
| Male gender, n (%) | 28 (46.7) | 22 (48.9) | 0.845 |
| BMI, mean (SD) | 28.9 (4.3) | 29.6 (6.3) | 0.95 |
| Chronic diseases and comorbidities | |||
| Any comorbidity, n (%) | 37 (61) | 32 (71) | 0.407 |
| Hypertension, n (%) | 21 (35) | 16 (36) | 1.0 |
| COPD, n (%) | 2 (3.3) | 1 (2.2) | 1.0 |
| Asthma, n (%) | 3 (5) | 2 (4.4) | 1.0 |
| Diabetes, n (%) | 5 (8.3) | 3 (6.7) | 1.0 |
| Coronary artery disease, n (%) | 7 (11.7) | 3 (6.7) | 0.510 |
| Cerebrovascular disease, n (%) | 2 (3.3) | 1 (2.2) | 1.0 |
| Tumor, n (%) | 1 (1.6) | 4 (8.8) | 0.389 |
| At Month 3 | At Month 6 | |||||
|---|---|---|---|---|---|---|
| R (n = 28) | LC (n = 27) | p-Value ** | R (n = 18) | LC (n = 28) | p-Value ** | |
| Age, mean (SD) (years) | 56 (17) | 56 (15) | 0.933 | 59 (17) | 62 (16) | 0.547 |
| Male gender, n (%) | 13 (46) | 17 (63) | 0.282 | 9 (50) | 14 (50) | 1.000 |
| BMI, mean (SD) | 28.1 (4.6) | 30.1 (4.1) | 0.069 | 28.3 (4.8) | 29.8 (4.5) | 0.291 |
| Chronic diseases and comorbidities | ||||||
| Any comorbidity, n (%) | 17 (60.7) | 16 (59.3) | 1.000 | 9 (50) | 22 (78.6) | 0.046 |
| Hypertension, n (%) | 9 (32.1) | 8 (29.6) | 1.000 | 5 (27.8) | 16 (57.1) | 0.072 |
| COPD, n (%) | 1 (3.6) | 1 (3.7) | 1.000 | 0 | 2 (7.1) | 0.513 |
| Asthma, n (%) | 2 (7.1) | 1 (3.7) | 1.000 | 1 (5.6) | 2 (7.1) | 1.000 |
| Diabetes, n (%) | 2 (7.1) | 3 (11.1) | 0.669 | 0 | 5 (17.9) | 0.140 |
| Coronary artery disease, n (%) | 4 (14.3) | 2 (7.4) | 0.669 | 2 (11.1) | 5 (17.9) | 0.688 |
| Cerebrovascular disease, n (%) | 2 (7.1) | 0 | 0.491 | 0 | 2 (7.1) | 0.513 |
| Tumor, n (%) | 0 | 4 (14.8) | 0.051 | 1 (5.6) | 3 (10.7) | 1.000 |
| Treatment in acute phase | ||||||
| Hospitalized, n (%) | 11 (39.3) | 16 (59.3) | 0.181 | 6 (33.3) | 19 (67.9) | 0.034 |
| Parenteral antibiotics, n (%) | 6 (21.4) | 12 (44.4) | 0.089 | 3 (16.7) | 13 (46.4) | 0.058 |
| Hydroxychloroquine, n (%) | 9 (32.1) | 13 (48.1) | 0.277 | 3 (16.7) | 17 (60.7) | 0.006 |
| Glucocorticoids, n (%) | 0 | 1 (3.7) | 0.491 | 0 | 1 (3.6) | 1.000 |
| Supplemental oxygen, n (%) | 10 (35.7) | 16 (59.3) | 0.108 | 5 (27.8) | 20 (71.4) | 0.006 |
| NIV, n (%) | 1 (3.6) | 6 (22.2) | 0.051 | 0 | 5 (17.9) | 0.140 |
| Invasive ventilation, n (%) | 0 | 6 (22.2) | 0.010 | 0 | 3 (10.7) | 0.270 |
| ICU admission, n (%) | 2(7.1) | 7 (25.9) | 0.078 | 1 (5) | 5 (17.9) | 0.380 |
| WHO group, n (%): | ||||||
| Mild | 8 (28.6) | 5 (18.6) | 0.528 | 7 (38.9) | 4 (14.3) | 0.08 |
| Moderate | 11 (39.3) | 6 (22.2) | 0.245 | 7 (38.9) | 4 (14.3) | 0.08 |
| Severe | 9 (32.1) | 9 (33.3) | 1.000 | 4 (22.2) | 16 (57.1) | 0.032 |
| Critical | 0 | 7 (25.9) | 0.004 | 0 | 4 (14.3) | 0.144 |
| Acute complications | ||||||
| Pneumonia, n (%) | 10 (35.7) | 16 (59.3) | 0.108 | 5 (27.8) | 20 (71.4) | 0.006 |
| Respiratory failure, n (%) | 8 (28.6) | 13 (48.1) | 0.112 | 4 (22.2) | 16 (57.1) | 0.032 |
| At Month 3 | At Month 6 | |||||||
|---|---|---|---|---|---|---|---|---|
| Laboratory Biomarkers, Median (IQR) | Non-COV (n = 36) | R (n = 28) | LC (n = 27) | p-Value ** | Non-COV *** (n = 36) | R (n = 18) | LC (n = 28) | p-Value ** |
| Leukocytes (E9/L) | 5.8 (4.9–7.0) | 5.7 (5.0–6.7) | 6.4 (4.8–7.0) | 0.482 | 5.8 (4.9–7.0) | 6.2 (5.4–6.9) | 6.0 (5.3–7.4) | 0.59 |
| Lymphocytes (E9/L) | 1.9 (1.6–2.1) | 1.9 (1.7–2.3) | 2.0 (1.6–2.6) | 0.640 | 1.9 (1.6–2.1) | 2.2 (1.5–2.6) | 2.1 (1.7–2.6) | 0.763 |
| Neutrophils (E9/L) | 3.2 (2.5–3.8) | 3.0 (2.6–3.8) | 3.2 (2.4–3.8) | 0.869 | 3.2 (2.5–3.8) | 3.4 (2.7–4.0) | 3.3 (3.0–4.2) | 0.763 |
| Thrombocytes (E9/L) | 217 (190–238) | 236 (208–278) | 242 (216–281) | 0.094 | 217 (190–238) | 250 (208–278) | 228 (204–261) | 0.131 |
| CRP (mg/L) | 1.5 (1–3.75) | 1.0 (1.0–3.0) | 1.0 (1.0–2.0) | 0.433 | 1.5 (1–3.75) | 2.0 (1.0–3.0) | 1.0 (1.0–2.8) | 0.128 |
| ALT (U/L) | 17.5 (12–23.75) | 23 (15–32) | 26 (19–34) | 0.028 **** | 17.5 (12–23.75) | 25 (17–38) | 21 (16–27) | 0.114 |
| Ferritin (µg/L) | 160 (75–245) | 140 (95–220) | 92 (56–234) | 0.302 | 160 (75–245) | 161 (70–204) | 129 (74–181) | 0.365 |
| NT-proBNP (ng/L) | 62 (37–112) | 64 (44–102) | 67 (36–99) | 0.663 | 62 (37–112) | 66 (34–150) | 65 (35–147) | 0.763 |
| Creatinine (µmol/L) | 70 (66–80) | 69 (57–84) | 71 (55–86) | 0.702 | 70 (66–80) | 72 (62–81) | 67 (58–80) | 0.763 |
| Bilirubin (µmol/L) | 8.7 (7.0–11.7) | 7.4 (6.0–9.9) | 8.3 (5.9–11.2) | 0.597 | 8.7 (7.0–11.7) | 7.4 (5.5–10.0) | 8.2 (7.0–12.0) | 0.763 |
| D-dimers (mg/L) | 0.31 (0.27–0.51) | 0.32 (0.27–0.63) | 0.32 (0.27–0.55) | 0.976 | 0.31 (0.27–0.51) | 0.37 (0.32–0.54) | 0.39 (0.29–0.56) | 0.663 |
| Comparable Subgroups (Group Sizes) | Model * | Independent Variables in the Final Model ** | Coefficient Estimate *** | Adjusted OR (CI95%) **** | Adjusted p-Value ***** | Model AUC (Sensitivity, Specificity) |
|---|---|---|---|---|---|---|
| LC vs. R (27/28) | M1 | CD5 | 0.66698 | 1.95 (1.025; 3.70) | 0.042 | 0.56 (0.64; 0.44) |
| M2 a | D-dimers | −18.3908 | 10−8 (10−21; 13510) | 0.196 | 0.30 (0.30; 0.37) | |
| AST | −1.5196 | 0.22 (0.04; 1.1) | 0.068 | |||
| Age | −0.7566 | 0.47 (0.18; 1.23) | 0.124 | |||
| Leucocytes | −0.6806 | 0.51 (0.24; 1.08) | 0.080 | |||
| Female gender | −0.5542 | 0.57 (0.28; 1.2) | 0.140 | |||
| ALT | 1.0891 | 2.97 (0.76; 11.7) | 0.118 | |||
| IL10RB | 0.9672 | 2.63 (0.89; 7.8) | 0.080 | |||
| TNFRSF9 | 0.8655 | 2.38 (0.81; 7.0) | 0.117 | |||
| IL6 | 0.7328 | 2.08 (0.93; 4.67) | 0.076 | |||
| M3 a | Ferritin | −0.61458 | 0.54 (0.26; 1.13) | 0.10 | 0.62 (0.59; 0.63) | |
| IL10RB | 0.79028 | 2.2 (1.07; 4.5) | 0.032 | |||
| BMI | 0.61753 | 1.85 (0.98; 3.53) | 0.06 | |||
| GEN-LC vs. R (15/28) | M4 b | TNF | −2.7214 | 0.066 (0.003; 1.51) | 0.090 | 0.60 (0.82; 0.33) |
| CXCL1 | −0.8787 | 0.42 (0.15; 1.17) | 0.098 | |||
| TRANCE | 2.0625 | 7.87 (1.21; 51.1) | 0.031 | |||
| TNFRSF9 | 1.6732 | 5.33 (0.82; 34.6) | 0.080 | |||
| IL10RB | 1.4862 | 4.42 (0.86; 22.6) | 0.075 | |||
| SLAMF1 | 0.9473 | 2.58 (0.78; 8.57) | 0.122 |
| Comparable Subgroups (Group Sizes) | Model * | Independent Variables in the Final Model ** | Coefficient Estimate *** | Adjusted OR (CI95%) *** | Adjusted p-Value **** | Model AUC (Sensitivity, Specificity) |
|---|---|---|---|---|---|---|
| LC vs. R (28/18) | M1 | MCP2 | −0.9241 | 0.40 (0.18; 0.87) | 0.020 | 0.70 (0.50; 0.71) |
| IL2RB | −0.6255 | 0.53 (0.26; 1.08) | 0.081 | |||
| M2 | MCP2 | −0.8227 | 0.44 (0.18; 1.1) | 0.078 | 0.73 (0.61; 0.75) | |
| WHO group | 0.8846 | 2.42 (1.14; 5.1) | 0.021 | |||
| GEN-LC vs. R (17/18) | M3 | MCP2 | −1.42641 | 0.24 (0.08; 0.75) | 0.0142 | 0.59 (0.55; 0.59) |
| TRAIL | −0.85045 | 0.43 (0.15; 1.22) | 0.1133 | |||
| uPA | 1.43955 | 4.22 (1.12; 15.9) | 0.0339 | |||
| M4 | TRAIL | −0.9255 | 0.40 (0.13; 1.17) | 0.095 | 0.83 (0.78; 0.88) | |
| WHO group | 2.4988 | 12.2 (2.35; 63) | 0.0029 | |||
| uPA | 1.3796 | 3.97 (1.06; 14.9) | 0.041 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kallaste, A.; Kisand, K.; Aart, A.; Salumets, A.; Kisand, K.; Peterson, P.; Lember, M. Long COVID and Biomarker Dysregulation—A Shift Toward Immune Exhaustion? Medicina 2025, 61, 996. https://doi.org/10.3390/medicina61060996
Kallaste A, Kisand K, Aart A, Salumets A, Kisand K, Peterson P, Lember M. Long COVID and Biomarker Dysregulation—A Shift Toward Immune Exhaustion? Medicina. 2025; 61(6):996. https://doi.org/10.3390/medicina61060996
Chicago/Turabian StyleKallaste, Anne, Kalle Kisand, Agnes Aart, Ahto Salumets, Kai Kisand, Pärt Peterson, and Margus Lember. 2025. "Long COVID and Biomarker Dysregulation—A Shift Toward Immune Exhaustion?" Medicina 61, no. 6: 996. https://doi.org/10.3390/medicina61060996
APA StyleKallaste, A., Kisand, K., Aart, A., Salumets, A., Kisand, K., Peterson, P., & Lember, M. (2025). Long COVID and Biomarker Dysregulation—A Shift Toward Immune Exhaustion? Medicina, 61(6), 996. https://doi.org/10.3390/medicina61060996






