Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Oral Sample Collection and DNA Extraction
2.3. Oral Investigation
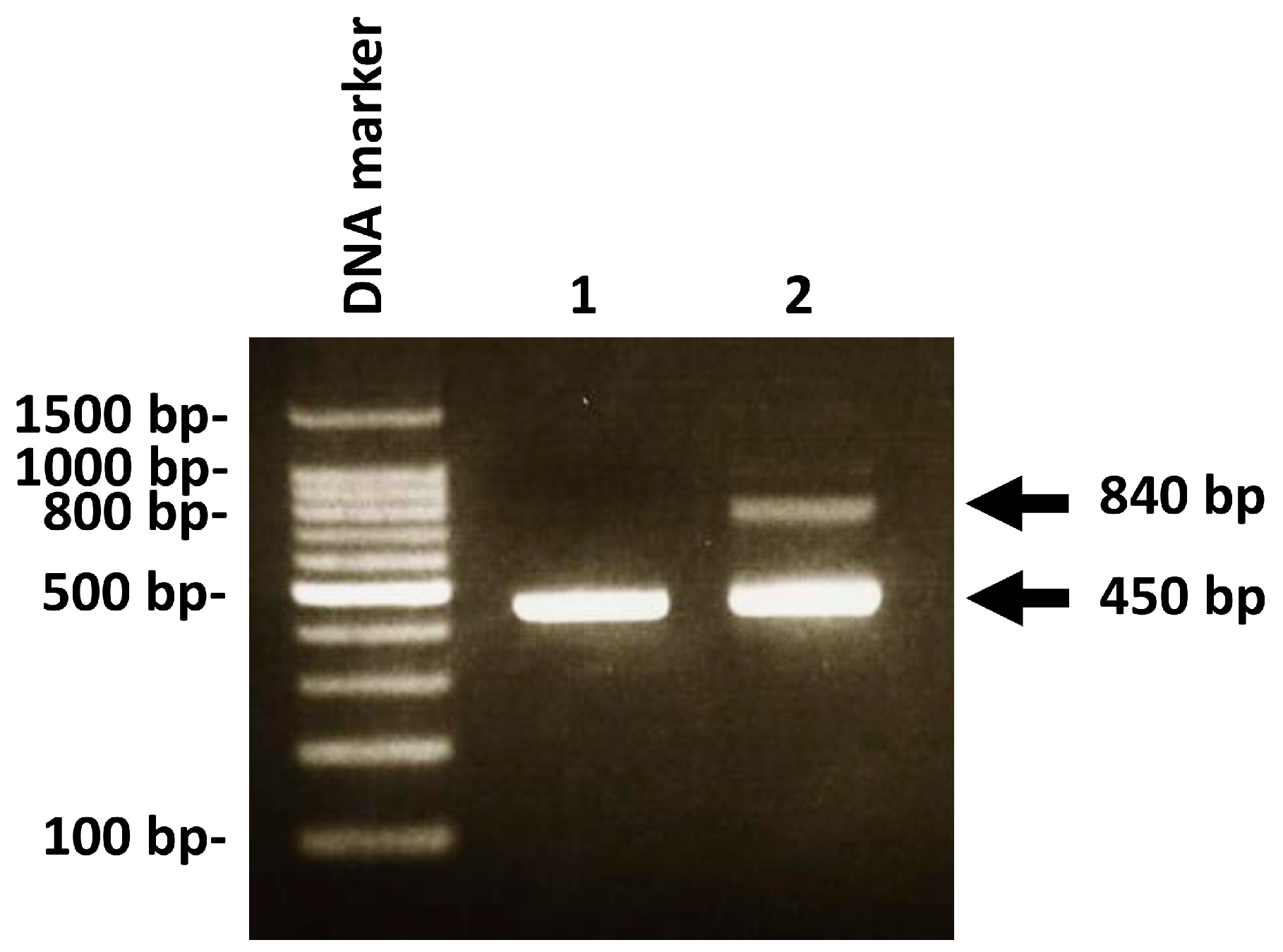
2.4. Detecting C. albicans DNA and P. gingivalis DNA by Real-Time PCR
2.5. PCR Analysis for Genotype Identification of C. albicans
2.6. Statistical Analysis
3. Results
3.1. Relationship between Oral C. albicans and Clinical Parameters
3.2. Relationship between Intron Insertion of C. albicans and Clinical Parameters
3.3. Relationship between Oral C. albicans/P. gingivalis and Clinical Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontol. 2000 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.; Haffajee, A.; Cugini, M.; Smith, C.; Kent, R.L. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Slots, J.; Slots, H. Periodontal herpesvirus morbidity and treatment. Periodontol. 2000 2019, 79, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Su, C.Y.; Shigeishi, H.; Murodumi, H.; Sugiyama, M.; Ohta, K.; Takemoto, T. Association of oral Epstein-Barr virus with periodontal health in Japanese adults. Exp. Ther. Med. 2021, 22, 767. [Google Scholar] [CrossRef]
- Nakamura, M.; Shigeishi, H.; Cheng-Yih, S.U.; Sugiyama, M.; Ohta, K. Oral human cytomegalovirus prevalence and its relationships with periodontitis and Porphyromonas gingivalis in Japanese adults: A cross-sectional study. J. Appl. Oral Sci. 2020, 28, e20200501. [Google Scholar] [CrossRef] [PubMed]
- Satala, D.; Gonzalez-Gonzalez, M.; Smolarz, M.; Surowiec, M.; Kulig, K.; Wronowska, E.; Zawrotniak, M.; Kozik, A.; Rapala-Kozik, M.; Karkowska-Kuleta, J. The Role of Candida albicans Virulence Factors in the Formation of Multispecies Biofilms with Bacterial Periodontal Pathogens. Front. Cell. Infect. Microbiol. 2022, 11, 765942. [Google Scholar] [CrossRef]
- Urzúa, B.; Hermosilla, G.; Gamonal, J.; Morales-Bozo, I.; Canals, M.; Barahona, S.; Cóccola, C.; Cifuentes, V. Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med. Mycol. 2008, 46, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Slots, J. Periodontal herpesviruses: Prevalence, pathogenicity, systemic risk. Periodontol. 2000 2015, 69, 28–45. [Google Scholar] [CrossRef]
- Imai, K.; Inoue, H.; Tamura, M.; Cueno, M.E.; Inoue, H.; Takeichi, O. The periodontal pathogen Porphyromonas gingivalis induces the Epstein-Barr virus lytic switch transactivator ZEBRA by histone modification. Biochimie 2012, 94, 839–846. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Bartnicka, D.; Zawrotniak, M.; Zielinska, G.; Kieronska, A.; Bochenska, O.; Ciaston, I.; Koziel, J.; Potempa, J.; Baster, Z.; et al. The activity of bacterial peptidylarginine deiminase is important during formation of dual-species biofilm by periodontal pathogen Porphyromonas gingivalis and opportunistic fungus Candida albicans. Pathog. Dis. 2018, 76, fty033. [Google Scholar] [CrossRef]
- McCullough, M.J.; Clemons, K.V.; Stevens, D.A. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 1999, 37, 417–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, M.; Watanabe, K.; Mikami, Y.; Yazawa, K.; Nishimura, K. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: Analysis reveals a new genotype of C. albicans with group I intron. J. Clin. Microbiol. 2001, 39, 4309–4315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantivitayakul, P.; Panpradit, N.; Maudcheingka, T.; Klaophimai, A.; Lapirattanakul, J. Genotyping of Candida albicans and Candida dubliniensis by 25S rDNA analysis shows association with virulence attributes in oral candidiasis. Arch. Oral Biol. 2019, 97, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.B.; Al Marzooq, F.; Himratul-Aznita, W.H.; Ahmed, H.M.A.; Samaranayake, L.P. Prevalence, virulence and antifungal activity of C. albicans isolated from infected root canals. BMC Oral Health 2020, 20, 347. [Google Scholar] [CrossRef] [PubMed]
- Nesse, W.; Abbas, F.; van der Ploeg, I.; Spijkervet, F.K.; Dijkstra, P.U.; Vissink, A. Periodontal inflamed surface area: Quantifying inflammatory burden. J. Clin. Periodontol. 2008, 35, 668–673. [Google Scholar] [CrossRef] [Green Version]
- Cutress, T.W.; Ainamo, J.; Sardo-Infirri, J. The community periodontal index of treatment needs (CPITN) procedure for population groups and individuals. Int. Dent. J. 1987, 37, 222–233. [Google Scholar]
- Hsu, M.C.; Chen, K.W.; Lo, H.J.; Chen, Y.C.; Liao, M.H.; Lin, Y.H.; Li, S.Y. Species identification of medically important fungi by use of real-time LightCycler PCR. J. Med. Microbiol. 2003, 52, 1071–1076. [Google Scholar] [CrossRef] [Green Version]
- Yasukawa, T.; Ohmori, M.; Sato, S. The relationship between physiologic halitosis and periodontopathic bacteria of the tongue and gingival sulcus. Odontology 2010, 98, 44–51. [Google Scholar] [CrossRef]
- Shigeishi, H.; Ohta, K.; Sugiyama, M. Prevalence and risk factors for oral HPV16/18 and candida in Japanese people without oral cancer or premalignant lesions. Oral Sci. Int. 2022; in press. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Wang, Y.; Jin, Y.; Wang, C. Heme Competition Triggers an Increase in the Pathogenic Potential of Porphyromonas gingivalis in Porphyromonas gingivalis-Candida albicans Mixed Biofilm. Front. Microbiol. 2020, 11, 596459. [Google Scholar] [CrossRef]
- Tamai, R.; Sugamata, M.; Kiyoura, Y. Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb. Pathog. 2011, 51, 250–254. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Könönen, E.; Paju, S.; Pussinen, P.J.; Hyvönen, M.; Di Tella, P.; Suominen-Taipale, L.; Knuuttila, M. Population-based study of salivary carriage of periodontal pathogens in adults. J. Clin. Microbiol. 2007, 45, 2446–2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.Y.; Shigeishi, H.; Nishimura, R.; Ohta, K.; Sugiyama, M. Detection of oral bacteria on the tongue dorsum using PCR amplification of 16S ribosomal RNA and its association with systemic disease in middle-aged and elderly patients. Biomed. Rep. 2019, 10, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigeishi, H.; Nakamura, M.; Oka, I.; Su, C.Y.; Yano, K.; Ishikawa, M.; Kaneyasu, Y.; Sugiyama, M.; Ohta, K. The Associations of Periodontopathic Bacteria and Oral Candida with Periodontal Inflamed Surface Area in Older Adults Receiving Supportive Periodontal Therapy. Diagnostics 2021, 11, 1397. [Google Scholar] [CrossRef] [PubMed]
| Clinical Factor (n) | C. albicans | p-Value | |
|---|---|---|---|
| (−) (n = 64) | (+) (n = 22) | ||
| Age | 69.7 ± 10.3 | 72.3 ± 9.4 | 0.43 |
| Age in years | |||
| 40–49 (4) | 4 (100%) | 0 (0%) | 0.64 |
| 50–59 (8) | 6 (75%) | 2 (25%) | |
| 60–69 (25) | 18 (72%) | 7 (28%) | |
| 70–79 (32) | 25 (78.1%) | 7 (21.9%) | |
| 80–89 (17) | 11 (64.7%) | 6 (35.3%) | |
| Gender | |||
| Male (25) | 15 (60%) | 10 (40%) | 0.06 |
| Female (61) | 49 (80.3%) | 12 (19.7%) | |
| Hypertension | |||
| No (69) | 53 (76.8%) | 16 (23.2%) | 0.36 |
| Yes (17) | 11 (64.7%) | 6 (35.3%) | |
| Diabetes | |||
| No (76) | 56 (73.7%) | 20 (26.3%) | 1.0 |
| Yes (10) | 8 (80%) | 2 (20%) | |
| Dyslipidemia | |||
| No (72) | 54 (75%) | 18 (25%) | 0.75 |
| Yes (14) | 10 (71.4%) | 4 (28.6%) | |
| Remaining teeth | 23.3 ± 5.8 | 21.9 ± 5.2 | 0.10 |
| Denture use | |||
| Non-user (55) | 44 (80%) | 11 (20%) | 0.13 |
| User (31) | 20 (64.5%) | 11 (35.5%) | |
| Plaque Control Record scores (%) | 20.3 ± 15.9 | 16.4 ± 9.1 | 0.49 |
| Probing depth | |||
| <4 mm (18) | 13 (72.2%) | 5 (27.8%) | 0.13 |
| ≥4 mm and <6 mm (42) | 35 (83.3%) | 7 (16.7%) | |
| ≥6 mm (26) | 16 (61.5%) | 10 (38.5%) | |
| Bleeding on probing (%) | 8.0 ± 8.9 | 8.4 ± 9.4 | 0.92 |
| ≥4 mm periodontal pocket with BOP | |||
| No (48) | 34 (70.8%) | 14 (29.2%) | 0.46 |
| Yes (38) | 30 (78.9%) | 8 (21.1%) | |
| ≥6 mm periodontal pocket with BOP | |||
| No (73) | 56 (76.7%) | 17 (23.3%) | 0.30 |
| Yes (13) | 8 (61.5%) | 5 (38.5%) | |
| PISA (mm2) | 93.6 ± 107.6 | 105.1 ± 143.9 | 0.66 |
| PESA (mm2) | 1023.0 ± 333.4 | 1007.5 ± 305.9 | 0.96 |
| Clinical Factor (n) | C. albicans (+) (n = 22) | p-Value | |
|---|---|---|---|
| Intron Insertion (−) (n = 12) | Intron Insertion (+) (n = 10) | ||
| Age | 69.4 ± 9.8 | 75.8 ± 7.8 | 0.16 |
| Age in years | |||
| 50–59 (2) | 2 (100%) | 0 (0%) | 0.27 |
| 60–69 (7) | 5 (71.4%) | 2 (28.6%) | |
| 70–79 (7) | 3 (42.9%) | 4 (57.1%) | |
| 80–89 (6) | 2 (33.4%) | 4 (66.7%) | |
| Gender | |||
| Male (10) | 5 (50%) | 5 (50%) | 1.0 |
| Female (12) | 7 (58.3%) | 5 (41.7%) | |
| Hypertension | |||
| No (16) | 9 (56.3%) | 7 (43.8%) | 1.0 |
| Yes (6) | 3 (50%) | 3 (50%) | |
| Diabetes | |||
| No (20) | 11 (55%) | 9 (45%) | 1.0 |
| Yes (2) | 1 (50%) | 1 (50%) | |
| Dyslipidemia | |||
| No (18) | 10 (55.6%) | 8 (44.4%) | 1.0 |
| Yes (4) | 2 (50%) | 2 (50%) | |
| Remaining teeth | 20.7 ± 6.5 | 23.4 ± 2.8 | 0.46 |
| Denture use | |||
| Non-user (11) | 5 (45.6%) | 6 (54.5%) | 0.67 |
| User (11) | 7 (63.6%) | 4 (36.4%) | |
| Plaque Control Record scores (%) | 20.7 ± 6.5 | 23.4 ± 2.8 | 0.67 |
| Probing depth | |||
| <4 mm (5) | 3 (60%) | 2 (40%) | 0.07 |
| ≥4 mm and <6 mm (7) | 6 (85.7%) | 1 (14.3%) | |
| ≥6 mm (10) | 3 (30%) | 7 (70%) | |
| Bleeding on probing (%) | 7.7 ± 10.1 | 9.2 ± 8.9 | 0.97 |
| ≥4 mm periodontal pocket with BOP | |||
| No (14) | 8 (57.2%) | 6 (42.9%) | 1.0 |
| Yes (8) | 4 (50%) | 4 (50%) | |
| ≥6 mm periodontal pocket with BOP | |||
| No (17) | 10 (58.8%) | 7 (41.2%) | 0.62 |
| Yes (5) | 2 (40%) | 3 (60%) | |
| PISA (mm2) | 80.1 ± 146.2 | 135.0 ± 142.7 | 0.58 |
| PESA (mm2) | 940.1 ± 342.1 | 1088.3 ± 1019.0 | 0.50 |
| Clinical Factor (n) | C. albicans/P. gingivalis | p-Value | |
|---|---|---|---|
| (−) (n = 74) | (+) (n = 12) | ||
| Age | 69.9 ± 10.0 | 73.1 ± 10.0 | 0.35 |
| Age in years | |||
| 40–49 (4) | 4 (100%) | 0 (0%) | 0.63 |
| 50–59 (8) | 7 (87.5%) | 1 (12.5%) | |
| 60–69 (25) | 21 (84%) | 4 (16%) | |
| 70–79 (32) | 29 (90.6%) | 3 (9.4%) | |
| 80–89 (17) | 13 (76.5%) | 4 (23.5%) | |
| Gender | |||
| Male (25) | 20 (80%) | 5 (20%) | 0.32 |
| Female (61) | 54 (88.5%) | 7 (11.5%) | |
| Hypertension | |||
| No (69) | 59 (85.5%) | 10 (14.5%) | 1.0 |
| Yes (17) | 15 (88.2%) | 2 (11.8%) | |
| Diabetes | |||
| No (76) | 65 (85.5%) | 11 (14.5%) | 1.0 |
| Yes (10) | 9 (90%) | 1 (10%) | |
| Dyslipidemia | |||
| No (72) | 60 (83.3%) | 12 (16.7%) | 0.20 |
| Yes (14) | 14 (100%) | 0 (0%) | |
| Remaining teeth | 23.3 ± 5.6 | 21.15 ± 6.3 | 0.16 |
| Denture use | |||
| Non-user (55) | 51 (92.7%) | 4 (7.3%) | 0.02 |
| User (31) | 23 (74.2%) | 8 (25.8%) | |
| Plaque Control Record scores (%) | 19.6 ± 15.0 | 17.0 ± 10.7 | 0.65 |
| Probing depth | |||
| <4 mm (18) | 14 (77.8%) | 4 (22.2%) | 0.06 |
| ≥4 mm and <6 mm (42) | 40 (95.2%) | 2 (4.8%) | |
| ≥6 mm (26) | 20 (76.9%) | 6 (23.1%) | |
| Bleeding on probing (%) | 7.7 ± 8.6 | 10.3 ± 11.2 | 0.52 |
| ≥4 mm periodontal pocket with BOP | |||
| No (48) | 42 (87.5%) | 6 (12.5%) | 0.76 |
| Yes (38) | 32 (84.2%) | 6 (15.8%) | |
| ≥6 mm periodontal pocket with BOP | |||
| No (73) | 66 (90.4%) | 7 (9.6%) | 0.02 |
| Yes (13) | 8 (61.5%) | 5 (38.5%) | |
| PISA (mm2) | 89.3 ± 106.9 | 141.5 ± 166.5 | 0.50 |
| PESA (mm2) | 1026.4 ± 318.2 | 973.2 ± 375.7 | 0.89 |
| Clinical Variables | Adjusted Odds Ratio | 95% Confidence Interval | p-Value |
|---|---|---|---|
| ≥6 mm periodontal pocket with BOP | 17.13 | 1.60–183.84 | 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oka, I.; Shigeishi, H.; Ohta, K. Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People. Medicina 2022, 58, 723. https://doi.org/10.3390/medicina58060723
Oka I, Shigeishi H, Ohta K. Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People. Medicina. 2022; 58(6):723. https://doi.org/10.3390/medicina58060723
Chicago/Turabian StyleOka, Iori, Hideo Shigeishi, and Kouji Ohta. 2022. "Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People" Medicina 58, no. 6: 723. https://doi.org/10.3390/medicina58060723
APA StyleOka, I., Shigeishi, H., & Ohta, K. (2022). Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People. Medicina, 58(6), 723. https://doi.org/10.3390/medicina58060723





