Investigation of Lonicera japonica Flos against Nonalcoholic Fatty Liver Disease Using Network Integration and Experimental Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening of Active Components in LJF
2.2. Target Prediction of Active Compounds in LJF
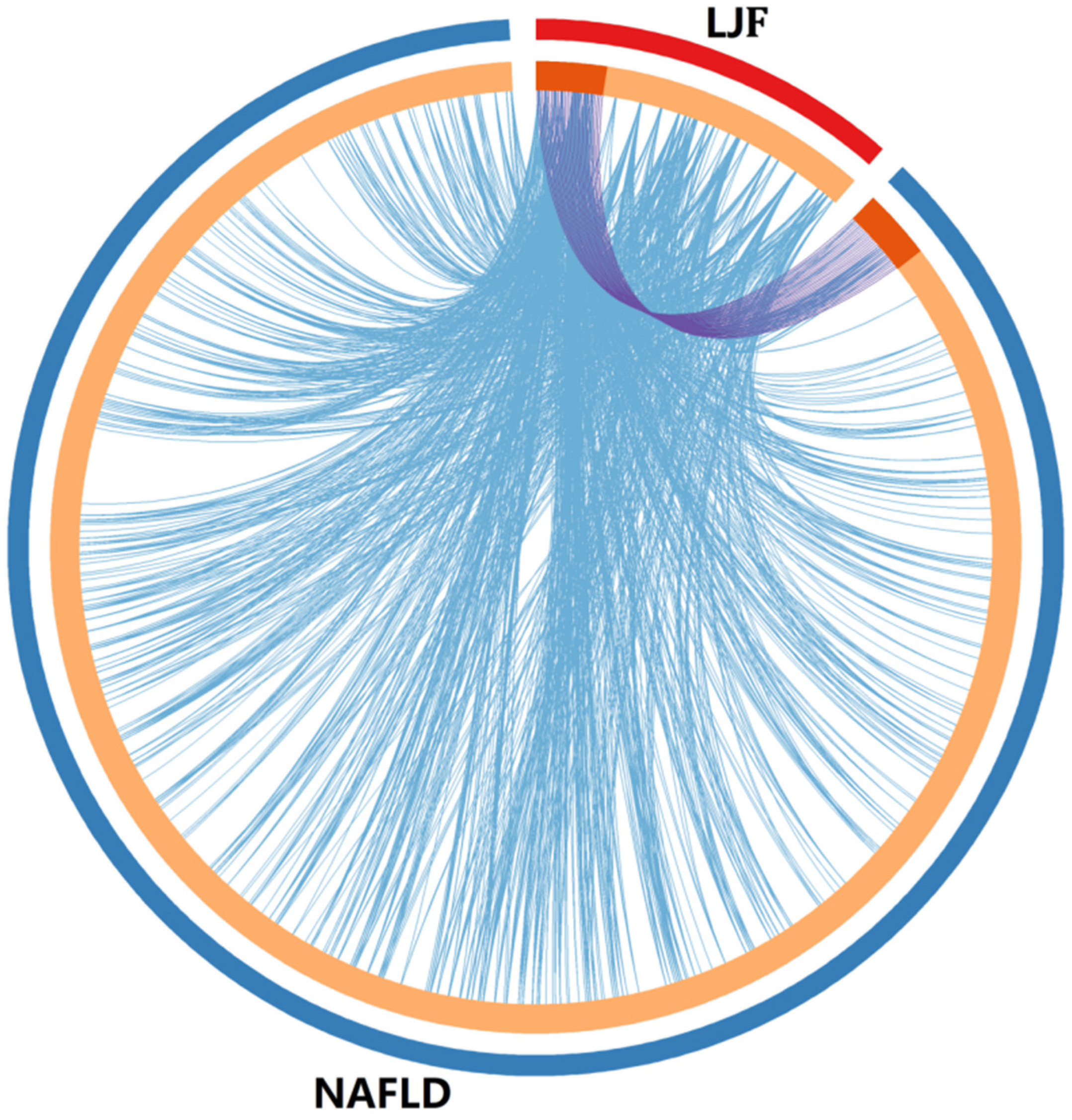
2.3. Collection of Potential Targets of LJF against NAFLD
2.4. Construction of Protein-Protein Interaction (PPI) Network for Potential Targets of LJF against NAFLD
2.5. GO Annotation and KEGG Pathways Enrichment Analysis
2.6. Acquisition Protein Function Information for Potential Targets of LJF against NAFLD
2.7. Construction of “Active Ingredient–Target–Pathway” Network for LJF against NAFLD
2.8. Molecular Docking Studies on the Key Active Ingredients with the Potential Targets
2.9. Lipid Accumulation Detection
2.10. Detection of ALT and AST Content in HepG2 Cells
2.11. TNF-α and CASP3 Expressions Were Detected in HepG2 Cells
2.12. Statistical Analysis
3. Results
3.1. Screening of LJF Active Components
3.2. Prediction of Target Genes
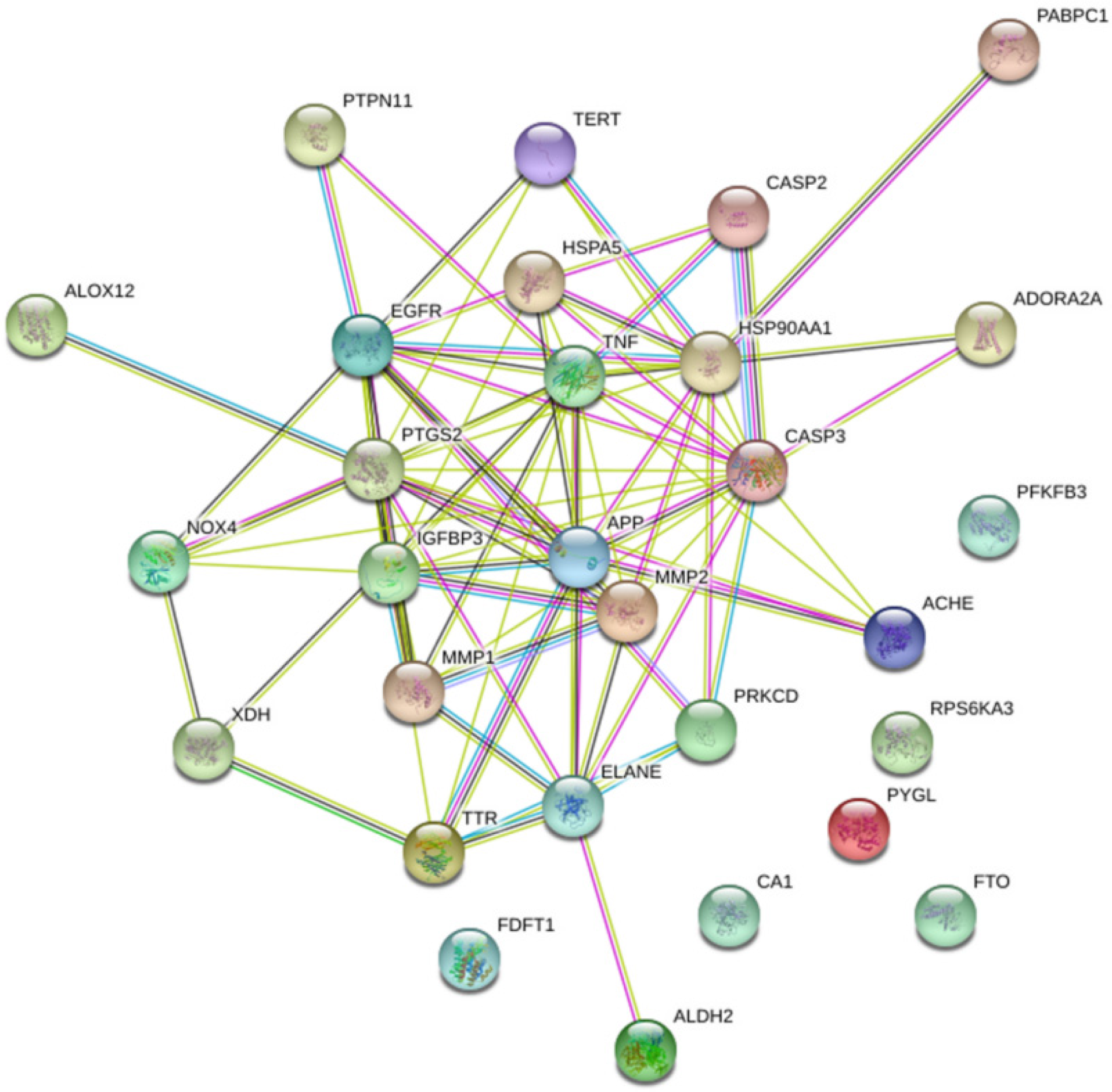
3.3. PPI Network for Potential Targets of LJF against NAFLD
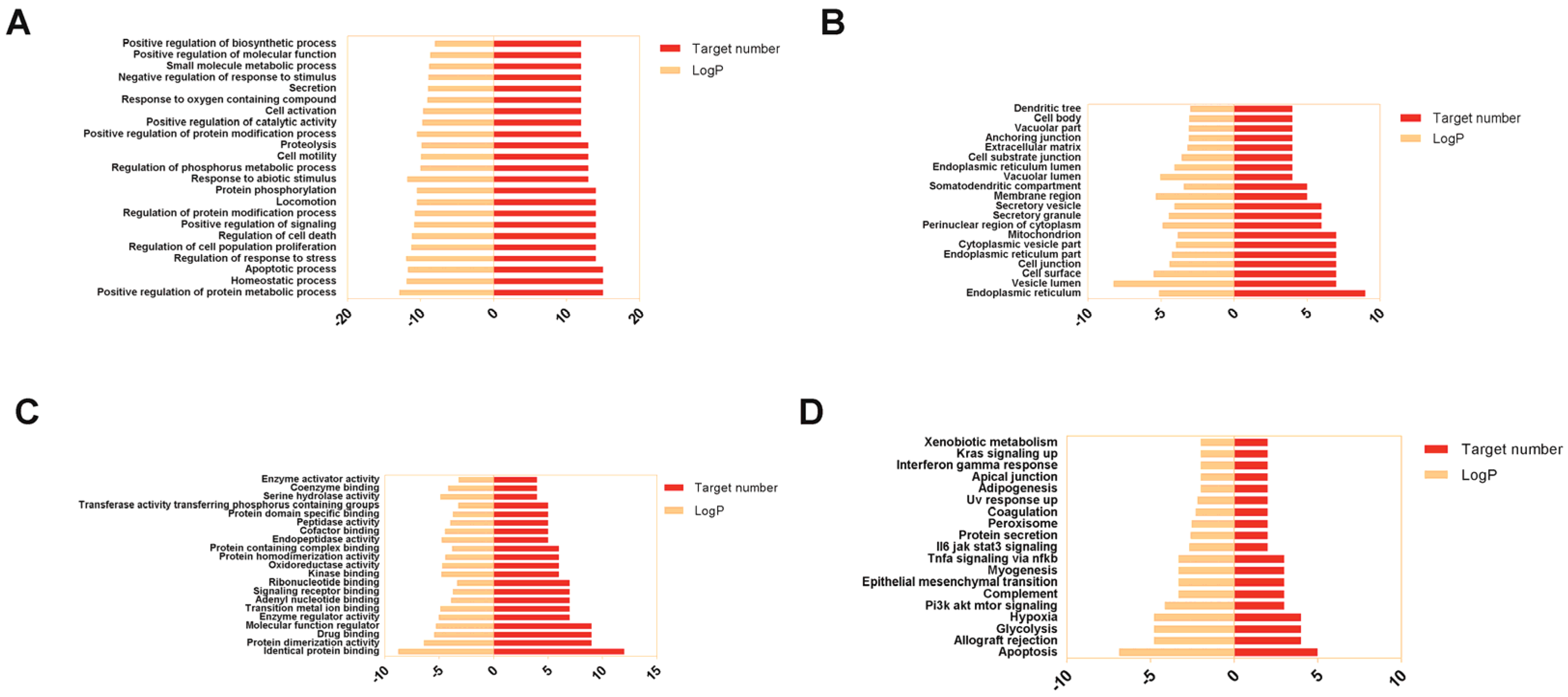
3.4. GO Annotation and KEGG Pathways Enrichment Analysis
3.5. Acquisition Protein Function Information for Potential Targets of LJF against NAFLD
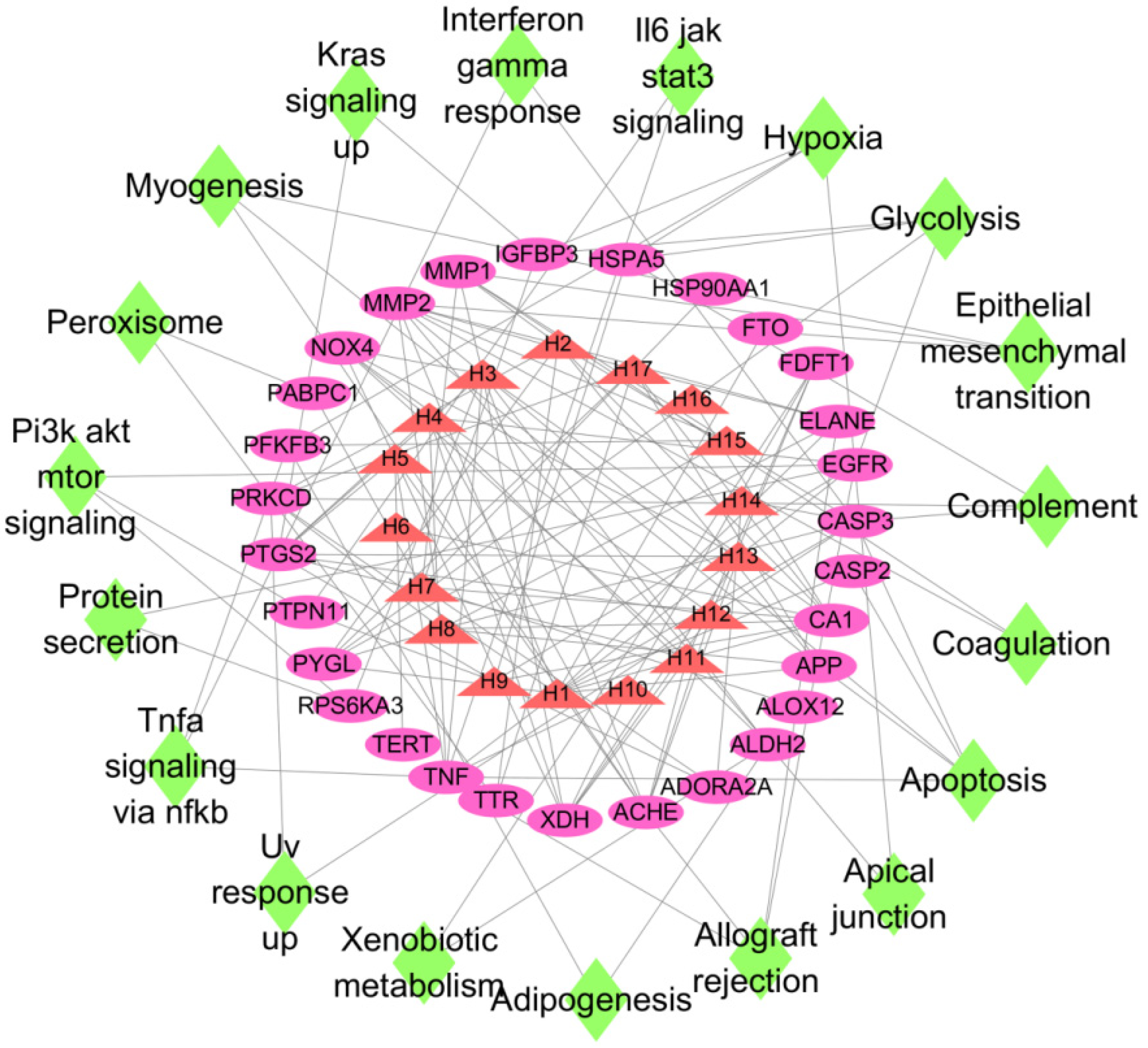
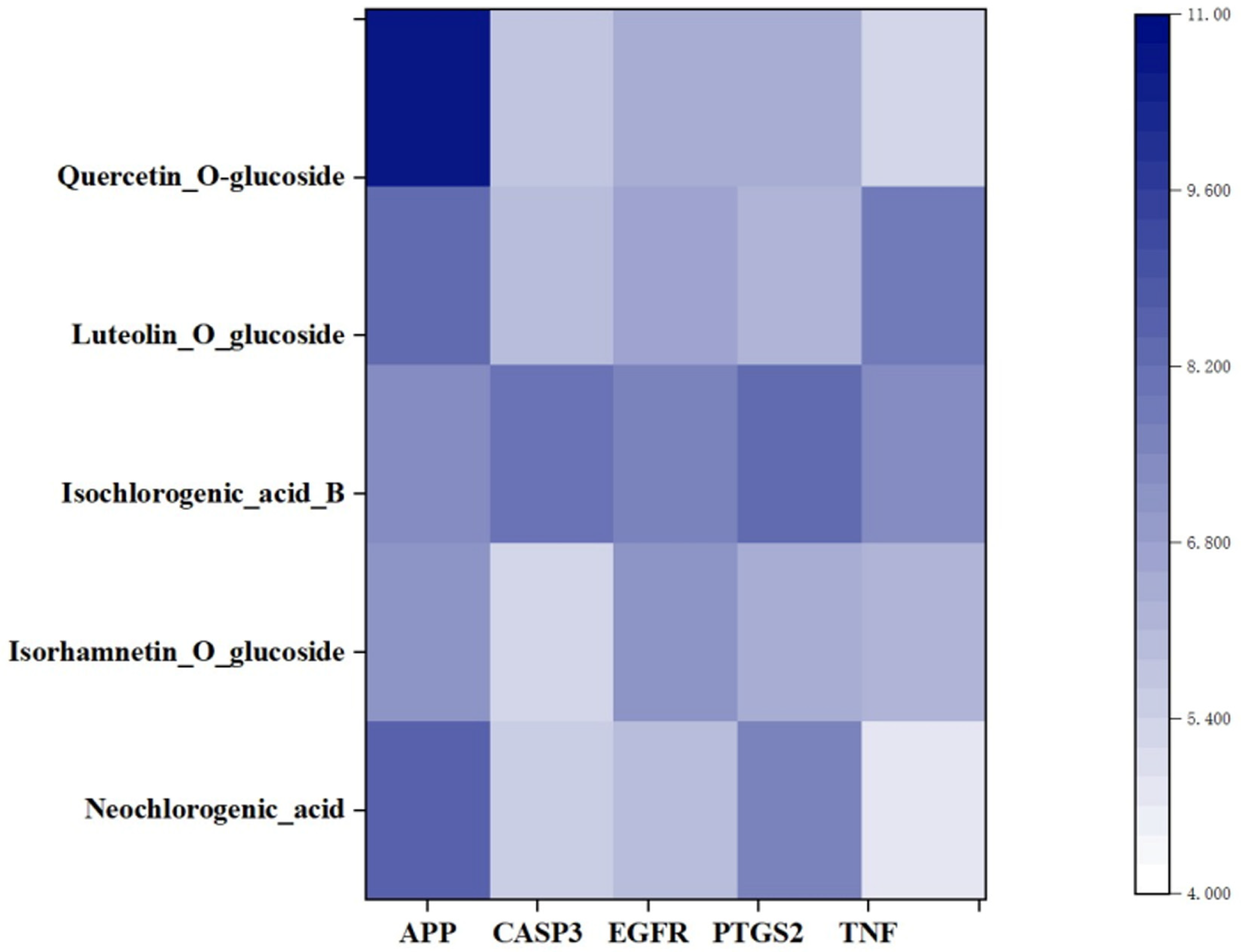
3.6. Construction of “Active Ingredient-Target-Pathway” Network for LJF against NAFLD
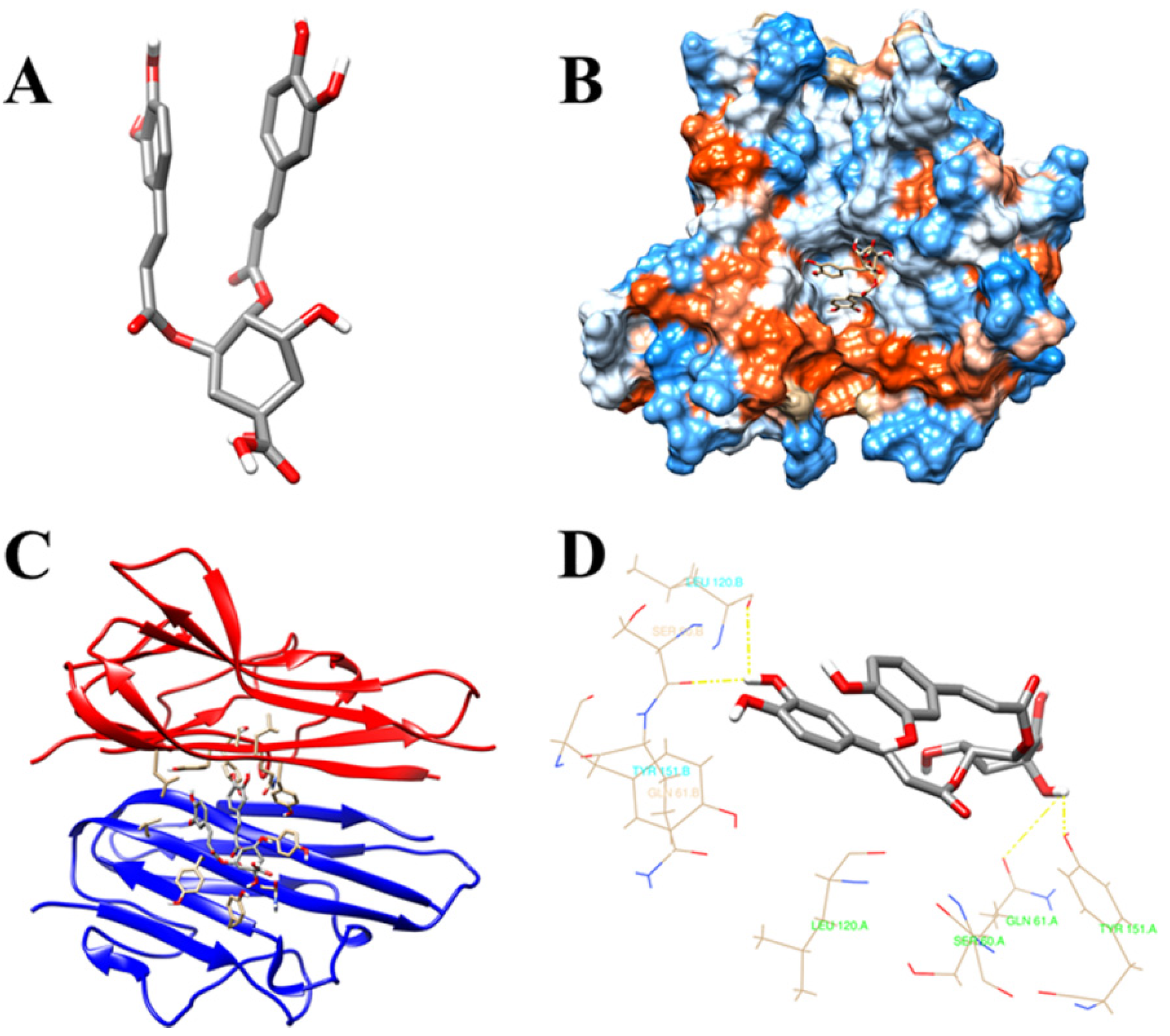
3.7. Molecular Docking Validation of the Key Ingredients and Target Proteins
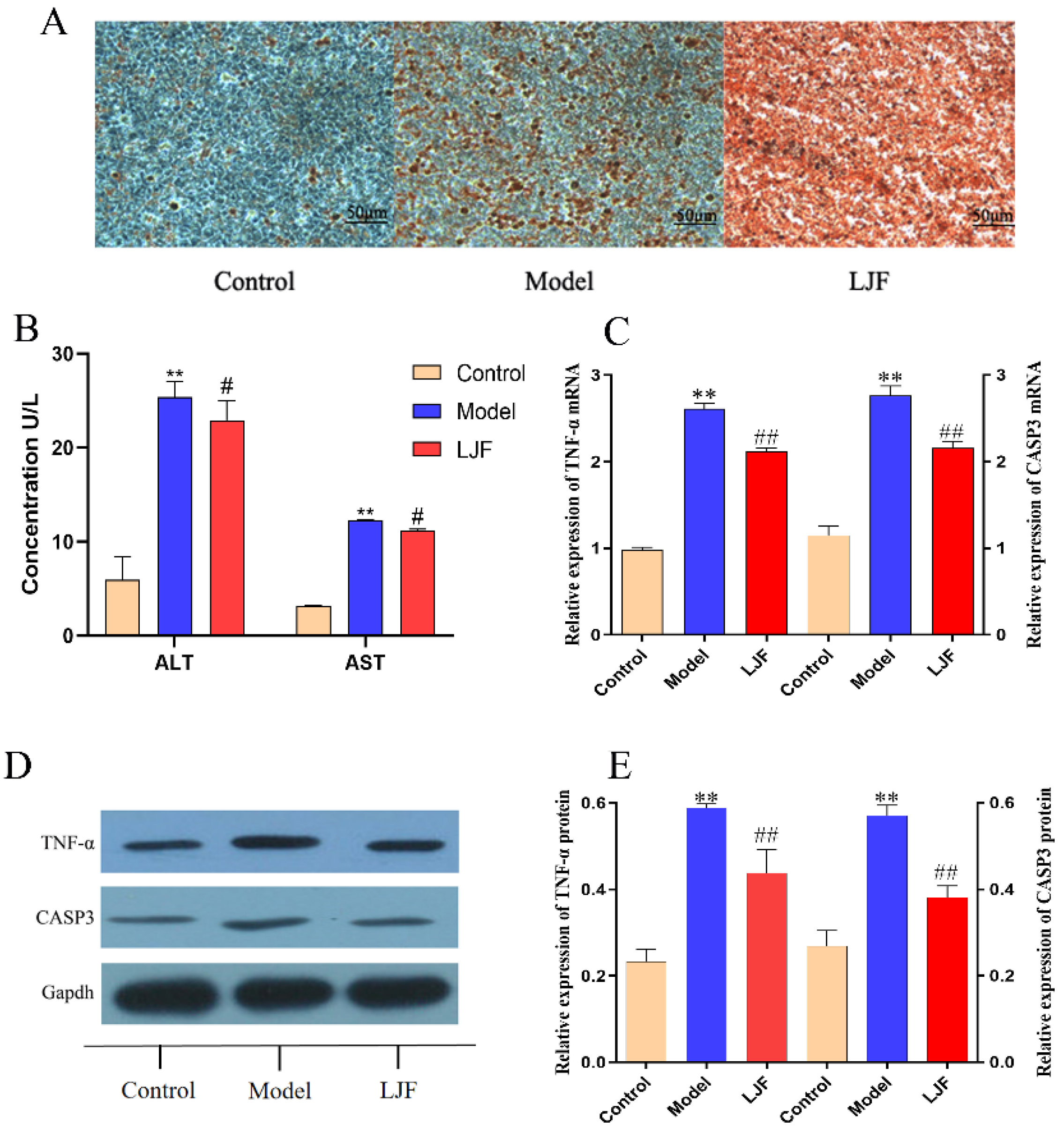
3.8. TNF-α and CASP3 Expression Were down Regulated in LJF-Treated NAFLD Model Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The Diagnosis and Management of Non-Alcoholic Fatty Liver Disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012, 55, 2005–2023. [Google Scholar] [CrossRef]
- Clark, J.M.; Brancati, F.L.; Diehl, A.M. Nonalcoholic fatty liver disease. Gastroenterology 2002, 122, 1649–1657. [Google Scholar] [CrossRef]
- Smith, C.J.; Ryckman, K.K. Epigenetic and Developmental Influences on the Risk of Obesity, Diabetes, and Metabolic Syndrome. Diabetes, Metab. Syndr. Obes. Targets Ther. 2015, 8, 295–302. [Google Scholar] [CrossRef]
- Loman, B.R.; Hernández-Saavedra, D.; An, R.; Rector, R.S. Prebiotic and Probiotic Treatment of Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Nutr. Rev. 2018, 76, 822–839. [Google Scholar] [CrossRef]
- Chander, K.; Negi, P.B.; Lola, B.; Julie, B.V.; Giovanni, T. Insights into the Molecular Targets and Emerging Pharmacotherapeutic Interventions for Nonalcoholic Fatty Liver Disease. Metabolism 2022, 126, 154925. [Google Scholar] [CrossRef]
- Fujimoto, M.; Tsuneyama, K.; Kinoshita, H.; Goto, H.; Takano, Y.; Selmi, C.; Keen, C.L.; Gershwin, M.E.; Shimada, Y. The Traditional Japanese Formula Keishibukuryogan Reduces Liver Injury and Inflammation in Patients with Nonalcoholic Fatty Liver Disease. Ann. N. Y. Acad. Sci. 2010, 1190, 151–158. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, J.; Song, H.; Yao, Z.; Ji, G. Extracts from Salvia-Nelumbinis Naturalis Alleviate Hepatosteatosis via Improving Hepatic Insulin Sensitivity. J. Transl. Med. 2014, 12, 236. [Google Scholar] [CrossRef][Green Version]
- Nam, Y.; Lee, J.M.; Wang, Y.; Ha, H.S.; Sohn, U.D. The Effect of Flos Lonicerae Japonicae Extract on Gastro-Intestinal Motility Function. J. Ethnopharmacol. 2016, 179, 280–290. [Google Scholar] [CrossRef]
- Shang, X.; Pan, H.; Li, M.; Miao, X.; Ding, H. Lonicera japonica Thunb.: Ethnopharmacology, Phytochemistry and Pharmacology of an Important Traditional Chinese Medicine. J. Ethnopharmacol. 2011, 138, 1–21. [Google Scholar] [CrossRef]
- Tzeng, T.-F.; Tzeng, Y.-C.; Cheng, Y.-J.; Liou, S.-S.; Liu, I.-M. The Ethanol Extract from Lonicera japonica Thunb. Regresses Nonalcoholic Steatohepatitis in a Methionine- and Choline-Deficient Diet-Fed Animal Model. Nutrients 2015, 7, 8670–8684. [Google Scholar] [CrossRef]
- Miao, H.; Zhang, Y.; Huang, Z.; Lu, B.; Ji, L. Lonicera japonica Attenuates Carbon Tetrachloride-Induced Liver Fibrosis in Mice: Molecular Mechanisms of Action. Am. J. Chin. Med. 2019, 47, 351–367. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, Y.; Li, Y.J.; Li, L.; Wang, S.H.; Xu, X.Y. The Immunomodulatory and the Protection of Experimental Liver Injury of Lonicera Macranthoides Hand: Mazz on Mice. Chin. Pharm. J. 2013, 48, 601–606. [Google Scholar]
- Hao, D.C.; Xiao, P.G. Network Pharmacology: A Rosetta Stone for Traditional Chinese Medicine. Drug Dev. Res. 2014, 75, 299–312. [Google Scholar] [CrossRef]
- Poornima, P.; Kumar, J.D.; Zhao, Q.; Blunder, M.; Efferth, T. Network Pharmacology of Cancer: From Understanding of Complex Interactomes to the Design of Multi-Target Specific Therapeutics from Nature. Pharmacol. Res. 2016, 111, 290–302. [Google Scholar] [CrossRef]
- Li, L.; Ma, S.; Wang, D.; Chen, L.; Wang, X. Plasma Metabolomics Analysis of Endogenous and Exogenous Metabolites in the Rat after Administration of Lonicerae japonicae Flos. Biomed. Chromatogr. 2020, 34, e4773. [Google Scholar] [CrossRef]
- Machado, D.; Girardini, M.; Viveiros, M.; Pieroni, M. Challenging the Drug-Likeness Dogma for New Drug Discovery in Tuberculosis. Front. Microbiol. 2018, 9, 1367. [Google Scholar] [CrossRef]
- Fong, S.Y.K.; Bauer-Brandl, A.; Brandl, M. Oral Bioavailability Enhancement through Supersaturation: An Update and Meta-Analysis. Expert Opin. Drug Deliv. 2017, 14, 403–426. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A Web Server for Target Prediction of Bioactive Small Molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D. GeneCards: Integrating Information about Genes, Proteins and Diseases. Trends Genet. 1997, 13, 163. [Google Scholar] [CrossRef]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.F.; et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Li, M.; Ying, M.; Zhao, R. Commentary: Efficacy and Safety of Chinese Herbal Medicine on Ovarian Cancer After Reduction Surgery and Adjuvant Chemotherapy: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 565812. [Google Scholar] [CrossRef]
- Yeung, W.-F.; Chung, K.-F.; Ng, K.-Y.; Yu, Y.-M.; Ziea, E.T.-C.; Ng, B.F.-L. A Systematic Review on the Efficacy, Safety and Types of Chinese Herbal Medicine for Depression. J. Psychiatr. Res. 2014, 57, 165–175. [Google Scholar] [CrossRef]
- Wang, Y.; Lou, X.-T.; Shi, Y.-H.; Tong, Q.; Zheng, G.-Q. Erxian Decoction, a Chinese Herbal Formula, for Menopausal Syndrome: An Updated Systematic Review. J. Ethnopharmacol. 2019, 234, 8–20. [Google Scholar] [CrossRef]
- Paterson, C.; Kennedy, C. Pharmacological Interventions for Treating Chronic Prostatitis/Chronic Pelvic Pain Syndrome. Res. Nurs. Health 2020, 43, 548–549. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, N.; Zhu, S.; Hu, X.; Chang, D.; Liu, J. Elucidation of the Mechanisms and Molecular Targets of Yiqi Shexue Formula for Treatment of Primary Immune Thrombocytopenia Based on Network Pharmacology. Front. Pharmacol. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Shi, T.; Wu, L.; Ma, W.; Ju, L.; Bai, M.; Chen, X.; Liu, S.; Yang, X.; Shi, J. Nonalcoholic Fatty Liver Disease: Pathogenesis and Treatment in Traditional Chinese Medicine and Western Medicine. Evid. -Based Complement. Altern. Med. 2020, 2020, 8749564. [Google Scholar] [CrossRef]
- Kakino, S.; Ohki, T.; Nakayama, H.; Yuan, X.; Otabe, S.; Hashinaga, T.; Wada, N.; Kurita, Y.; Tanaka, K.; Hara, K.; et al. Pivotal Role of TNF-α in the Development and Progression of Nonalcoholic Fatty Liver Disease in a Murine Model. Horm. Metab. Res. 2018, 50, 80–87. [Google Scholar] [CrossRef]
- Afonina, I.S.; Müller, C.; Martin, S.J.; Beyaert, R. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity 2015, 42, 991–1004. [Google Scholar] [CrossRef]
- Hirsova, P.; Gores, G.J. Death Receptor-Mediated Cell Death and Proinflammatory Signaling in Nonalcoholic Steatohepatitis. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 17–27. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Gores, G.J. Death Receptor-Mediated Apoptosis and the Liver. J. Hepatol. 2002, 37, 400–410. [Google Scholar] [CrossRef]
- Hsieh, P.-S.; Jin, J.-S.; Chiang, C.-F.; Chan, P.-C.; Chen, C.-H.; Shih, K.-C. COX-2-Mediated Inflammation in Fat Is Crucial for Obesity-Linked Insulin Resistance and Fatty Liver. Obesity 2009, 17, 1150–1157. [Google Scholar] [CrossRef]
- Fuchs, B.C.; Hoshida, Y.; Fujii, T.; Wei, L.; Yamada, S.; Lauwers, G.Y.; McGinn, C.M.; DePeralta, D.K.; Chen, X.; Kuroda, T.; et al. Epidermal Growth Factor Receptor Inhibition Attenuates Liver Fibrosis and Development of Hepatocellular Carcinoma. Hepatology 2014, 59, 1577–1590. [Google Scholar] [CrossRef]
- Min, H.K.; Maruyama, H.; Jang, B.K.; Shimada, M.; Mirshahi, F.; Ren, S.; Oh, Y.; Puri, P.; Sanyal, A.J. Suppression of IGF Binding Protein-3 by Palmitate Promotes Hepatic Inflammatory Responses. FASEB J. 2016, 30, 4071–4082. [Google Scholar] [CrossRef]
- Pinçon, A.; de Montgolfier, O.; Akkoyunlu, N.; Daneault, C.; Pouliot, P.; Villeneuve, L.; Lesage, F.; Levy, B.I.; Thorin-Trescases, N.; Thorin, É.; et al. Non-Alcoholic Fatty Liver Disease, and the Underlying Altered Fatty Acid Metabolism, Reveals Brain Hypoperfusion and Contributes to the Cognitive Decline in APP/PS1 Mice. Metabolites 2019, 9, 104. [Google Scholar] [CrossRef]
- Yokomori, H.; Oda, M.; Ando, W.; Inagaki, Y.; Okazaki, I. Hepatic Progenitor Cell Expansion in Early-Stage Nonalcoholic Steatohepatitis: Evidence from Immunohistochemistry and Immunoelectron Microscopy of Matrix Metalloproteinase-1. Med Mol. Morphol. 2017, 50, 238–242. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Luedde, T. Apoptosis and Necroptosis in the Liver: A Matter of Life and Death. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 738–752. [Google Scholar] [CrossRef]
- Mesarwi, O.A.; Shin, M.-K.; Bevans-Fonti, S.; Schlesinger, C.; Shaw, J.; Polotsky, V.Y. Hepatocyte Hypoxia Inducible Factor-1 Mediates the Development of Liver Fibrosis in a Mouse Model of Nonalcoholic Fatty Liver Disease. PLoS ONE 2016, 11, e0168572. [Google Scholar] [CrossRef]
- Yan, S.; Huda, N.; Khambu, B.; Yin, X.-M. Relevance of Autophagy to Fatty Liver Diseases and Potential Therapeutic Applications. Amino Acids 2017, 49, 1965–1979. [Google Scholar] [CrossRef]
- Zeng, J.; Zhu, B.; Su, M. Autophagy Is Involved in Acetylshikonin Ameliorating Non-Alcoholic Steatohepatitis through AMPK/mTOR Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 1645–1650. [Google Scholar] [CrossRef]
- Xu, C.; Chen, Y.; Xu, L.; Miao, M.; Li, Y.; Yu, C. Serum Complement C3 Levels Are Associated with Nonalcoholic Fatty Liver Disease Independently of Metabolic Features in Chinese Population. Sci. Rep. 2016, 6, 23279. [Google Scholar] [CrossRef]
- Syn, W.K.; Jung, Y.; Omenetti, A.; Abdelmalek, M.; Guy, C.D.; Yang, L.; Wang, J.; Witek, R.P.; Fearing, C.M.; Pereira, T.D.A.; et al. Hedgehog-Mediated Epithelial-to-Mesenchymal Transition and Fibrogenic Repair in Nonalcoholic Fatty Liver Disease. Gastroenterology 2009, 137, 1478–1488.e8. [Google Scholar] [CrossRef]
- Singh, J.; Kallwitz, E.R. Spotlight on Impactful Research: Relationship Between Relative Skeletal Muscle Mass and Nonalcoholic Fatty Liver Disease: A 7-Year Longitudinal Study. Clin. Liver Dis. 2020, 15, 213–214. [Google Scholar] [CrossRef]
| NO. | Name | Chemical Formula | 2D Structure |
|---|---|---|---|
| H1 | Neochlorogenic acid | C16H18O9 | 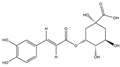 |
| H2 | Isochlorogenic acid B | C25H24O12 | 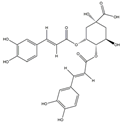 |
| H3 | Quercetin-O-glucoside | C21H20O12 | 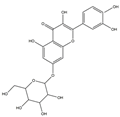 |
| H4 | Luteolin-O-glucoside | C21H20O11 | 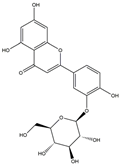 |
| H5 | Isorhamnetin-O-glucoside | C22H20O12 | 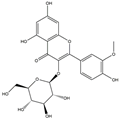 |
| H6 | Loganic acid | C16H24O10 | 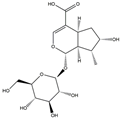 |
| H7 | Loganin | C17H26O10 | 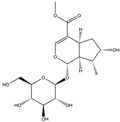 |
| H8 | Secoxyloganin | C17H24O11 | 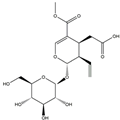 |
| H9 | Vogeloside | C17H24O10 | 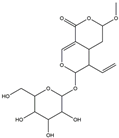 |
| H10 | Apigenin-glucuronide-sulfate | C21H18O14S | 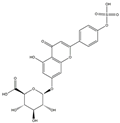 |
| H11 | Isorhamnetin-glucuronide-sulfate | C22H22O16S | 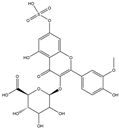 |
| H12 | Apigenin-glucuronide | C21H18O11 | 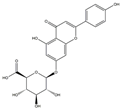 |
| H13 | 5-Hydroxy-6-methoxyindole glucuronide | C15H17NO8 | 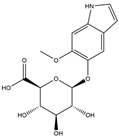 |
| H14 | Dihydrocaffeic acid-sulfate | C9H10O7S | 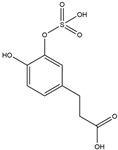 |
| H15 | Dihydroferulic acid-sulfate | C10H12O7S |  |
| H16 | Catechol sulfate | C6H6O5S | 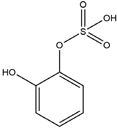 |
| H17 | Feruloylquinic acid | C17H20O9 | 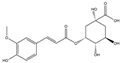 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.-Y.; Zhao, P.; Yan, P.-Z.; Li, J.; Zhao, D.-S. Investigation of Lonicera japonica Flos against Nonalcoholic Fatty Liver Disease Using Network Integration and Experimental Validation. Medicina 2022, 58, 1176. https://doi.org/10.3390/medicina58091176
Sun C-Y, Zhao P, Yan P-Z, Li J, Zhao D-S. Investigation of Lonicera japonica Flos against Nonalcoholic Fatty Liver Disease Using Network Integration and Experimental Validation. Medicina. 2022; 58(9):1176. https://doi.org/10.3390/medicina58091176
Chicago/Turabian StyleSun, Chun-Yong, Pan Zhao, Pei-Zheng Yan, Jia Li, and Dong-Sheng Zhao. 2022. "Investigation of Lonicera japonica Flos against Nonalcoholic Fatty Liver Disease Using Network Integration and Experimental Validation" Medicina 58, no. 9: 1176. https://doi.org/10.3390/medicina58091176
APA StyleSun, C.-Y., Zhao, P., Yan, P.-Z., Li, J., & Zhao, D.-S. (2022). Investigation of Lonicera japonica Flos against Nonalcoholic Fatty Liver Disease Using Network Integration and Experimental Validation. Medicina, 58(9), 1176. https://doi.org/10.3390/medicina58091176







