FICTION Technique—A Candidate for the Assessment of HER2 Status in Breast Invasive Carcinomas
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Group
2.2. Routine Microscopic Examination of Invasive Breast Carcinomas (Standard IHC and ISH Ancillary Testing)
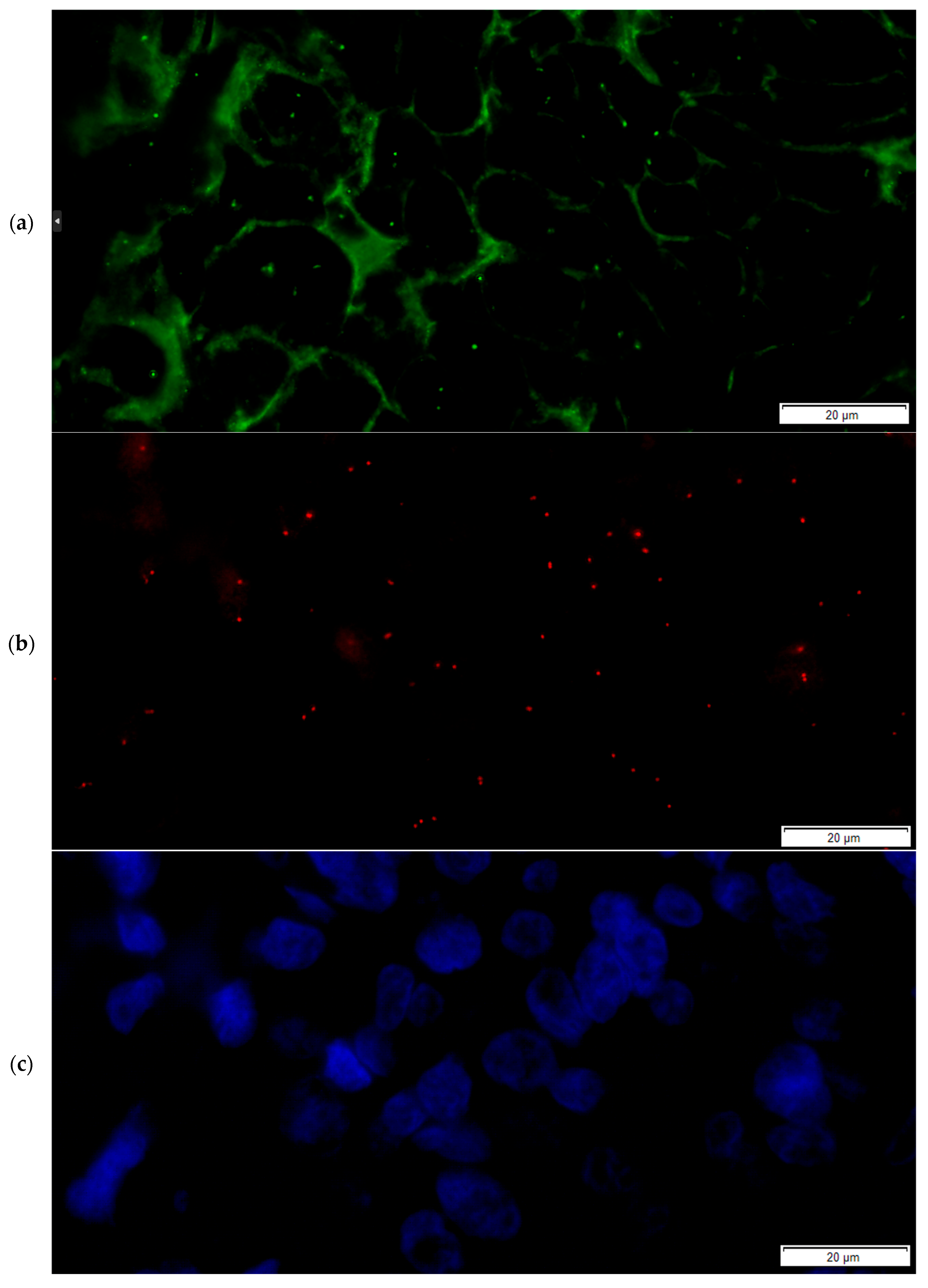
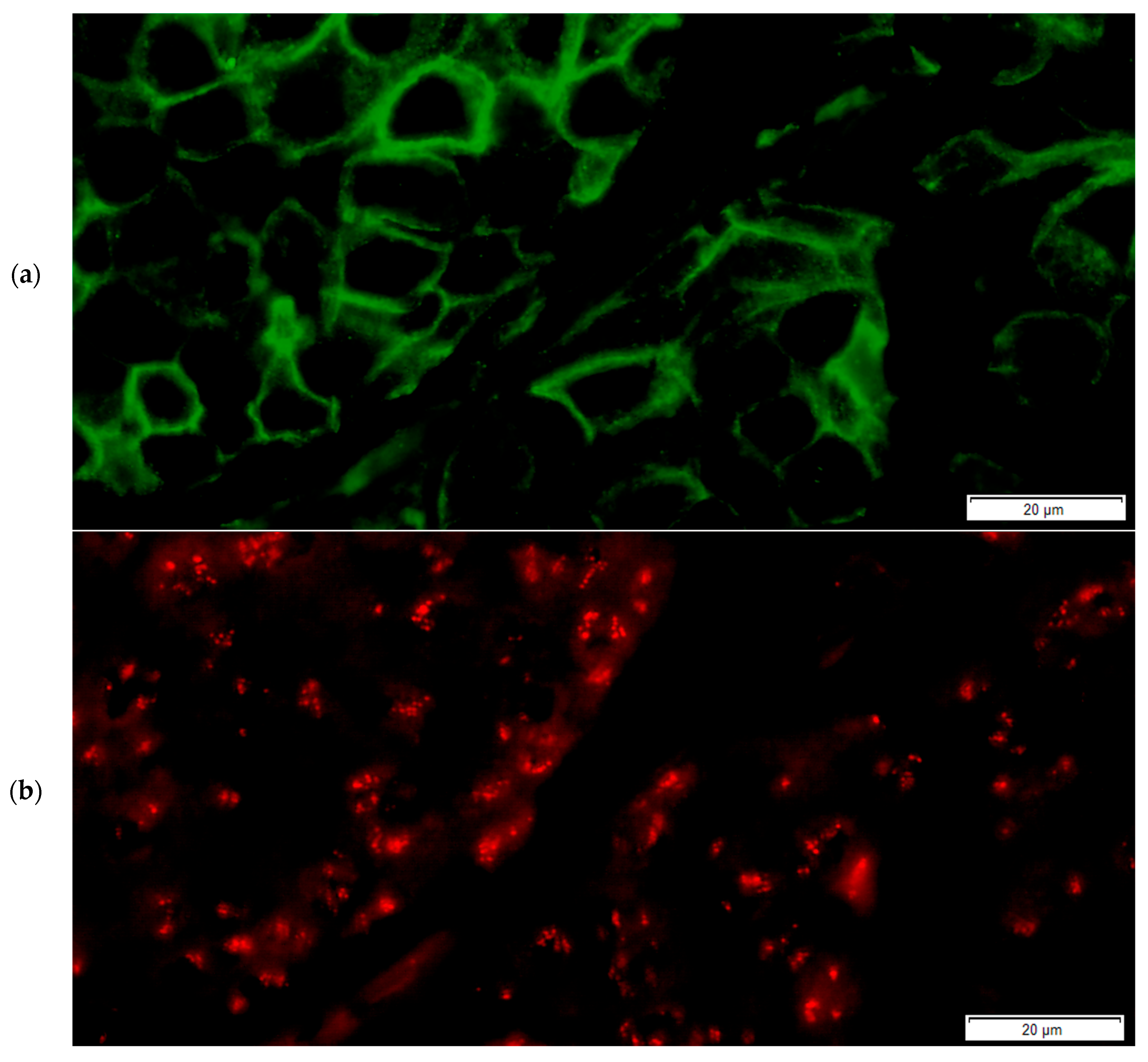
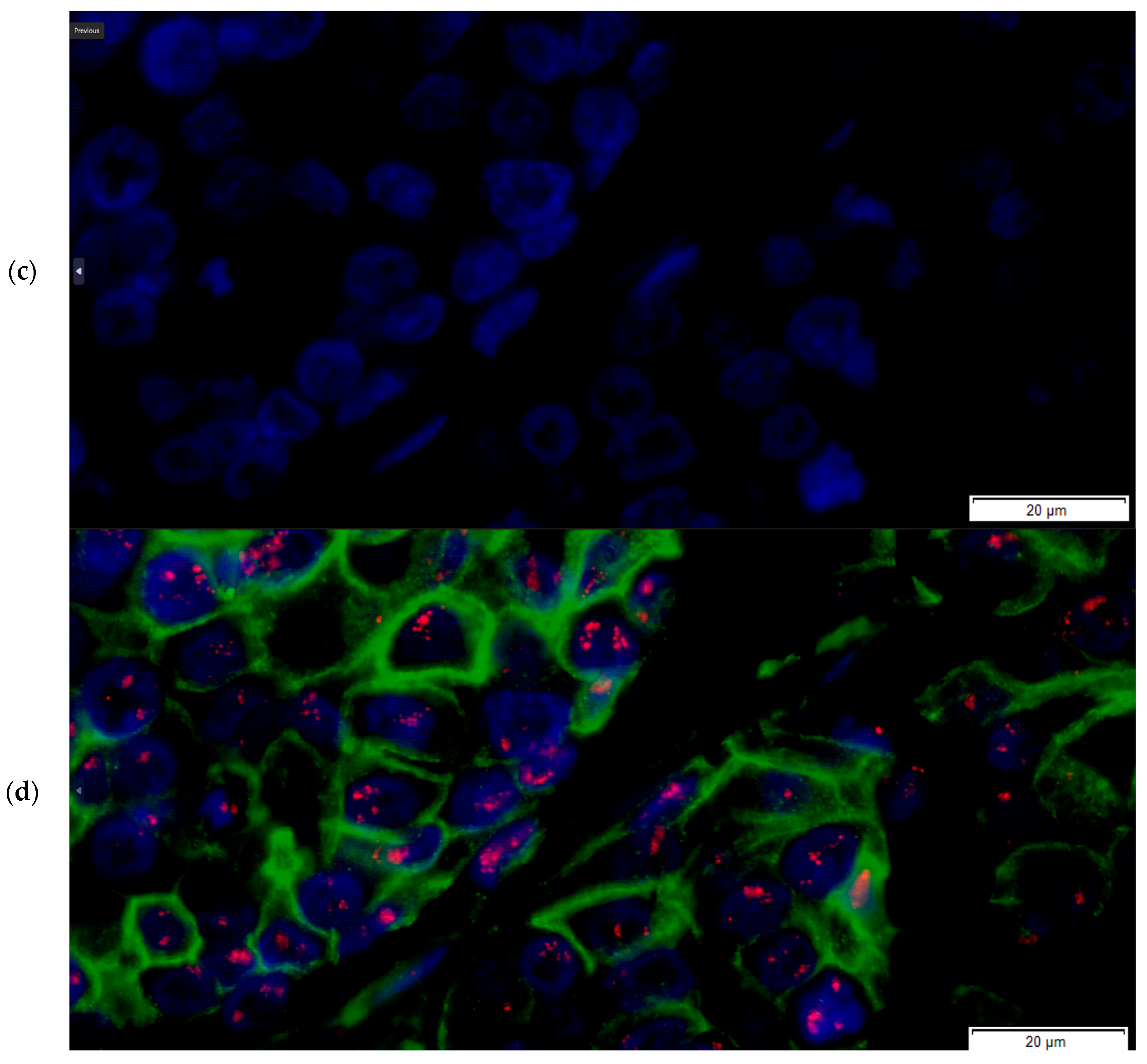
2.3. The FICTION Staining Protocol
- From the paraffin blocks, sections were cut at 4 μm and spread on silane-coated slides,
- After sectioning, the slides were incubated for at least 2 h at 56 °C or overnight at 37 °C for aging,
- The slides were immersed in three consecutive baths of xylene, with 5 min for each bath,
- The slides were then passed through two baths of absolute ethyl alcohol, with one minute in each bath,
- The slides were washed in tap water,
- The slides were placed in a steel holder, which was then placed in a Coplin jar containing citrate buffer solution, pH = 6, 0.01 M,
- The jar was boiled using a pressure cooker at 100 °C for 20 min,
- The pressure cooker was cooled under cold tap water and the steam gradually released,
- The lid of the pressure cooker was opened,
- The slides were moved to a Coplin jar with distilled water for 5 min,
- The slides were then dried at room temperature for 10 min,
- The slides were then immersed for 10 min in acetone,
- Washing of the slides was performed in 100 mL of PN buffer [13],
- Dehydration of the slides was performed in 70%, 85% and 96% ethanol, and the duration for each bath was 2 min at room temperature,
- The slides were dried at room temperature for 5 min,
- The slides were then hybridized by applying a 10 µL hybridization probe (PathVysion LSI HER-2/neu SpectrumOrange/CEP 17 SpectrumGreen Probes, Abbot molecular, Abbott Park, IL, USA),
- The slides were next covered with a Ø 10 mm × 10 mm coating glass,
- Sealing of the entire hybridization region was performed using Fixogum™ rubber cement,
- The slides were transferred to a hybridization unit (Thermobrite, Abbot molecular, Abbott Park, IL, USA), where denaturation was performed for 7 min at 75 °C,
- The hybridization process was performed for 16 h (overnight) at 37 °C in the hybridization unit,
- The next day, two Coplin jars containing wash buffer (2X SSC/0.3% NP-40) were prepared; one was immersed in a water bath at 72 °C, and the other jar was left at room temperature,
- Using tweezers, the Fixogum™ rubber cement and the glass coverslips were carefully removed; then, the excess probe was washed in the Coplin jar containing wash buffer solution at 72 °C for a duration of 2 min and 15 s, and the slides were then moved to the room temperature Coplin jar,
- The slides were washed with 100 mL of PN buffer,
- The slides were incubated with 100 µL of the primary antibody solution for immunofluorescence staining of the section and incubated for 30 min away from light (recombinant anti-ErbB2/HER2 antibody [clone CAL27] (ab237715), rabbit monoclonal, dilution 1:50, Abcam, Abcam Limited, Cambridge, UK),
- The slides were then washed with 100 mL of PN buffer,
- Incubation with 100 µL of the secondary antibody solution for 30 min (goat anti-rabbit IgG H&L (FITC) (ab6717), 1:10 dilution) followed,
- The slides were washed in a Coplin jar with 2 × SSC for 10 min,
- The slides were then completely air dried,
- Nuclear counterstaining and mounting were performed using DAPI counterstain (4,6-diamidino-2-phenylindole) in phenylenediamine dihydrochloride, glycerol and buffer,
- The slides were then coverslipped and sealed and stored for at least 30 min at 4 °C before the examination.
2.4. Preparation of the PN Buffer Solution [13]
2.5. Comparing Standard IHC + ISH Assays to the FICTION Technique
3. Results
3.1. The Study Group Histology Examination and Standard IHC and FISH Testing
3.2. Comparison of Standard IHC Her2 Testing Results and IF Results in the FICTION Assay
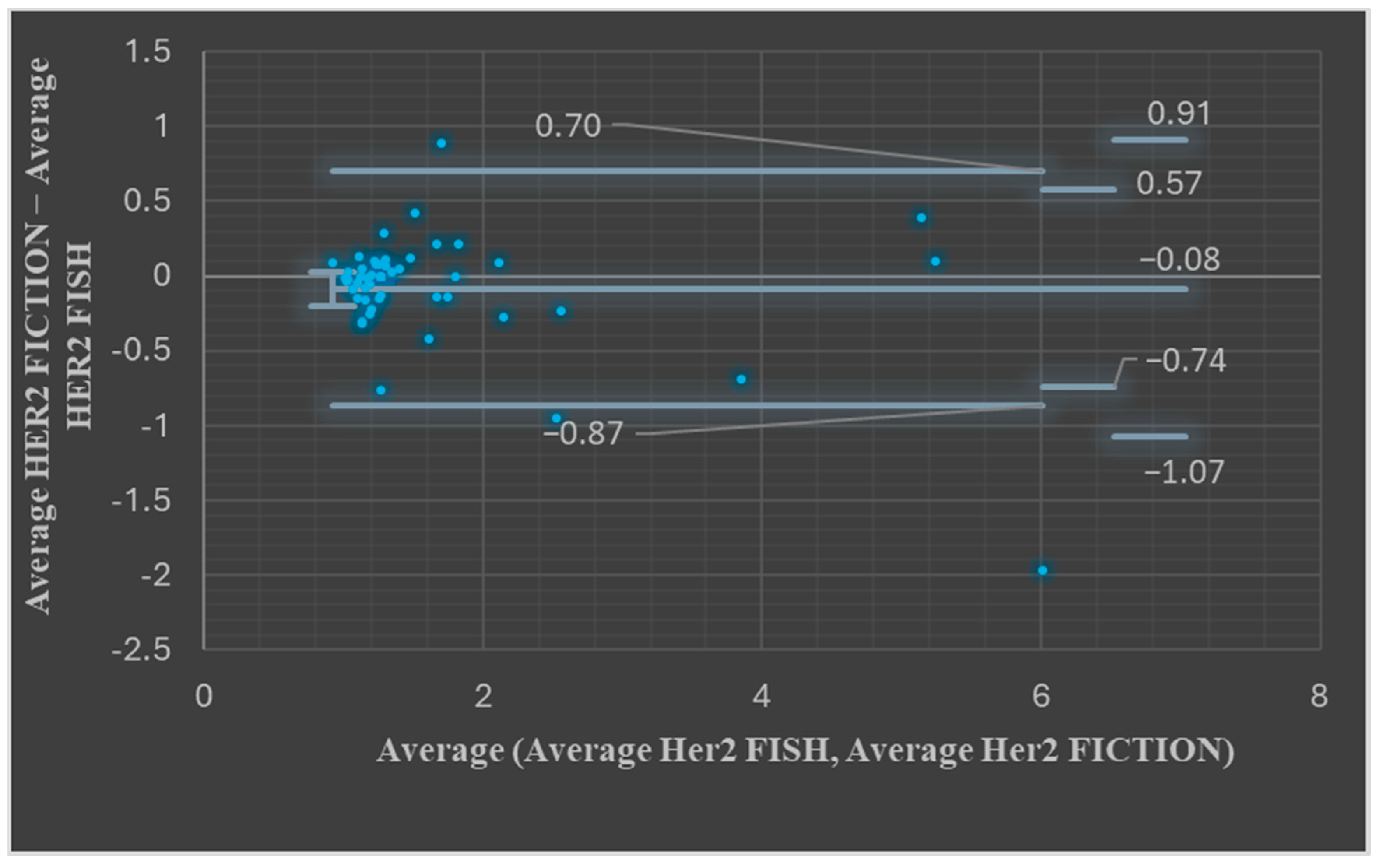
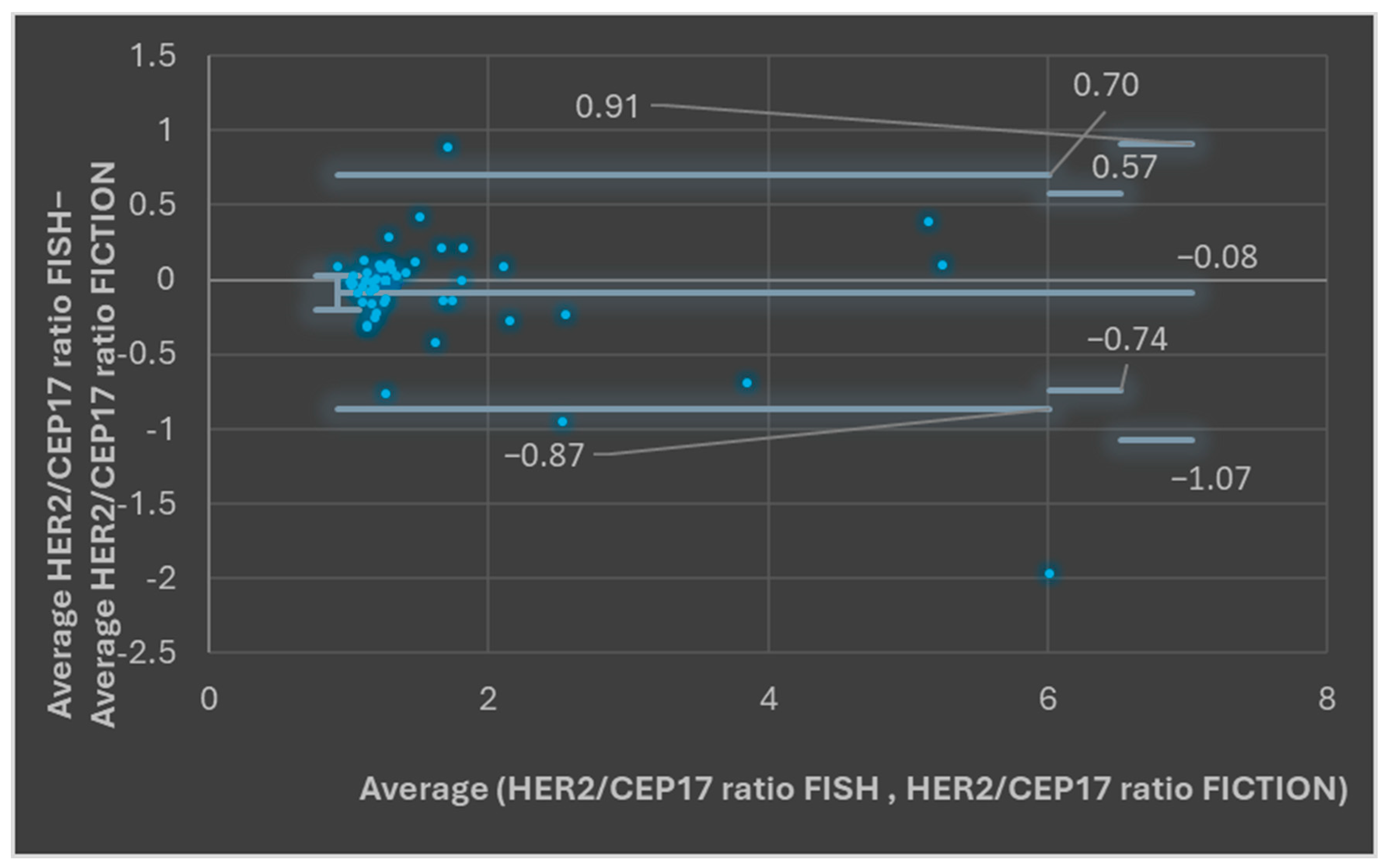
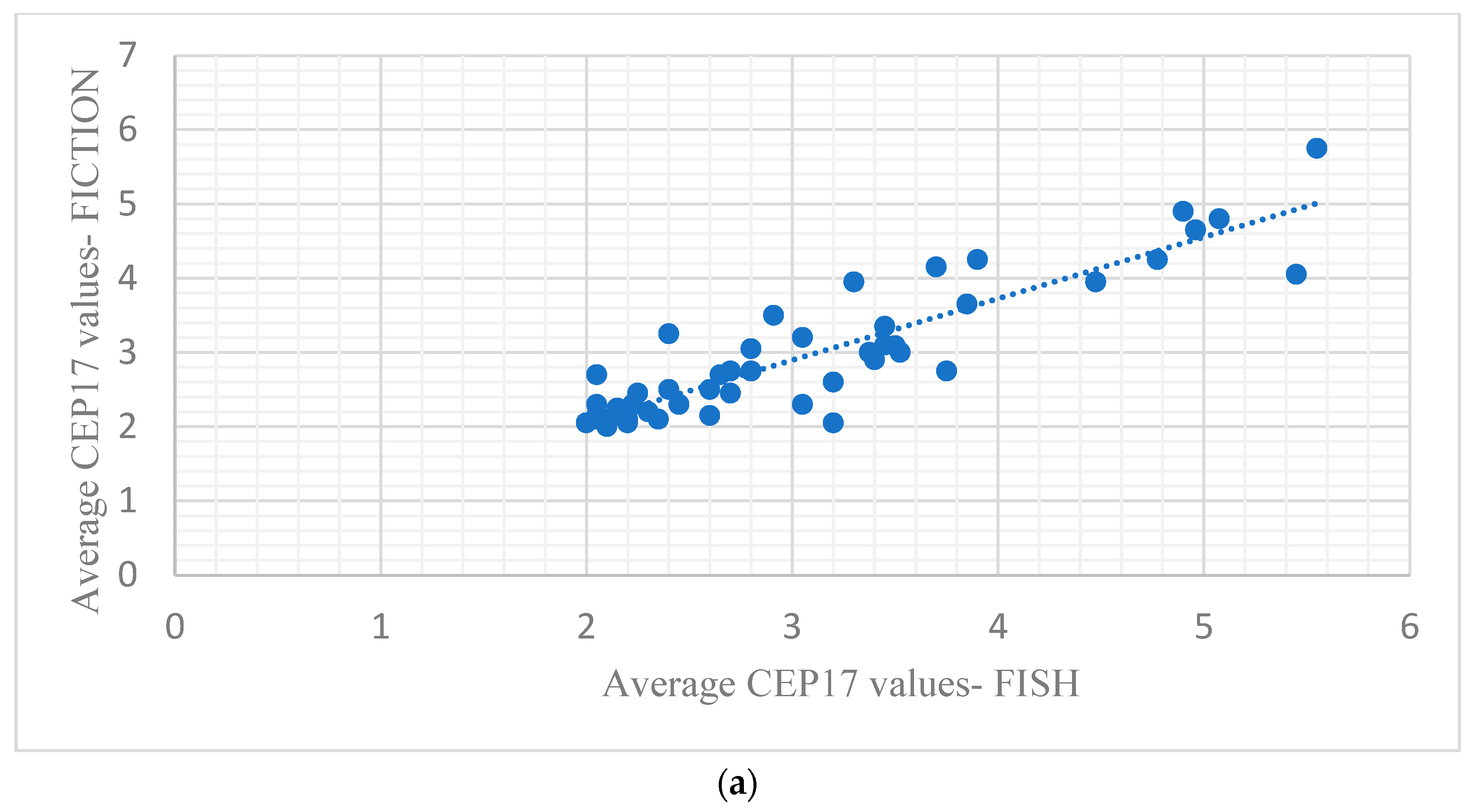
3.3. Comparison of the Results of ISH Using the FISH Technique and ISH Results in the FICTION Assay
3.4. Comparison of the Results of ASCO/CAP Groups Using the FISH Technique and FICTION Assay and of the Final Binary Classification (Interpretation) of HER2 Status
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LOA | Limit(s) of agreement |
| IBC | Invasive breast carcinoma |
| CEP17 | Centromeric enumeration probe for chromosome 17 |
| FISH | Fluorescent in situ hybridization |
| FICTION | Fluorescence immunophenotyping and interphase cytogenetics as a tool for the investigation of neoplasms |
| IHC | Immunohistochemistry |
| ISH | In situ hybridization |
| FITC | Fluorescein isothiocyanate |
| TRITC | Tetramethylrhodamine |
| DAPI | 4′,6-diamidino-2-phenylindole |
References
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Somerfield, M.R.; Dowsett, M.; Hammond, M.E.H.; Hayes, D.F.; McShane, L.M.; Saphner, T.J.; Spears, P.A.; Allison, K.H. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology–College of American Pathologists Guideline Update. Arch. Pathol. Lab. Med. 2023, 147, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S. Trastuzumab deruxtecan in previously treated HER2-low advanced breast cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef]
- Fernandez, A.I.; Liu, M.; Bellizzi, A.; Brock, J.; Fadare, O.; Hanley, K.; Harigopal, M.; Jorns, J.M.; Kuba, M.G.; Ly, A.; et al. Examination of Low ERBB2 Protein Expression in Breast Cancer Tissue. JAMA Oncol. 2022, 8, 607–610. [Google Scholar] [CrossRef]
- Moutafi, M.; Robbins, C.J.; Yaghoobi, V.; Fernandez, A.I.; Martinez-Morilla, S.; Xirou, V.; Bai, Y.; Song, Y.; Gaule, P.; Krueger, J.; et al. Quantitative measurement of HER2 expression to subclassify ERBB2 unamplified breast cancer. Lab. Investig. 2022, 102, 1101–1108. [Google Scholar] [CrossRef]
- Curigliano, G.; Hu, X.; Dent, R.A.; Yonemori, K.; Barrios, C.H.; O’Shaughnessy, J.; Wildiers, H.; Zhang, Q.; Im, S.-A.; Saura, C.; et al. Trastuzumab deruxtecan (T-DXd) vs physician’s choice of chemotherapy (TPC) in patients (pts) with hormone receptor-positive (HR+), human epidermal growth factor receptor 2 (HER2)-low or HER2-ultralow metastatic breast cancer (mBC) with prior endocrine therapy (ET): Primary results from DESTINY-Breast06 (DB-06). J. Clin. Oncol. 2024, 42, LBA1000. [Google Scholar] [CrossRef]
- Bardia, A.; Hu, X.; Dent, R.; Yonemori, K.; Barrios, C.H.; O’Shaughnessy, J.A.; Wildiers, H.; Pierga, J.-Y.; Zhang, Q.; Saura, C. Trastuzumab deruxtecan after endocrine therapy in metastatic breast cancer. N. Engl. J. Med. 2024, 391, 2110–2122. [Google Scholar] [CrossRef] [PubMed]
- Gatta, L.B.; Incardona, P.; Cadei, M.; Grigolato, P.; Simoncelli, S.; Balzarini, P. Simultaneous fluorescence immunophenotyping and Her-2/neu genotyping (FICTION) in breast carcinoma candidates to target therapy. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 413–420. [Google Scholar] [CrossRef]
- Giefing, M.; Siebert, R. FISH and FICTION to detect chromosomal aberrations in lymphomas. Methods Mol. Biol. 2013, 971, 227–244. [Google Scholar] [CrossRef]
- Lottner, C.; Schwarz, S.; Diermeier, S.; Hartmann, A.; Knuechel, R.; Hofstaedter, F.; Brockhoff, G. Simultaneous detection of HER2/neu gene amplification and protein overexpression in paraffin-embedded breast cancer. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2005, 205, 577–584. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Breast Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019; Volume 2, ISBN-13 978-92-832-4500-1; Available online: https://publications.iarc.fr/581 (accessed on 12 March 2025).
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J. Estrogen and progesterone receptor testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists guideline update. Arch. Pathol. Lab. Med. 2020, 144, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Weber-Matthiesen, K.; Winkemann, M.; Müller-Hermelink, A.; Schlegelberger, B.; Grote, W. Simultaneous fluorescence immunophenotyping and interphase cytogenetics: A contribution to the characterization of tumor cells. J. Histochem. Cytochem. 1992, 40, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Carkeet, A. A Review of the Use of Confidence Intervals for Bland-Altman Limits of Agreement in Optometry and Vision Science. Optom. Vision Sci. 2020, 97, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S. Novel and simple method of double-detection using fluorescence in situ hybridization and fluorescence immunostaining of formalin-fixed paraffin-embedded tissue sections. Oncol. Lett. 2018, 15, 1084–1088. [Google Scholar] [CrossRef]
- Tubbs, R.; Pettay, J.; Skacel, M.; Powell, R.; Stoler, M.; Roche, P.; Hainfeld, J. Gold-facilitated in situ hybridization: A bright-field autometallographic alternative to fluorescence in situ hybridization for detection of Her-2/neu gene amplification. Am. J. Pathol. 2002, 160, 1589–1595. [Google Scholar] [CrossRef]
- Yoder, B.; Rybicki, L.; Myles, J.; Sreenan, J.; Roche, P.; Powell, R.; Hainfeld, J.; Grogan, T.; Tubbs, R. Analytical Validation and Interobserver Reproducibility of EnzMet GenePro. Am. J. Surg. Pathol. 2005, 29, 1505–1511. [Google Scholar]
- Ni, R.; Mulligan, A.M.; Have, C.; O’Malley, F.P. PGDS, a novel technique combining chromogenic in situ hybridization and immunohistochemistry for the assessment of ErbB2 (HER2/neu) status in breast cancer. Appl. Immunohistochem. Mol. Morphol. 2007, 15, 316–324. [Google Scholar] [CrossRef]
- Reisenbichler, E.S.; Horton, D.; Rasco, M.; Andea, A.; Hameed, O. Evaluation of dual immunohistochemistry and chromogenic in situ hybridization for HER2 on a single section. Am. J. Clin. Pathol. 2012, 137, 102–110. [Google Scholar] [CrossRef]
- Nitta, H.; Kelly, B.D.; Padilla, M.; Wick, N.; Brunhoeber, P.; Bai, I.; Singh, S.; Ranger-Moore, J.; Bieniarz, C.; Tsuda, H.; et al. A gene-protein assay for human epidermal growth factor receptor 2 (HER2): Brightfield tricolor visualization of HER2 protein, the HER2 gene, and chromosome 17 centromere (CEN17) in formalin-fixed, paraffin-embedded breast cancer tissue sections. Diagn. Pathol. 2012, 7, 60. [Google Scholar] [CrossRef]
- Stålhammar, G.; Farrajota, P.; Olsson, A.; Silva, C.; Hartman, J.; Elmberger, G. Gene protein detection platform—A comparison of a new human epidermal growth factor receptor 2 assay with conventional immunohistochemistry and fluorescence in situ hybridization platforms. Ann. Diagn. Pathol. 2015, 19, 203–210. [Google Scholar] [CrossRef]
- Robbins, C.; He, M.; Khaimova, R.; Bates, K.; Chan, N.N.N.; Liebler, D.; Fulton, R.; Rimm, D. Abstract PO3-13-11: Quantitative Multiplex Immunofluorescence Assay for TROP2 and HER2 Expression in Breast Cancer: Towards Guiding Patient Selection for Antibody Drug Conjugate Therapies. Cancer Res. 2024, 84, PO3-13-11. [Google Scholar] [CrossRef]
- Tseng, Y.-F.; Li, Y.-C.; Lee, Y.-H.; Hu, H.-W.; Zhang, M.-S.; Hung, T.-C.; Lien, H.-C. Correlation of in situ HER2 RNA expression with HER2 immunohistochemistry and fluorescence in situ hybridization categories in breast cancer. Arch. Pathol. Lab. Med. 2024, 148, e48–e56. [Google Scholar] [CrossRef]
- Li, X.; Lee, J.-H.; Gao, Y.; Zhang, J.; Bates, K.M.; Rimm, D.L.; Zhang, H.; Smith, G.H.; Lawson, D.; Meisel, J. Correlation of HER2 protein level with mRNA level quantified by RNAscope in breast cancer. Mod. Pathol. 2024, 37, 100408. [Google Scholar] [CrossRef]
- Memon, R.; Prieto Granada, C.N.; Harada, S.; Winokur, T.; Reddy, V.; Kahn, A.G.; Siegal, G.P.; Wei, S. Discordance Between Immunohistochemistry and In Situ Hybridization to Detect HER2 Overexpression/Gene Amplification in Breast Cancer in the Modern Age: A Single Institution Experience and Pooled Literature Review Study. Clin. Breast Cancer 2022, 22, e123–e133. [Google Scholar] [CrossRef]





| IHC +FISH | FICTION | p Value | |
|---|---|---|---|
| Her2 protein expression(categorial) | 83.67% agreement | <0.0001 | |
| Cohen kappa 0.533 | |||
| Her2 protein expression(percentage of tumor cells) | 60.71% mean expression | 73.77% mean expression | =0.00026 |
| 91.83% between upper and lower LOA (45/49 samples) | >0.05 | ||
| CEP17—ISH | 2.95 average CEP17 signals | 3.06 average CEP 17 signals | >0.05 |
| 91.83% between upper and lower LOA (45/49 samples) | >0.05 | ||
| Pearson correlation coefficient = 0.90 | <0.001 | ||
| HER2—ISH | 5.05 average HER2 signals | 5.25 average HER2 signals | >0.05 |
| 95.91% between upper and lower LOA (47/49) | >0.05 | ||
| Pearson correlation coefficient = 0.96 | <0.001 | ||
| HER2: CEP 17—ISH | 1.78 average HER2/CEP 17 ratio | 1.66 average HER2/CEP 17 ratio | >0.05 |
| 93.87% between upper and lower LOA (47/49 samples) | >0.05 | ||
| Pearson correlation coefficient = 0.95 | <0.001 | ||
| ASCO/CAP ISH groups | 85.7% group agreement | ||
| Cohen’s kappa coefficient = 0.785 | <0.0001 | ||
| Final category (Her2 positive/Her2 negative) | 100% agreement Cohen’s kappa coefficient = 1 | <0.0001 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fetica, B.; Blaga, M.L.; Trifa, A.P.; Bocean, C.M.; Balacescu, O.; Fulop, A.; Pop, B. FICTION Technique—A Candidate for the Assessment of HER2 Status in Breast Invasive Carcinomas. Medicina 2025, 61, 1069. https://doi.org/10.3390/medicina61061069
Fetica B, Blaga ML, Trifa AP, Bocean CM, Balacescu O, Fulop A, Pop B. FICTION Technique—A Candidate for the Assessment of HER2 Status in Breast Invasive Carcinomas. Medicina. 2025; 61(6):1069. https://doi.org/10.3390/medicina61061069
Chicago/Turabian StyleFetica, Bogdan, Mihaiela Luminita Blaga, Adrian Pavel Trifa, Cosmina Maria Bocean, Ovidiu Balacescu, Annamaria Fulop, and Bogdan Pop. 2025. "FICTION Technique—A Candidate for the Assessment of HER2 Status in Breast Invasive Carcinomas" Medicina 61, no. 6: 1069. https://doi.org/10.3390/medicina61061069
APA StyleFetica, B., Blaga, M. L., Trifa, A. P., Bocean, C. M., Balacescu, O., Fulop, A., & Pop, B. (2025). FICTION Technique—A Candidate for the Assessment of HER2 Status in Breast Invasive Carcinomas. Medicina, 61(6), 1069. https://doi.org/10.3390/medicina61061069







