Explainable Boosting Machines Identify Key Metabolomic Biomarkers in Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Dataset, and Sample Size
2.2. Data Preprocessing
2.3. Biostatistical Data Analysis
2.4. Machine Learning Algorithms and Performance Evaluation
2.5. Global and Local Explanations with Explainable Boosting Machine
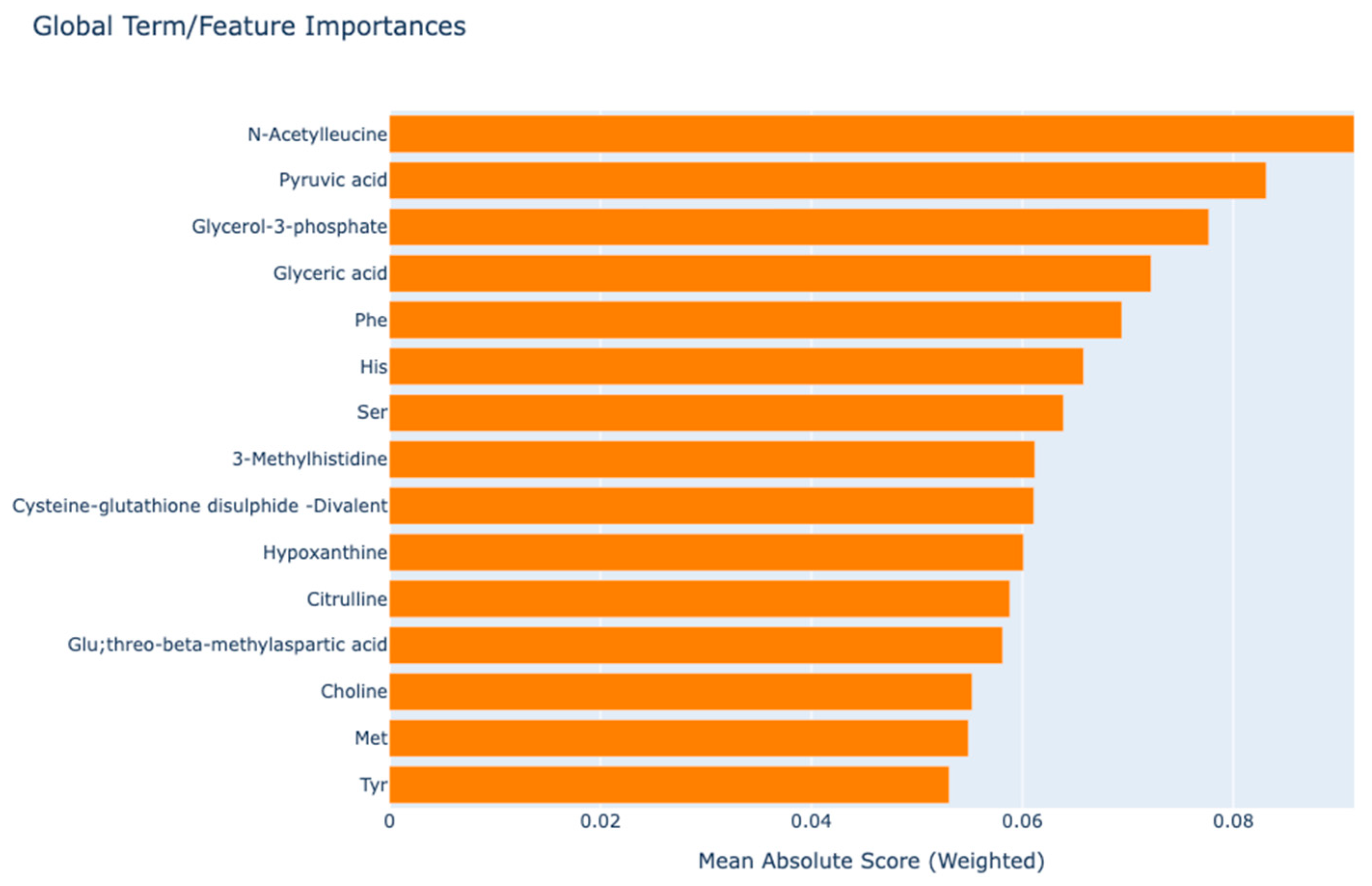
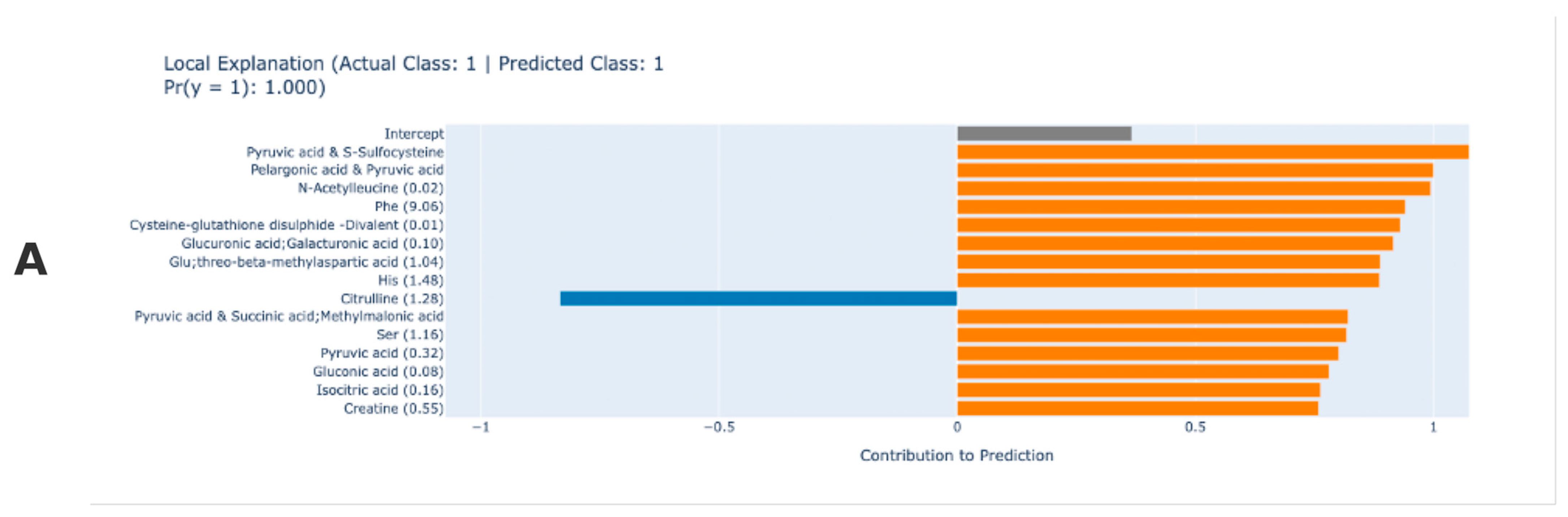
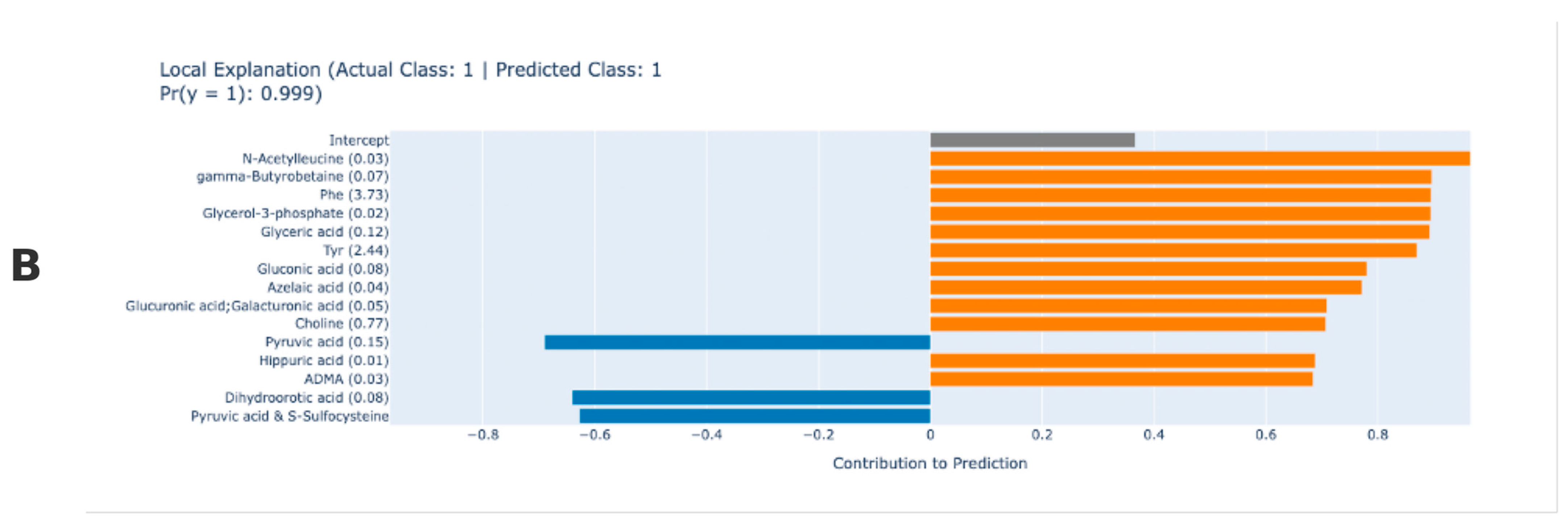
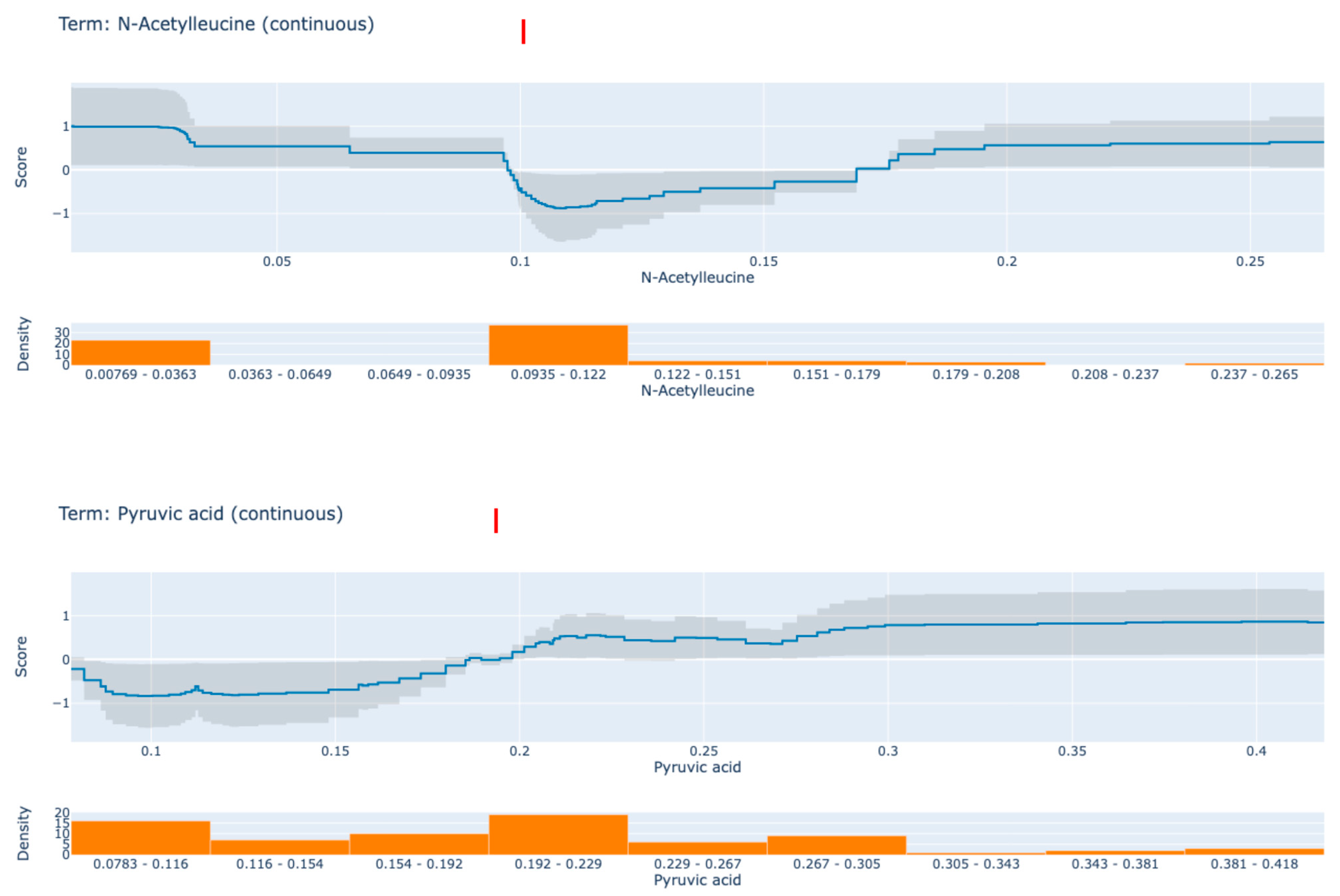
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajdan, N.; Srivastava, B.; Sharma, K. Rheumatoid Arthritis: Treatments Available at Present. In Nanotherapeutics for Inflammatory Arthritis; CRC Press: Boca Raton, FL, USA, 2024; pp. 243–295. [Google Scholar]
- Safiri, S.; Kolahi, A.A.; Hoy, D.; Smith, E.; Bettampadi, D.; Mansournia, M.A.; Almasi-Hashiani, A.; Ashrafi-Asgarabad, A.; Moradi-Lakeh, M.; Qorbani, M.; et al. Global, regional and national burden of rheumatoid arthritis 1990–2017: A systematic analysis of the Global Burden of Disease study 2017. Ann. Rheum. Dis. 2019, 78, 1463–1471. [Google Scholar] [PubMed]
- Ruzzon, F.; Adami, G. Environment and arthritis. Clin. Exp. Rheumatol. 2024, 42, 1343–1349. [Google Scholar] [CrossRef]
- Srivastava, S.; Rasool, M. Genetics, epigenetics and autoimmunity constitute a Bermuda triangle for the pathogenesis of rheumatoid arthritis. Life Sci. 2024, 357, 123075. [Google Scholar] [CrossRef] [PubMed]
- Mondillo, G.; Colosimo, S.; Perrotta, A.; Frattolillo, V.; Gicchino, M.F. Unveiling Artificial Intelligence’s Power: Precision, Personalization, and Progress in Rheumatology. J. Clin. Med. 2024, 13, 6559. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Y.; Chen, Y.; Luo, D.; Xu, K.; Zhang, L. Artificial intelligence for predicting treatment responses in autoimmune rheumatic diseases: Advancements, challenges, and future perspectives. Front. Immunol. 2024, 15, 1477130. [Google Scholar] [CrossRef]
- Cai, X.; Jin, J.; Ye, H.; Xiang, X.; Luo, L.; Li, J. Altered serum metabolome is associated with disease activity and immune responses in rheumatoid arthritis. Clin. Rheumatol. 2024, 43, 3669–3678. [Google Scholar]
- Manik, M.; Nilima, S.; Mahmud, M.; Sharmin, S.; Hasan, R. Discovering Disease Biomarkers in Metabolomics via Big Data Analytics. Am. J. Stat. Actuar. Sci. 2024, 4, 35–49. [Google Scholar] [CrossRef]
- Fan, W.; Pi, Z.; Kong, K.; Qiao, H.; Jin, M.; Chang, Y.; Zhang, J.; Li, H. Analyzing the impact of heavy metal exposure on osteoarthritis and rheumatoid arthritis: An approach based on interpretable machine learning. Front. Nutr. 2024, 11, 1422617. [Google Scholar]
- Liu, F.; Ye, J.; Wang, S.; Li, Y.; Yang, Y.; Xiao, J.; Jiang, A.; Lu, X.; Zhu, Y. Identification and Verification of Novel Biomarkers Involving Rheumatoid Arthritis with Multimachine Learning Algorithms: An In Silicon and In Vivo Study. Mediat. Inflamm. 2024, 2024, 3188216. [Google Scholar] [CrossRef]
- Akshitha, K.; Roopashree, R.; Kodipalli, A.; Rao, T. Utilizing Explainable AI Methodologies: LIME and SHAP, for the Classification of Natural Disasters Through Machine Learning Algorithms. In Proceedings of the 2024 Parul International Conference on Engineering and Technology (PICET), Waghodia, India, 3–4 May 2024; pp. 1–5. [Google Scholar]
- Sekaran, K.; Alsamman, A.M.; George Priya Doss, C.; Zayed, H. Bioinformatics investigation on blood-based gene expressions of Alzheimer’s disease revealed ORAI2 gene biomarker susceptibility: An explainable artificial intelligence-based approach. Metab. Brain Dis. 2023, 38, 1297–1310. [Google Scholar] [CrossRef]
- Rezk, N.G.; Alshathri, S.; Sayed, A.; El-Din Hemdan, E.; El-Behery, H. XAI-Augmented Voting Ensemble Models for Heart Disease Prediction: A SHAP and LIME-Based Approach. Bioengineering 2024, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Rychkov, D.; Neely, J.; Oskotsky, T.; Yu, S.; Perlmutter, N.; Nititham, J.; Carvidi, A.; Krueger, M.; Gross, A.; Criswell, L.; et al. Cross-Tissue Transcriptomic Analysis Leveraging Machine Learning Approaches Identifies New Biomarkers for Rheumatoid Arthritis. Front. Immunol. 2020, 12, 638066. [Google Scholar] [CrossRef] [PubMed]
- Dudek, G.; Sakowski, S.; Brzezińska, O.; Sarnik, J.; Budlewski, T.; Dragan, G.; Poplawska, M.; Poplawski, T.; Bijak, M.; Makowska, J. Machine learning-based prediction of rheumatoid arthritis with development of ACPA autoantibodies in the presence of non-HLA genes polymorphisms. PLoS ONE 2024, 19, e0300717. [Google Scholar] [CrossRef]
- Sasaki, C.; Hiraishi, T.; Oku, T.; Okuma, K.; Suzumura, K.; Hashimoto, M.; Ito, H.; Aramori, I.; Hirayama, Y. Metabolomic approach to the exploration of biomarkers associated with disease activity in rheumatoid arthritis. PLoS ONE 2019, 14, e0219400. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Gormley, I.C.; Fan, Y.; Gallagher, W.M.; Brennan, L. MetSizeR: Selecting the optimal sample size for metabolomic studies using an analysis based approach. BMC Bioinform. 2013, 14, 338. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Royston, P.; White, I.R. Multiple Imputation by Chained Equations (MICE): Implementation in Stata. J. Stat. Softw. 2011, 45, 1–20. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Zhou, Z. Ensemble Methods: Foundations and Algorithms; Chapman & Hall/CRC: Boca Raton, FL, USA, 2012. [Google Scholar]
- Naghibi, S.A.; Moghaddam, D.D.; Kalantar, B.; Pradhan, B.; Kisi, O. A comparative assessment of GIS-based data mining models and a novel ensemble model in groundwater well potential mapping. J. Hydrol. 2017, 548, 471–483. [Google Scholar] [CrossRef]
- Estevez, P.A.; Tesmer, M.; Perez, C.A.; Zurada, J.M. Normalized Mutual Information Feature Selection. IEEE Trans. Neural Netw. 2009, 20, 189–201. [Google Scholar] [CrossRef]
- Hatwell, J.; Gaber, M.M.; Azad, R.M.A. Ada-WHIPS: Explaining AdaBoost classification with applications in the health sciences. BMC Med. Inform. Decis. Mak. 2020, 20, 250. [Google Scholar] [CrossRef] [PubMed]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 1–9. [Google Scholar]
- Rufo, D.D.; Debelee, T.G.; Ibenthal, A.; Negera, W.G. Diagnosis of Diabetes Mellitus Using Gradient Boosting Machine (LightGBM). Diagnostics 2021, 11, 1714. [Google Scholar] [CrossRef] [PubMed]
- Yagin, F.H.; Al-Hashem, F.; Ahmad, I.; Ahmad, F.; Alkhateeb, A. Pilot-Study to Explore Metabolic Signature of Type 2 Diabetes: A Pipeline of Tree-Based Machine Learning and Bioinformatics Techniques for Biomarkers Discovery. Nutrients 2024, 16, 1537. [Google Scholar] [CrossRef]
- Naveed, S.; Husnain, M. A drug recommendation system based on response prediction: Integrating gene expression and K-mer fragmentation of drug SMILES using LightGBM. Intell. Med. 2025, 11, 100206. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Sharma, M.; Donaldson, K.A. Explainable Boosting Machines for Slope Failure Spatial Predictive Modeling. Remote Sens. 2021, 13, 4991. [Google Scholar] [CrossRef]
- Sarica, A.; Quattrone, A.; Quattrone, A. Explainable boosting machine for predicting Alzheimer’s disease from MRI hippocampal subfields. In Proceedings of the 2021 International Conference on Brain Informatics, Virtual, 17–19 September 2021; pp. 341–350. [Google Scholar]
- El-Mihoub, T.A.; Nolle, L.; Stahl, F. Explainable boosting machines for network intrusion detection with features reduction. In Proceedings of the International Conference on Innovative Techniques and Applications of Artificial Intelligence, Cambridge, UK, 13–15 December 2022; pp. 280–294. [Google Scholar]
- Körner, A.; Sailer, B.; Sari-Yavuz, S.; Haeberle, H.A.; Mirakaj, V.; Bernard, A.; Rosenberger, P.; Koeppen, M. Explainable Boosting Machine approach identifies risk factors for acute renal failure. Intensiv. Care Med. Exp. 2024, 12, 55. [Google Scholar] [CrossRef]
- Liu, G.; Sun, B. Concrete compressive strength prediction using an explainable boosting machine model. Case Stud. Constr. Mater. 2023, 18, e01845. [Google Scholar] [CrossRef]
- Assel, M.; Sjoberg, D.D.; Vickers, A.J. The Brier score does not evaluate the clinical utility of diagnostic tests or prediction models. Diagn. Progn. Res. 2017, 1, 19. [Google Scholar] [CrossRef]
- Jiang, Y.; Pan, Q.; Liu, Y.; Evans, S. A statistical review: Why average weighted accuracy, not accuracy or AUC? Biostat. Epidemiol. 2021, 5, 267–286. [Google Scholar] [CrossRef]
- Monaghan, T.F.; Rahman, S.N.; Agudelo, C.W.; Wein, A.J.; Lazar, J.M.; Everaert, K.; Dmochowski, R.R. Foundational Statistical Principles in Medical Research: Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value. Medicina 2021, 57, 503. [Google Scholar] [CrossRef] [PubMed]
- Nori, H.; Caruana, R.; Bu, Z.; Shen, J.H.; Kulkarni, J. Accuracy, interpretability, and differential privacy via explainable boosting. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 8227–8237. [Google Scholar]
- Mahmoudian, A.; Bypour, M.; Kioumarsi, M. Explainable Boosting Machine Learning for Predicting Bond Strength of FRP Rebars in Ultra High-Performance Concrete. Computation 2024, 12, 202. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, L.; Bathon, J.M.; England, B.R.; St. Clair, E.W.; Arayssi, T.; Carandang, K.; Deane, K.D.; Genovese, M.; Huston, K.K.; Kerr, G.; et al. 2021 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 1108–1123. [Google Scholar] [CrossRef]
- Hu, S.; Lin, Y.; Tang, Y.; Zhang, J.; He, Y.; Li, G.; Li, L.; Cai, X. Targeting dysregulated intracellular immunometabolism within synovial microenvironment in rheumatoid arthritis with natural products. Front. Pharmacol. 2024, 15, 1403823. [Google Scholar] [CrossRef]
- Smallwood, M.J.; Nissim, A.; Knight, A.R.; Whiteman, M.; Haigh, R.; Winyard, P.G. Oxidative stress in autoimmune rheumatic diseases. Free Radic. Biol. Med. 2018, 125, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Smolen, J.S. Diagnosis and management of rheumatoid arthritis: A review. JAMA 2018, 320, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C.Y.; Rao, D.A.; Kelly, S.; Goodman, S.M.; Tabechian, D.; Hughes, L.B.; Salomon-Escoto, K.; et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019, 20, 928–942. [Google Scholar] [CrossRef]
- Okano, T.; Saegusa, J.; Takahashi, S.; Ueda, Y.; Morinobu, A. Immunometabolism in rheumatoid arthritis. Immunol. Med. 2018, 41, 89–97. [Google Scholar] [CrossRef]
- Luo, T.-T.; Wu, Y.-J.; Yin, Q.; Chen, W.-G.; Zuo, J. The Involvement of Glucose and Lipid Metabolism Alteration in Rheumatoid Arthritis and Its Clinical Implication. J. Inflamm. Res. 2023, 16, 1837–1852. [Google Scholar] [PubMed]
- Cai, W.; Yu, Y.; Zong, S.; Wei, F. Metabolic reprogramming as a key regulator in the pathogenesis of rheumatoid arthritis. Inflamm. Res. 2020, 69, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Turunen, S.; Huhtakangas, J.; Nousiainen, T.; Valkealahti, M.; Melkko, J.; Risteli, J.; Lehenkari, P. Rheumatoid arthritis antigens homocitrulline and citrulline are generated by local myeloperoxidase and peptidyl arginine deiminases 2, 3 and 4 in rheumatoid nodule and synovial tissue. Arthritis Res. Ther. 2016, 18, 239. [Google Scholar] [CrossRef]
- Kaore, S.N.; Amane, H.S.; Kaore, N.M. Citrulline: Pharmacological perspectives and its role as an emerging biomarker in future. Fundam. Clin. Pharmacol. 2013, 27, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Hormesis: Why it is important to toxicology and toxicologists. Environ. Toxicol. Chem. 2008, 27, 1451–1474. [Google Scholar] [CrossRef]
- Sokolove, J.; Strand, V. Rheumatoid arthritis classification criteria: It’s finally time to move on! Bull. NYU Hosp. Jt. Dis. 2010, 68, 232–238. [Google Scholar]
- Kaore, S.N.; Kaore, N.M. Arginine and citrulline as nutraceuticals: Efficacy and safety in diseases. In Nutraceuticals; Academic Press: Cambridge, MA, USA, 2021; pp. 925–944. [Google Scholar]
- Sharma, K.; Saini, N.; Hasija, Y. Identifying the mitochondrial metabolism network by integration of machine learning and explainable artificial intelligence in skeletal muscle in type 2 diabetes. Mitochondrion 2024, 74, 101821. [Google Scholar] [CrossRef]
- Villafañe, J.H.; Valdes, K.; Pedersini, P.; Berjano, P. Osteoarthritis: A call for research on central pain mechanism and personalized prevention strategies. Clin. Rheumatol. 2019, 38, 583–584. [Google Scholar] [CrossRef]
- Pedersini, P.; Turroni, S.; Villafañe, J.H. Gut microbiota and physical activity: Is there an evidence-based link? Sci. Total Environ. 2020, 727, 138648. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., III; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D. 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
| Metabolite Name | Control | RA | p-Value | ES |
|---|---|---|---|---|
| Median (Min–Max) | Median (Min–Max) | |||
| 1-Methyladenosine | 0.01 (0.007–0.013) | 0.009 (0.006–0.019) | 0.269 | |
| 1-Methylnicotinamide | 0.018 (0.008–0.036) | 0.016 (0.009–0.068) | 0.099 | |
| 2-Amino-2-(hydroxymethyl)-1,3-propanediol | 0.181 (0.063–1.071) | 0.088 (0.054–1.168) | <0.001 | 0.54 |
| 2-Aminoadipic acid | 0.021 (0.014–0.03) | 0.023 (0.014–0.049) | 0.092 | |
| 2-Aminobutyric acid | 0.369 (0.221–0.473) | 0.355 (0.207–0.699) | 0.823 | |
| 2-Hydroxybutyric acid; 2-Hydroxyisobutyric acid | 0.228 (0.117–0.287) | 0.246 (0.129–0.522) | 0.032 | 0.60 |
| 2-Hydroxypentanoic acid | 0.097 (0.059–0.165) | 0.091 (0.055–0.344) | 0.400 | |
| 2-Oxoglutaric acid | 0.086 (0.067–0.108) | 0.087 (0.057–0.123) | 0.491 | |
| 2-Oxoisovaleric acid; 4-Oxovaleric acid | 0.092 (0.073–0.122) | 0.098 (0.06–0.15) | 0.010 | 0.43 |
| 3-(2-Hydroxyphenyl)propionic acid; 3-Phenyllactic acid; Tropic acid;3-Ethoxybenzoic acid; 3-(4-Hydroxyphenyl)propionic acid | 0.031 (0.024–0.062) | 0.03 (0.016–0.087) | 0.211 | |
| 3-Hydroxybutyric acid | 0.244 (0.1–0.422) | 0.157 (0.072–1.025) | 0.006 | 0.01 |
| 3-Indoxylsulfuric acid | 0.073 (0.032–0.183) | 0.085 (0.017–0.659) | 0.873 | |
| 3-Methylhistidine | 0.132 (0.081–0.375) | 0.207 (0.062–0.81) | 0.001 | 0.66 |
| 3-Phenylpropionic acid | 0.032 (0.028–0.046) | 0.03 (0.023–0.085) | 0.175 | |
| 4-Methyl-2-oxopentanoic acid; 3-Methyl-2-oxovaleric acid | 0.552 (0.409–0.81) | 0.537 (0.283–0.952) | 0.851 | |
| 5-Oxoproline | 0.174 (0.132–0.218) | 0.16 (0.108–0.22) | 0.016 | 0.51 |
| ADMA | 0.023 (0.02–0.029) | 0.027 (0.016–0.038) | 0.002 | 0.57 |
| ADP;dGDP | 0.017 (0.009–0.054) | 0.012 (0.003–0.08) | 0.154 | |
| Ala | 5.065 (2.838–7.465) | 5.521 (3.686–11.994) | 0.035 | 0.35 |
| Allantoin | 0.113 (0.088–0.138) | 0.113 (0.064–0.158) | 0.334 | |
| Arg | 2.676 (1.857–3.427) | 2.556 (1.191–4.405) | 0.318 | |
| Asn | 0.699 (0.61–0.975) | 0.726 (0.481–1.234) | 0.449 | |
| Asp | 0.064 (0.045–0.083) | 0.064 (0.04–0.356) | 0.672 | |
| Azelaic acid | 0.024 (0.02–0.035) | 0.026 (0.01–0.058) | 0.004 | 0.57 |
| Benzoic acid | 0.034 (0.027–0.047) | 0.037 (0.024–0.077) | 0.170 | |
| beta-Ala | 0.044 (0.027–0.067) | 0.046 (0.026–0.175) | 0.218 | |
| Betaine | 1.775 (1.066–2.106) | 1.603 (0.922–2.975) | 0.453 | |
| Carnitine | 2.812 (2.112–3.388) | 2.531 (1.701–3.355) | 0.008 | 0.61 |
| Cholic acid | 0.024 (0.009–0.183) | 0.029 (0.006–0.183) | 0.272 | |
| Choline | 1.023 (0.715–1.317) | 0.87 (0.535–2.133) | 0.007 | 0.30 |
| Citric acid | 1.692 (1.37–2.293) | 1.6 (1.018–2.827) | 0.520 | |
| Citrulline | 0.823 (0.626–1.246) | 0.676 (0.196–1.826) | 0.002 | 0.46 |
| Creatine | 1.647 (0.811–2.923) | 1.389 (0.267–5.287) | 0.021 | 0.35 |
| Creatinine | 1.623 (1.339–1.833) | 1.52 (1.021–5.961) | 0.171 | |
| Cys | 0.098 (0.065–0.123) | 0.104 (0.051–0.173) | 0.110 | |
| Cysteine-glutathione disulphide | 0.032 (0.016–0.042) | 0.023 (0.01–0.048) | <0.001 | 0.84 |
| Cysteine-glutathione disulphide-Divalent | 0.052 (0.03–0.085) | 0.036 (0.014–0.065) | <0.001 | 1.44 |
| Cystine | 1.128 (0.987–1.503) | 1.25 (0.756–1.754) | 0.068 | |
| Decanoic acid | 0.056 (0.035–0.125) | 0.055 (0.022–0.175) | 0.652 | |
| Dihydroorotic acid | 0.131 (0.085–0.145) | 0.13 (0.085–0.165) | 0.430 | |
| gamma-Aminobutyric acid | 0.051 (0.02–0.107) | 0.045 (0.012–0.237) | 0.332 | |
| gamma-Butyrobetaine | 0.126 (0.084–0.199) | 0.092 (0.055–0.183) | <0.001 | 1.07 |
| Gln | 11.709 (9.18–15.218) | 11.016 (7.37–14.963) | 0.013 | 0.52 |
| Glu;threo-beta-methylaspartic acid | 0.518 (0.376–0.857) | 0.773 (0.222–2.049) | <0.001 | 0.72 |
| Gluconic acid | 0.047 (0.041–0.059) | 0.054 (0.032–0.263) | 0.001 | 0.41 |
| Glucose 6-phosphate; Fructose 6-phosphate; Glucose 1-phosphate | 0.052 (0.027–0.091) | 0.036 (0.012–0.113) | <0.001 | 0.71 |
| Glucuronic acid; Galacturonic acid | 0.032 (0.018–0.033) | 0.032 (0.013–0.133) | 0.035 | 0.54 |
| Gly | 2.866 (1.632–5.633) | 2.054 (1.211–3.48) | <0.001 | 1.27 |
| Glyceric acid | 0.064 (0.043–0.082) | 0.078 (0.04–0.133) | <0.001 | 0.99 |
| Glycerol-3-phosphate | 0.011 (0.008–0.019) | 0.018 (0.006–0.039) | <0.001 | 1.14 |
| Glycocholic acid | 0.021 (0.006–0.115) | 0.016 (0.005–0.115) | 0.085 | |
| Glycolic acid | 0.066 (0.04–0.076) | 0.062 (0.039–0.113) | 0.386 | |
| Glyoxylic acid | 0.02 (0.014–0.026) | 0.02 (0.013–0.032) | 0.259 | |
| Guanidinosuccinic acid | 0.015 (0.009–0.023) | 0.014 (0.008–0.089) | 0.994 | |
| Guanidoacetic acid | 0.064 (0.037–0.097) | 0.047 (0.03–0.11) | <0.001 | 0.76 |
| Hippuric acid | 0.035 (0.014–0.132) | 0.029 (0.008–0.162) | 0.045 | 0.18 |
| His | 2.006 (1.81–2.576) | 1.854 (1.172–2.888) | 0.001 | 0.65 |
| Homoarginine or N6.N6.N6-Trimethyllysine | 0.05 (0.031–0.068) | 0.062 (0.032–0.097) | 0.005 | 0.50 |
| Homovanillic acid | 0.014 (0.011–0.024) | 0.017 (0.012–0.036) | <0.001 | 0.54 |
| Hydroxyproline | 0.174 (0.082–0.352) | 0.186 (0.116–0.889) | 0.404 | |
| Hypoxanthine | 0.061 (0.016–0.111) | 0.049 (0.022–0.166) | 0.004 | 0.26 |
| Ile | 2.74 (1.796–4.963) | 2.731 (1.732–6.114) | 0.392 | |
| Indole-3-acetic acid | 0.048 (0.033–0.073) | 0.046 (0.029–0.145) | 0.728 | |
| Isethionic acid | 0.02 (0.012–0.038) | 0.019 (0.011–0.084) | 0.185 | |
| Isocitric acid | 0.082 (0.057–0.109) | 0.091 (0.055–0.189) | 0.007 | 0.49 |
| Kynurenine | 0.051 (0.038–0.06) | 0.047 (0.027–0.145) | 0.388 | |
| Lactic acid | 5.526 (3.405–8.157) | 6.551 (3.506–14.422) | 0.004 | 0.54 |
| Lauric acid | 0.139 (0.064–0.203) | 0.089 (0.045–0.46) | 0.028 | 0.03 |
| Leu | 5.258 (3.7–8.267) | 5.091 (2.734–9.618) | 0.548 | |
| Lys | 3.802 (2.864–6.097) | 4.281 (2.786–8.892) | 0.169 | |
| Malic acid | 0.063 (0.034–0.096) | 0.059 (0.034–0.13) | 0.491 | |
| Met | 0.36 (0.288–0.413) | 0.318 (0.18–0.798) | 0.016 | 0.04 |
| Methionine sulfoxide | 0.078 (0.047–0.119) | 0.088 (0.043–0.141) | 0.152 | |
| Mucic acid; Glucaric acid | 0.029 (0.017–0.047) | 0.034 (0.013–0.057) | 0.214 | |
| N.N-Dimethylglycine | 0.082 (0.059–0.138) | 0.108 (0.055–0.229) | <0.001 | 0.60 |
| N5-Ethylglutamine | 0.063 (0.044–0.124) | 0.068 (0.04–0.919) | 0.408 | |
| N6.N6.N6-Trimethyllysine | 0.047 (0.033–0.068) | 0.047 (0.033–0.083) | 0.388 | |
| N-Acetyl-beta-alanine; N-Acetyl-beta-alanine | 0.017 (0.011–0.028) | 0.02 (0.01–0.034) | 0.021 | 0.27 |
| N-Acetyleucine | 0.105 (0.098–0.163) | 0.03 (0.008–0.265) | <0.001 | 0.67 |
| N-Acetylneuraminic acid | 0.084 (0.068–0.128) | 0.08 (0.047–0.14) | 0.400 | |
| O-Acetylcarnitine | 1.069 (0.58–1.344) | 0.824 (0.412–1.684) | <0.001 | 0.42 |
| Octanoic acid | 0.069 (0.051–0.143) | 0.065 (0.039–0.155) | 0.398 | |
| Ornithine | 1.299 (0.758–1.901) | 1.194 (0.647–2.414) | 0.392 | |
| Pelargonic acid | 0.07 (0.061–0.094) | 0.08 (0.054–0.138) | 0.007 | 0.66 |
| Phe | 2.22 (1.992–2.857) | 2.876 (1.511–9.056) | <0.001 | 0.72 |
| Pipecolic acid | 0.047 (0.033–0.549) | 0.059 (0.03–0.511) | 0.131 | |
| Pro | 3.809 (1.937–8.386) | 4.831 (2.252–9.739) | 0.007 | 0.37 |
| Pyruvic acid | 0.124 (0.085–0.273) | 0.228 (0.078–0.418) | <0.001 | 1.58 |
| Quinic acid | 0.038 (0.024–0.074) | 0.036 (0.014–0.15) | 0.101 | 0.39 |
| Sarcosine | 0.06 (0.046–0.119) | 0.061 (0.03–0.186) | 0.834 | |
| SDMA | 0.022 (0.018–0.028) | 0.024 (0.016–0.084) | 0.037 | 0.38 |
| Ser | 1.836 (1.45–2.595) | 1.524 (0.781–3.111) | <0.001 | 0.72 |
| S-Sulfocysteine | 0.007 (0.006–0.011) | 0.008 (0.005–0.014) | 0.153 | |
| Succinic acid; Methylmalonic acid | 0.057 (0.035–0.074) | 0.049 (0.027–0.073) | <0.001 | 0.63 |
| Taurine | 0.459 (0.342–2.117) | 0.441 (0.265–3.031) | 0.408 | |
| Thr | 2.163 (1.828–3.16) | 2.19 (1.45–3.282) | 0.652 | |
| Threonic acid | 0.214 (0.086–0.26) | 0.23 (0.077–0.455) | <0.001 | 0.78 |
| Trimethylamine N-oxide | 0.033 (0.015–0.393) | 0.053 (0.019–1.444) | 0.091 | |
| Trp | 1.541 (1.236–1.879) | 1.636 (0.921–2.904) | 0.138 | |
| Tyr | 1.711 (1.363–1.989) | 1.977 (0.917–3.397) | 0.001 | 0.65 |
| Urea | 32.355 (21.09–47.464) | 35.811 (21.836–70.115) | 0.006 | 0.40 |
| Uric acid | 1.911 (1.381–2.553) | 2.005 (1.237–3.465) | 0.408 | |
| Uridine | 0.102 (0.086–0.168) | 0.096 (0.074–0.212) | 0.285 | |
| Val | 6.531 (5.305–11.255) | 7.277 (4.631–12.402) | 0.146 |
| Model | Metric | Value | BCI * (95%) |
|---|---|---|---|
| EBM | Accuracy | 0.847 | (0.776–0.918) |
| F1-score | 0.851 | (0.781–0.922) | |
| Sensitivity | 0.878 | (0.752–0.954) | |
| Specificity | 0.816 | (0.68–0.912) | |
| Positive predictive value | 0.827 | (0.697–0.918) | |
| Negative predictive value | 0.87 | (0.737–0.951) | |
| AUC | 0.901 | (0.847–0.955) | |
| Brier score | 0.129 | (0.109–0.153) | |
| LightGBM | Accuracy | 0.806 | (0.728–0.884) |
| F1-score | 0.812 | (0.735–0.889) | |
| Sensitivity | 0.837 | (0.703–0.927) | |
| Specificity | 0.776 | (0.634–0.882) | |
| Positive predictive value | 0.788 | (0.653–0.889) | |
| Negative predictive value | 0.826 | (0.686–0.922) | |
| AUC | 0.866 | (0.806–0.926) | |
| Brier score | 0.146 | (0.133–0.185) | |
| AdaBoost | Accuracy | 0.776 | (0.693–0.858) |
| F1-score | 0.784 | (0.703–0.866) | |
| Sensitivity | 0.816 | (0.68–0.912) | |
| Specificity | 0.735 | (0.589–0.851) | |
| Positive predictive value | 0.755 | (0.617–0.862) | |
| Negative predictive value | 0.8 | (0.654–0.904) | |
| AUC | 0.838 | (0.775–0.902) | |
| Brier score | 0.187 | (0.172–0.208) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yagin, F.H.; Colak, C.; Algarni, A.; Algarni, A.; Al-Hashem, F.; Ardigò, L.P. Explainable Boosting Machines Identify Key Metabolomic Biomarkers in Rheumatoid Arthritis. Medicina 2025, 61, 833. https://doi.org/10.3390/medicina61050833
Yagin FH, Colak C, Algarni A, Algarni A, Al-Hashem F, Ardigò LP. Explainable Boosting Machines Identify Key Metabolomic Biomarkers in Rheumatoid Arthritis. Medicina. 2025; 61(5):833. https://doi.org/10.3390/medicina61050833
Chicago/Turabian StyleYagin, Fatma Hilal, Cemil Colak, Abdulmohsen Algarni, Ali Algarni, Fahaid Al-Hashem, and Luca Paolo Ardigò. 2025. "Explainable Boosting Machines Identify Key Metabolomic Biomarkers in Rheumatoid Arthritis" Medicina 61, no. 5: 833. https://doi.org/10.3390/medicina61050833
APA StyleYagin, F. H., Colak, C., Algarni, A., Algarni, A., Al-Hashem, F., & Ardigò, L. P. (2025). Explainable Boosting Machines Identify Key Metabolomic Biomarkers in Rheumatoid Arthritis. Medicina, 61(5), 833. https://doi.org/10.3390/medicina61050833










