CD74 and HLA-DRA in Cervical Carcinogenesis: Potential Targets for Antitumour Therapy
Abstract
:1. Introduction
2. Materials and Methods
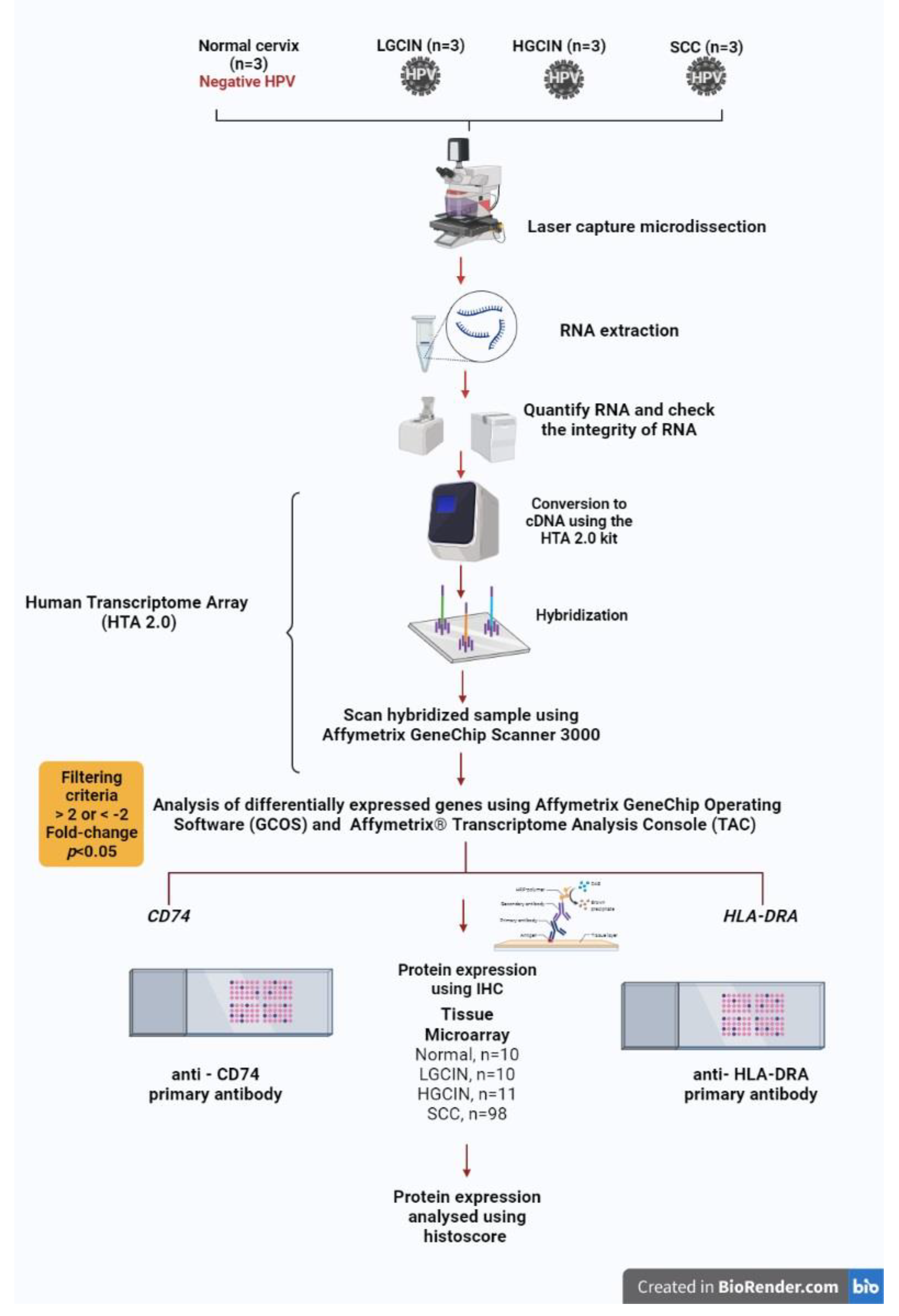
2.1. Tissue Samples, RNA Extraction, Transcriptome Array
2.2. Immunohistochemical Expression of CD74 and HLA-DRA Proteins in Tissue Microarrays
2.3. Statistical Analysis
3. Results
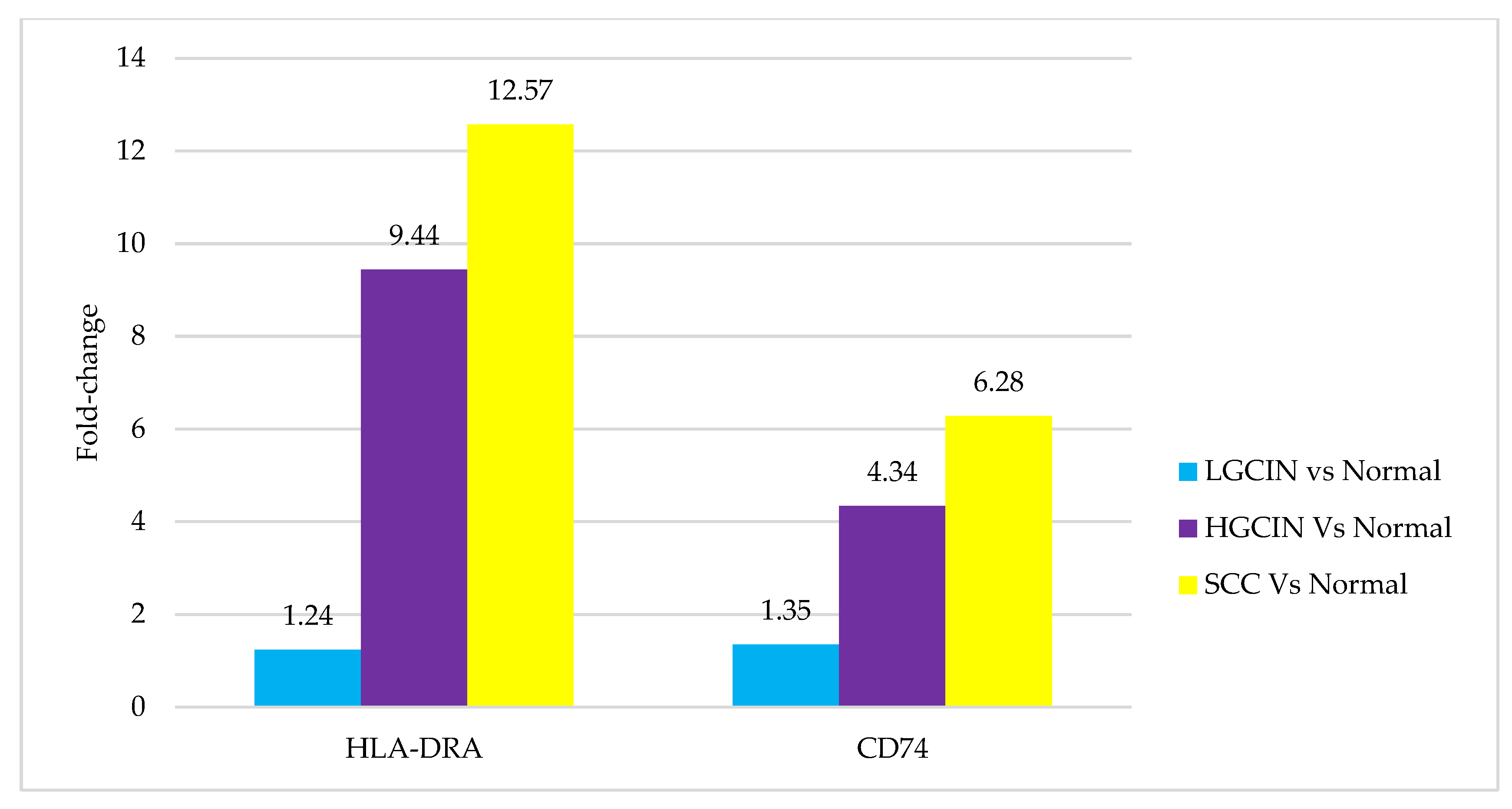
3.1. Transcriptomics Profile
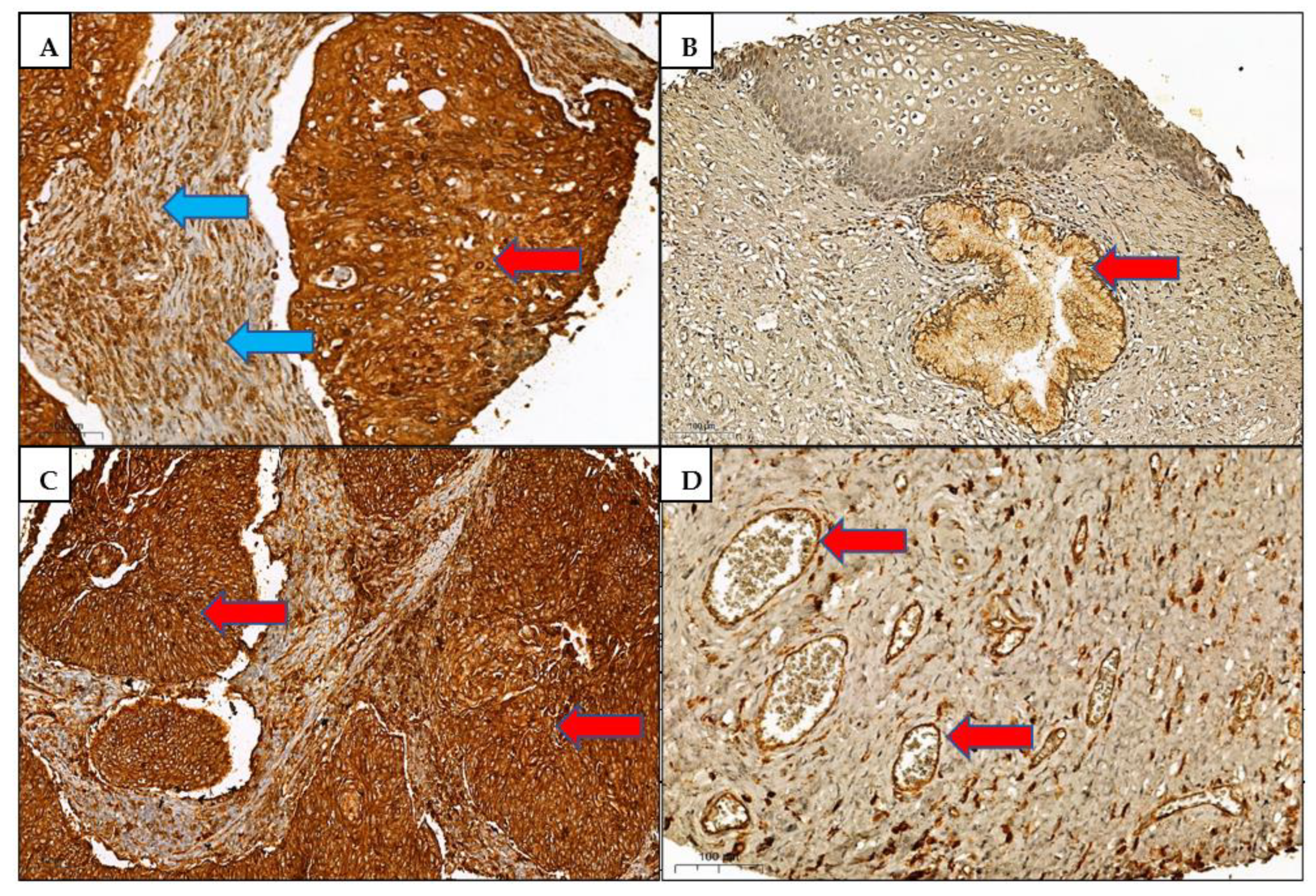
3.2. Immunohistochemistry
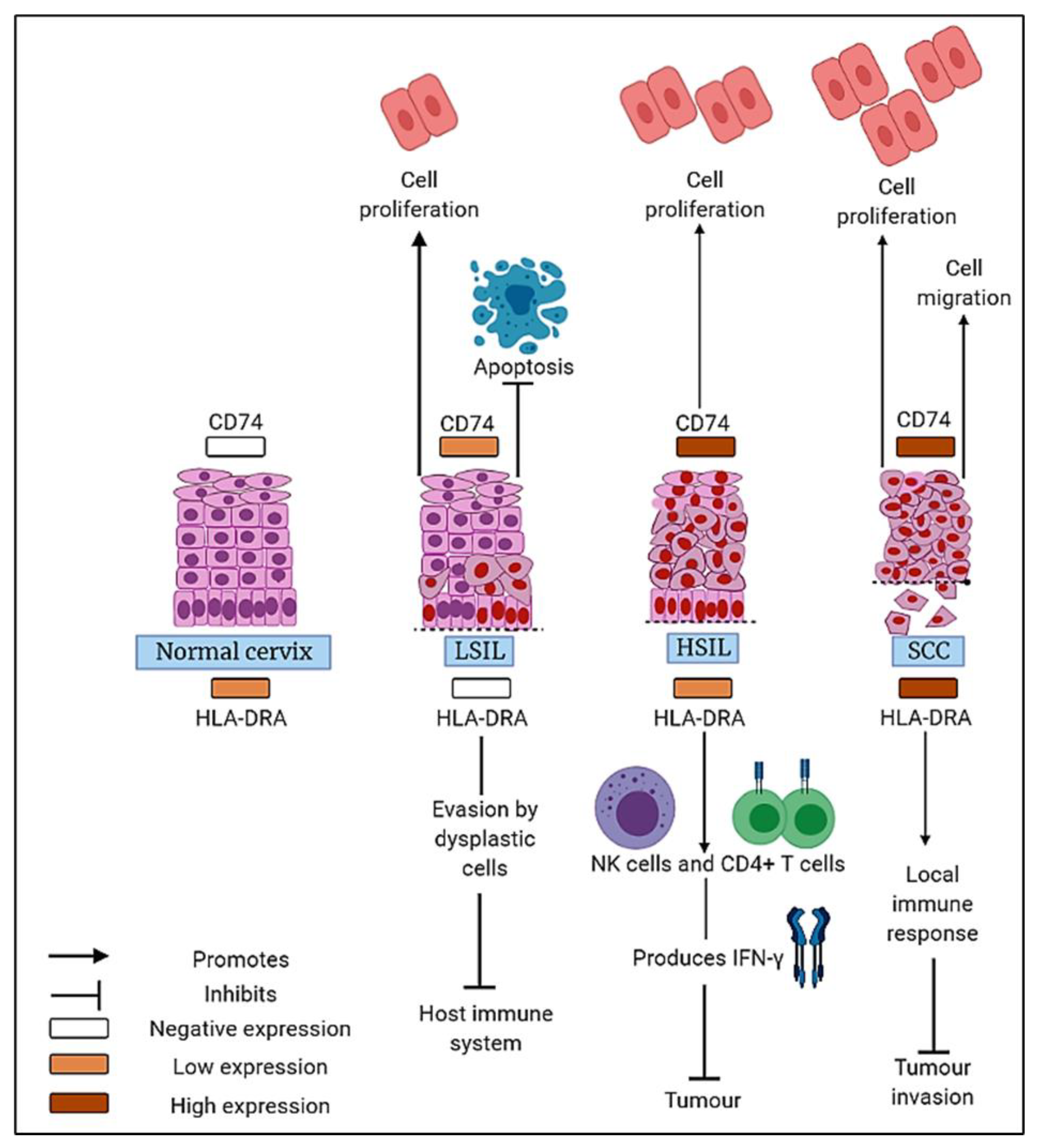
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Mallmann, P.; Mallmann, C. Neoadjuvant and Adjuvant Chemotherapy of Cervical Cancer. Oncol. Res. Treat. 2016, 39, 522–524. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key molecular events in cervical cancer development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class i and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Park, I.A.; Hwang, S.-H.; Song, I.H.; Heo, S.-H.; Kim, Y.-A.; Bang, W.S.; Park, H.S.; Lee, M.; Gong, G.; Lee, H.J. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS ONE 2017, 12, e0182786. [Google Scholar] [CrossRef]
- Da Silva, G.B.R.F.; Silva, T.G.A.; Duarte, R.A.; Neto, N.L.; Carrara, H.H.A.; Donadi, E.A.; Gonçalves, M.A.G.; Soares, E.G.; Soares, C.P. Expression of the Classical and Nonclassical HLA Molecules in Breast Cancer. Int. J. Breast Cancer 2013, 2013, 250435. [Google Scholar] [CrossRef] [Green Version]
- Michel, S.; Linnebacher, M.; Alcaniz, J.; Voss, M.; Wagner, R.; Dippold, W.; Becker, C.; Doeberitz, M.V.K.; Ferrone, S.; Kloor, M. Lack of HLA class II antigen expression in microsatellite unstable colorectal carcinomas is caused by mutations in HLA class II regulatory genes. Int. J. Cancer 2010, 127, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Warabi, M.; Kitagawa, M.; Hirokawa, K. Loss of MHC class II expression is associated with a decrease of tumor-infiltrating T cells and an increase of metastatic potential of colorectal cancer: Immunohistological and histopathological analyses as compared with normal colonic mucosa and adenomas. Pathol. Res. Pract. 2000, 196, 807–815. [Google Scholar] [CrossRef]
- Kübler, K.; Arndt, P.F.; Wardelmann, E.; Krebs, D.; Kuhn, W.; Van Der Ven, K. HLA-class II haplotype associations with ovarian cancer. Int. J. Cancer 2006, 119, 2980–2985. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Bordeaux, J.M.; Kim, J.-Y.; Vaupel, C.A.; Rimm, D.L.; Ho, T.H.; Joseph, R.W.; Daud, A.; Conry, R.M.; Gaughan, E.M.; et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin. Cancer Res. 2018, 24, 5250–5260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Morrison, I.E.G.; O’Toole, P.; Metodieva, G.; Navarrete, C.V.; Gomez, J.; Miranda-Sayago, J.M.; Cherry, R.J.; Metodiev, M.; Fernandez, N. Interaction of HLA-DR and CD74 at the cell surface of antigen-presenting cells by single particle image analysis. FASEB J. 2012, 26, 4886–4896. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Lin, J.-Y. Recent advances of cluster of differentiation 74 in cancer. World J. Immunol. 2014, 4, 174. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, Y.; Li, N.; Liu, X.; Dong, J. CD74: A potential novel target for triple-negative breast cancer. Tumour Biol. 2012, 33, 2273–2277. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, Y.; Lee, J.H.; Kim, Y.S. CD74 expression is increased in high-grade, invasive urothelial carcinoma of the bladder. Int. J. Urol. 2013, 20, 251–255. [Google Scholar] [CrossRef] [Green Version]
- Rossi, H.A.; Liu, Q.; Banner, B.; Hsieh, C.C.; Savas, L.; Savarese, D. The prognostic value of invariant chain (Ii) and Her-2/neu expression in curatively resected colorectal cancer. Cancer J. 2002, 8, 268–275. [Google Scholar] [CrossRef]
- Matsuura, S.; Shinmura, K.; Kamo, T.; Igarashi, H.; Maruyama, K.; Tajima, M.; Ogawa, H.; Tanahashi, M.; Niwa, H.; Funai, K.; et al. CD74-ROS1 fusion transcripts in resected non-small cell lung carcinoma. Oncol. Rep. 2013, 30, 1675–1680. [Google Scholar] [CrossRef]
- Sabbatino, F.; Liguori, L.; Polcaro, G.; Salvato, I.; Caramori, G.; Salzano, F.A.; Casolaro, V.; Stellato, C.; Col, J.D.; Pepe, S. Role of human leukocyte antigen system as a predictive biomarker for checkpoint-based immunotherapy in cancer patients. Int. J. Mol. Sci. 2020, 21, 7295. [Google Scholar] [CrossRef]
- Seliger, B.; Kloor, M.; Ferrone, S. HLA class II antigen-processing pathway in tumors: Molecular defects and clinical relevance. Oncoimmunology 2017, 6, e1171447. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, K.; Takenouchi, T.; Shimada, H.; Tomonaga, T.; Hayashi, H.; Shioya, A.; Komatsu, A.; Matsubara, H.; Ochiai, T. Strong HLA-DR antigen expression on cancer cells relates to better prognosis of colorectal cancer patients: Possible involvement of c-myc suppression by interferon-γ in situ. Cancer Sci. 2006, 97, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Matoba, K.; Iizuka, N.; Gondo, T.; Ishihara, T.; Yamada-Okabe, H.; Tamesa, T.; Takemoto, N.; Hashimoto, K.; Sakamoto, K.; Miyamoto, T.; et al. Tumor HLA-DR expression linked to early intrahepatic recurrence of hepatocellular carcinoma. Int. J. Cancer 2005, 115, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Daltoé, R.D.; Madeira, K.P.; de Araújo, K.L.; Lyra, P.C.M., Jr.; Tessarollo, N.G.; de Souza, M.L.M.; dos Santos, D.Z.; de Carvalho, A.A.; Rotstein, C.; Lima, R.; et al. Clinical Relevance of Co-Expression of HLA-DR α, CD74/II and NFkB in Epithelial Ovarian Cancer: Identification of Prognostic and Therapeutical Biomarker Profile. SL Clin. Med. Oncol. 2020, 2, 1–11. [Google Scholar]
- Chamuleau, M.E.D.; Ossenkoppele, G.J.; van de Loosdrecht, A.A. MHC class II molecules in tumour immunology: Prognostic marker and target for immune modulation. Immunobiology 2006, 211, 619–625. [Google Scholar] [CrossRef]
- Cheng, R.J.; Deng, W.G.; Niu, C.B.; Li, Y.Y.; Fu, Y. Expression of macrophage migration inhibitory factor and CD74 in cervical squamous cell carcinoma. Int. J. Gynecol. Cancer 2011, 21, 1004–1012. [Google Scholar] [CrossRef]
- Cuthbert, R.J.; Wilson, J.M.; Scott, N.; Coletta, P.L.; Hull, M.A. Differential CD74 (major histocompatibility complex Class II invariant chain) expression in mouse and human intestinal adenomas. Eur. J. Cancer 2009, 45, 1654–1663. [Google Scholar] [CrossRef]
- Datta, M.W.; Shahsafaei, A.; Nadler, L.M.; Freeman, G.J.; Dorfman, D.M. Expression of MHC class II-associated invariant chain (Ii;CD74) in thymic epithelial neoplasms. Appl. Immunohistochem. Mol. Morphol. 2000, 8, 210–215. [Google Scholar] [CrossRef]
- Gold, D.V.; Stein, R.; Burton, J.; Goldenberg, D.M. Enhanced expression of CD74 in gastrointestinal cancers and benign tissues. Int. J. Clin. Exp. Pathol. 2011, 4, 1–12. [Google Scholar]
- Shi, X.; Leng, L.; Wang, T.; Wang, W.; Du, X.; Li, J.; McDonald, C.; Chen, Z.; Murphy, J.W.; Lolis, E.; et al. CD44 Is the Signaling Component of the Macrophage Migration Inhibitory Factor-CD74 Receptor Complex. Immunity 2006, 25, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Gil-Yarom, N.; Radomir, L.; Sever, L.; Kramer, M.P.; Lewinsky, H.; Bornstein, C.; Blecher-Gonen, R.; Barnett-Itzhaki, Z.; Mirkin, V.; Friedlander, G.; et al. CD74 is a novel transcription regulator. Proc. Natl. Acad. Sci. USA 2017, 114, 562–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramaniam, S.D.; Wong, K.K.; Oon, C.E.; Balakrishnan, V.; Kaur, G. Comparative transcriptomic profiling in HPV-associated cervical carcinogenesis: Implication of MHC class II and immunoglobulin heavy chain genes. Life Sci. 2020, 256, 118026. [Google Scholar] [CrossRef] [PubMed]
- Rangel, L.B.; Agarwal, R.; Sherman-Baust, C.A.; de Mello-Coelho, V.; Pizer, E.S.; Ji, H.; Taub, D.D.; Morin, P.J. Anomalous expression of the HLA-DR α and β chains in ovarian and other cancers. Cancer Biol. Ther. 2004, 3, 1021–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Liang, J.; Wu, Z.; Shan, X.; Qiao, H.; Jiang, T. Expression of HLA-DR genes in gliomas: Correlation with clinicopathological features and prognosis. Chin. Neurosurg. J. 2017, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Roemer, M.G.; Redd, R.A.; Cader, F.Z.; Pak, C.J.; Abdelrahman, S.; Ouyang, J.; Sasse, S.; Younes, A.; Fanale, M.; Santoro, A.; et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J. Clin. Oncol. 2018, 36, 942–950. [Google Scholar] [CrossRef]
- Rimsza, L.M.; Roberts, R.A.; Miller, T.P.; Unger, J.M.; LeBlanc, M.; Braziel, R.M.; Weisenberger, D.D.; Chan, W.C.; Muller-Hermelink, H.K.; Jaffe, E.S.; et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: A follow-up study from the Leukemia and Lymphoma Molecular. Blood 2004, 103, 4251–4258. [Google Scholar] [CrossRef] [Green Version]
- Yazawa, T.; Kamma, H.; Fujiwara, M.; Matsui, M.; Horiguchi, H.; Satoh, H.; Fujimoto, M.; Yokoyama, K.; Ogata, T. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J. Pathol. 1999, 187, 191–199. [Google Scholar] [CrossRef]
- Cycon, K.A.; Mulvaney, K.; Rimsza, L.M.; Persky, D.; Murphy, S.P. Histone deacetylase inhibitors activate CIITA and MHC class II antigen expression in diffuse large B-cell lymphoma. Immunology 2013, 140, 259–272. [Google Scholar] [CrossRef]
- Ishigami, S.; Natsugoe, S.; Tokuda, K.; Nakajo, A.; Iwashige, H.; Aridome, K.; Hokita, S.; Aikou, T. chain expression in gastric cancer. Cancer Lett. 2001, 168, 87–91. [Google Scholar] [CrossRef]
- Yaprak, M.; Erdogan, G.; Aricic, G.; Ozcan, B.; Mesci, A.; Dınckan, A.; Erdogan, O.; Arici, C. Prognostic Value of CD74 and HLA-DR Expressions in Invasive Ductal Breast Cancer. Adv. Breast Cancer Res. 2015, 4, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Qiu, G.; Jiang, Z.; Von Hofe, E.; Humphreys, R.E. Genetic modulation of tumor antigen presentation. Trends Biotechnol. 2000, 18, 167–172. [Google Scholar] [CrossRef]
| Percentage of Positive Cells | Score | Staining Intensity | Score |
|---|---|---|---|
| <1% positive cells | 0 | No staining | 0 |
| 1–25% positive cells | 1 | Weak staining | 1 |
| 26–50% positive cells | 2 | Moderate staining | 2 |
| 51–75% positive cells | 3 | Strong staining | 3 |
| ≥75% positive cells | 4 | ||
| Final score = percentage of positive cells score × staining intensity score. Final histoscore: 0 = negative. 1 to 3/12 = low expression. ≥4/12 = high expression. | |||
| Histoscore | ||||||
|---|---|---|---|---|---|---|
| Histological Group | Negative n (%) | Low n (%) | High n (%) | Total n (%) | Comparison between All Histological Groups p Value | Histological Group vs. Normal Cervix p Value |
| CD74 protein | ||||||
| Normal cervix | 4 (57.1) | 2 (28.6) | 1 (14.3) | 7 (100) | 0.0001 | |
| LGCIN HGCIN | 4 (40.0) 1 (10.0) | 5 (50.0) 0 (0.0) | 1 (10.0) 9 (90.0) | 10 (100) 10 (100) | 0.798 0.006 | |
| SCC | 5 (5.3) | 7 (7.4) | 83 (87.4) | 95 (100) | 0.0001 | |
| Total | 14 (11.5) | 14 (11.5) | 94 (77.0) | 122 (100) | ||
| HLA-DRA protein | ||||||
| Normal cervix | 0 (0.0) | 6 (85.7) | 1 (14.3) | 7 (100) | 0.0001 | |
| LGCIN HGCIN | 8 (80.0) 2 (20.0) | 1 (10.0) 5 (50.0) | 1 (10.0) 3 (30.0) | 10 (100) 10 (100) | 0.001 0.464 | |
| SCC | 6 (6.3) | 5 (5.3) | 84 (88.4) | 95 (100) | 0.0001 | |
| Total | 16 (13.1) | 17 (13.9) | 89 (73.0) | 122 (100) | ||
| Parameters | CD74 Histoscore | Total n (%) | p Value | HLA-DRA Histoscore | Total n (%) | p Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative n (%) | Low n (%) | High n (%) | Negative n (%) | Low n (%) | High n (%) | |||||
| Age (years) <50 | 4 (5.8) | 8 (11.6) | 57 (82.6) | 69 (100) | 0.083 | 10 (14.5) | 10 (14.5) | 49 (71.0) | 69 (100) | 0.838 |
| ≥50 | 10 (18.9) | 4 (7.5) | 39 (73.6) | 53 (100) | 6 (11.3) | 7 (13.2) | 40 (75.5) | 53 (100) | ||
| CIN grade LGCIN | 4 (40.0) | 5 (50.0) | 1 (10.0) | 10 (100) | 0.001 | 8 (80.0) | 1 (10.0) | 1 (10.0) | 10 (100) | 0.030 |
| HGCIN | 1 (10.0) | 0 (0.0) | 9 (90.0) | 10 (100) | 2 (20.0) | 5 (50.0) | 3 (30.0) | 10 (100) | ||
| SCC stage Stage I | 0 (0.0) | 2 (6.7) | 28 (93.3) | 30 (100) | 0.790 | 1 (3.3) | 0 (0.0) | 29 (96.7) | 30 (100) | 0.753 |
| Stage IA | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 1 (100) | 1 (100) | ||
| Stage IB | 3 (15.0) | 2 (10.0) | 15 (75.0) | 20 (100) | 2 (10.0) | 2 (10.0) | 16 (80.0) | 20 (100) | ||
| Stage IC | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 1 (100) | 1 (100) | ||
| Stage II | 0 (0.0) | 1 (16.7) | 5 (83.3) | 6 (100) | 0 (0.0) | 0 (0.0) | 6 (100) | 6 (100) | ||
| Stage IIA | 1 (5.3) | 1 (5.3) | 17 (89.5) | 19 (100) | 1 (5.3) | 1 (5.3) | 17 (89.5) | 19 (100) | ||
| Stage IIB | 1 (8.3) | 1 (8.3) | 10 (83.3) | 12 (100) | 2 (16.7) | 1 (8.3) | 9 (75.0) | 12 (100) | ||
| Stage III | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (100) | 1 (3.3) | 0 (0.0) | 29 (96.7) | 30 (100) | ||
| Stage IIIB | 0 (0.0) | 0 (0.0) | 4 (100.0) | 4 (100) | 0 (0.0) | 0 (0.0) | 1 (100) | 1 (100) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balakrishnan, C.K.; Tye, G.J.; Balasubramaniam, S.D.; Kaur, G. CD74 and HLA-DRA in Cervical Carcinogenesis: Potential Targets for Antitumour Therapy. Medicina 2022, 58, 190. https://doi.org/10.3390/medicina58020190
Balakrishnan CK, Tye GJ, Balasubramaniam SD, Kaur G. CD74 and HLA-DRA in Cervical Carcinogenesis: Potential Targets for Antitumour Therapy. Medicina. 2022; 58(2):190. https://doi.org/10.3390/medicina58020190
Chicago/Turabian StyleBalakrishnan, Carol K., Gee Jun Tye, Shandra Devi Balasubramaniam, and Gurjeet Kaur. 2022. "CD74 and HLA-DRA in Cervical Carcinogenesis: Potential Targets for Antitumour Therapy" Medicina 58, no. 2: 190. https://doi.org/10.3390/medicina58020190
APA StyleBalakrishnan, C. K., Tye, G. J., Balasubramaniam, S. D., & Kaur, G. (2022). CD74 and HLA-DRA in Cervical Carcinogenesis: Potential Targets for Antitumour Therapy. Medicina, 58(2), 190. https://doi.org/10.3390/medicina58020190






