Anti-CXCL8 Autoantibody: A Potential Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Differential Expression Gene Analysis
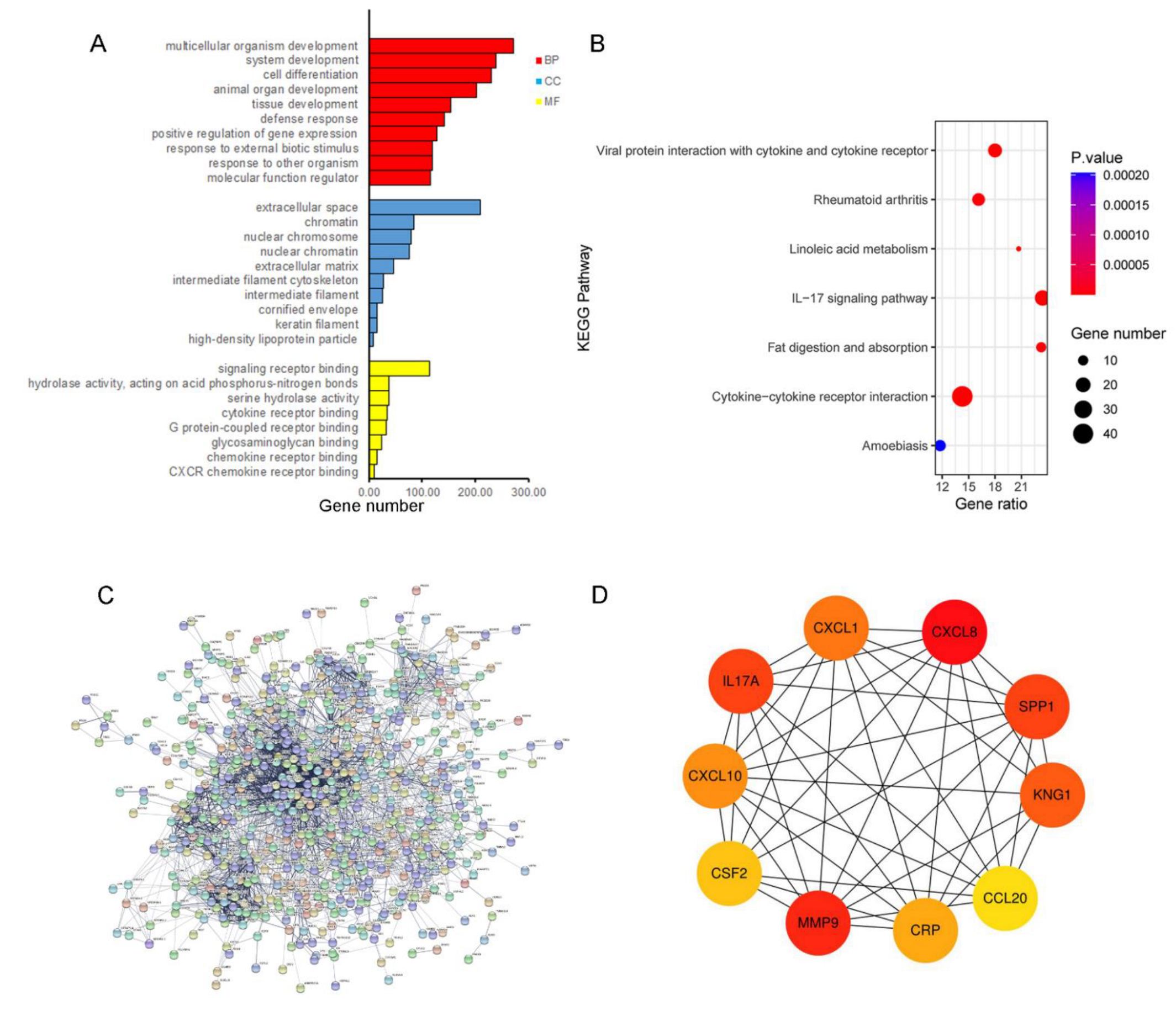
2.2. Functional Annotation and Hub Genes Screening
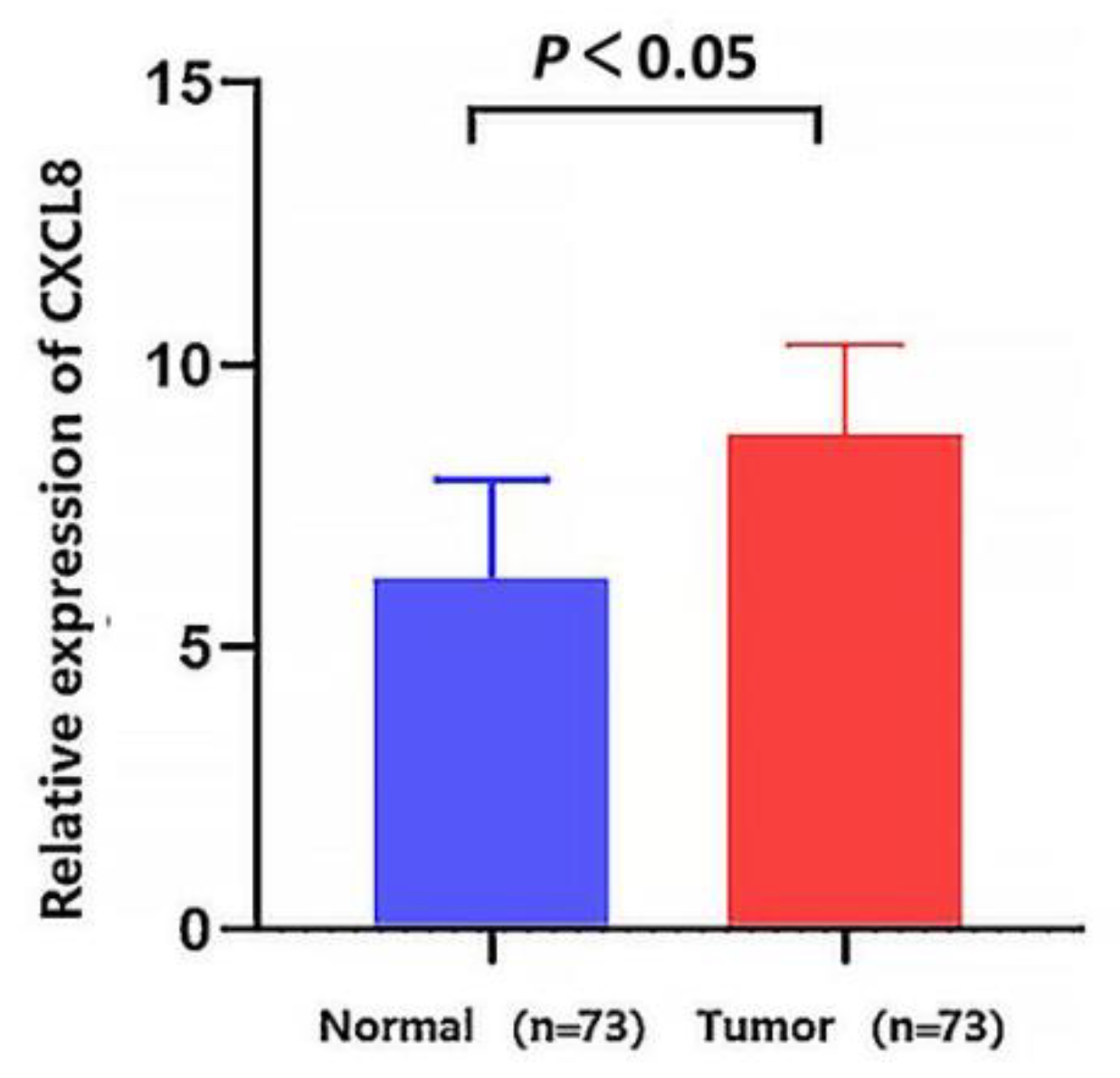
2.3. CXCL8 mRNA Expression Validation
2.4. Study Population
2.5. Enzyme-Linked Immunosorbent Assay
2.6. Statistical Analysis
3. Results
3.1. Identification of DEGs
3.2. Functional Annotation and PPI Analysis for the Up-Regulated Genes
3.3. Validation mRNA Expression of CXCL8
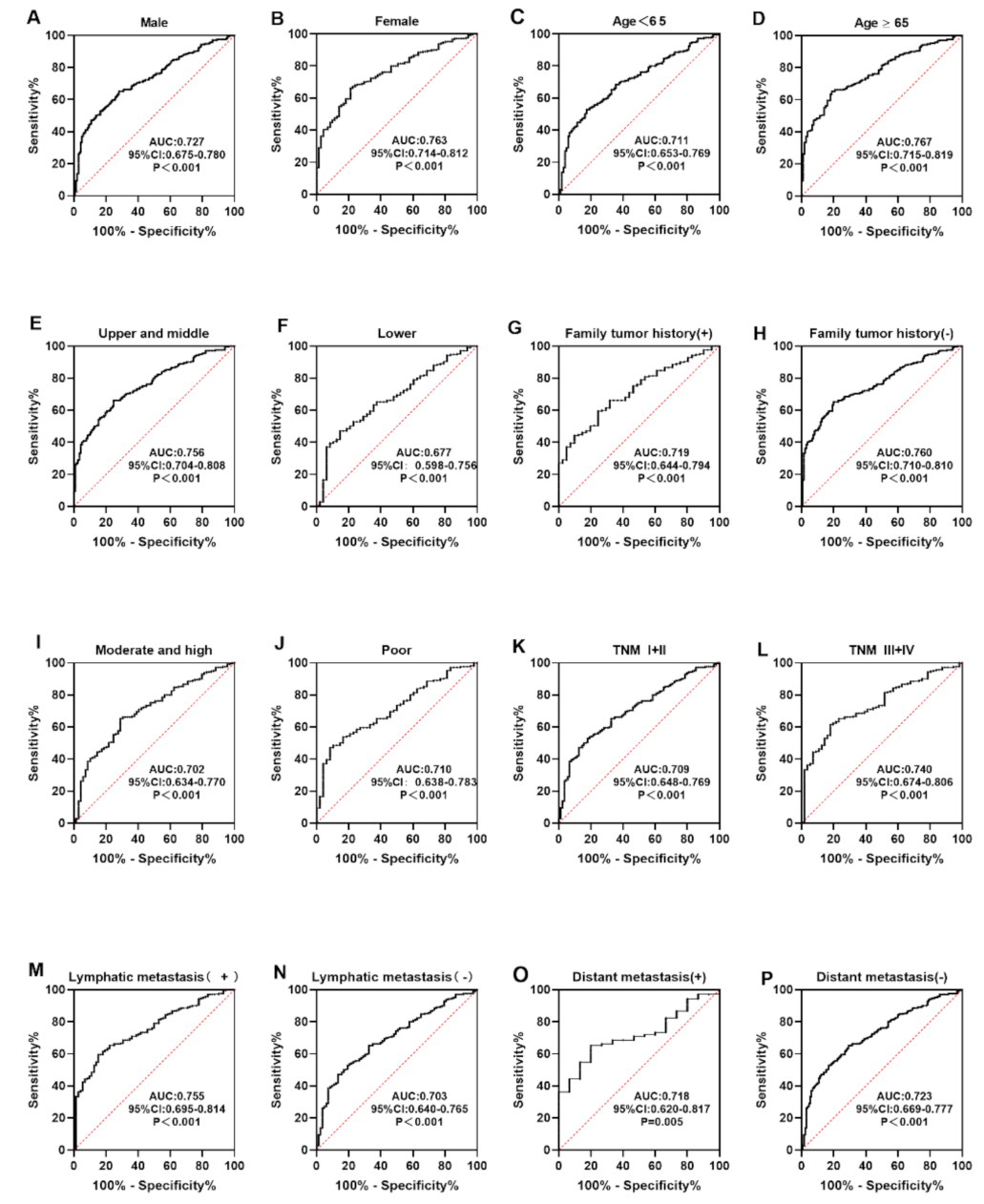
3.4. Level and Diagnostic Value of Anti-CXCL8 Autoantibody
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal cancer. Nat. Rev. Dis. Primers 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Pennathur, A.; Gibson, M.K.; Jobe, B.A.; Luketich, J.D. Oesophageal carcinoma. Lancet 2013, 381, 400–412. [Google Scholar] [CrossRef]
- Wang, G.Q.; Jiao, G.G.; Chang, F.B.; Fang, W.H.; Song, J.X.; Lu, N.; Lin, D.M.; Xie, Y.Q.; Yang, L. Long-term results of operation for 420 patients with early squamous cell esophageal carcinoma discovered by screening. Ann. Thorac. Surg. 2004, 77, 1740–1744. [Google Scholar] [CrossRef]
- Bird-Lieberman, E.L.; Fitzgerald, R.C. Early diagnosis of oesophageal cancer. Br. J. Cancer 2009, 101, 1–6. [Google Scholar] [CrossRef]
- Lao-Sirieix, P.; Fitzgerald, R.C. Screening for oesophageal cancer. Nat. Rev. Clin. Oncol. 2012, 9, 278–287. [Google Scholar] [CrossRef]
- Liang, H.; Fan, J.H.; Qiao, Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017, 14, 33–41. [Google Scholar]
- Codipilly, D.C.; Qin, Y.; Dawsey, S.M.; Kisiel, J.; Topazian, M.; Ahlquist, D.; Iyer, P.G. Screening for esophageal squamous cell carcinoma: Recent advances. Gastrointest. Endosc. 2018, 88, 413–426. [Google Scholar] [CrossRef]
- Tan, E.M.; Zhang, J. Autoantibodies to tumor-associated antigens: Reporters from the immune system. Immunol. Rev. 2008, 222, 328–340. [Google Scholar] [CrossRef]
- Heo, C.K.; Bahk, Y.Y.; Cho, E.W. Tumor-associated autoantibodies as diagnostic and prognostic biomarkers. BMB Rep. 2012, 45, 677–685. [Google Scholar] [CrossRef]
- Wang, M.; Liu, F.; Pan, Y.; Xu, R.; Li, F.; Liu, A.; Yang, H.; Duan, L.; Shen, L.; Wu, Q.; et al. Tumor-associated autoantibodies in ESCC screening: Detecting prevalent early-stage malignancy or predicting future cancer risk? EBioMedicine 2021, 73, 103674. [Google Scholar] [CrossRef]
- Xu, Y.W.; Peng, Y.H.; Chen, B.; Wu, Z.Y.; Wu, J.Y.; Shen, J.H.; Zheng, C.P.; Wang, S.H.; Guo, H.P.; Li, E.M.; et al. Autoantibodies as potential biomarkers for the early detection of esophageal squamous cell carcinoma. Am. J. Gastroenterol. 2014, 109, 36–45. [Google Scholar] [CrossRef]
- Li, C.Y.; Zhang, W.W.; Xiang, J.L.; Wang, X.H.; Li, J.; Wang, J.L. Identification of microRNAs as novel biomarkers for esophageal squamous cell carcinoma: A study based on The Cancer Genome Atlas (TCGA) and bioinformatics. Chin. Med. J. 2019, 132, 2213–2222. [Google Scholar] [CrossRef]
- Bhushan, A.; Singh, A.; Kapur, S.; Borthakar, B.B.; Sharma, J.; Rai, A.K.; Kataki, A.C.; Saxena, S. Identification and Validation of Fibroblast Growth Factor 12 Gene as a Novel Potential Biomarker in Esophageal Cancer Using Cancer Genomic Datasets. OMICS 2017, 21, 616–631. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, Z.; Zhang, Z.; Zhao, D.; Xu, L.; Zhang, L. RNA-Associated Co-expression Network Identifies Novel Biomarkers for Digestive System Cancer. Front. Genet. 2021, 12, 659788. [Google Scholar] [CrossRef]
- Zhang, H.F.; Qin, J.J.; Ren, P.F.; Shi, J.X.; Xia, J.F.; Ye, H.; Wang, P.; Song, C.H.; Wang, K.J.; Zhang, J.Y. A panel of autoantibodies against multiple tumor-associated antigens in the immunodiagnosis of esophageal squamous cell cancer. Cancer Immunol. Immunother. 2016, 65, 1233–1242. [Google Scholar] [CrossRef]
- Ohashi, S.; Miyamoto, S.; Kikuchi, O.; Goto, T.; Amanuma, Y.; Muto, M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology 2015, 149, 1700–1715. [Google Scholar] [CrossRef]
- Lin, H.N.; Chen, L.Q.; Shang, Q.X.; Yuan, Y.; Yang, Y.S. A meta-analysis on surgery with or without postoperative radiotherapy to treat squamous cell esophageal carcinoma. Int. J. Surg. 2020, 80, 184–191. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, L.; Tu, M.; Yin, X.; Cai, L.; Zhang, S.; Yu, L.; Pan, X.; Huang, Y. Development of a panel of autoantibody against NSG1 with CEA, CYFRA21-1, and SCC-Ag for the diagnosis of esophageal squamous cell carcinoma. Clin. Chim. Acta 2021, 520, 126–132. [Google Scholar] [CrossRef]
- Wang, X.B.; Jiang, X.R.; Yu, X.Y.; Wang, L.; He, S.; Feng, F.Y.; Guo, L.P.; Jiang, W.; Lu, S.H. Macrophage inhibitory factor 1 acts as a potential biomarker in patients with esophageal squamous cell carcinoma and is a target for antibody-based therapy. Cancer Sci. 2014, 105, 176–185. [Google Scholar] [CrossRef]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. How the evolution of multicellularity set the stage for cancer. Br. J. Cancer 2018, 118, 145–152. [Google Scholar] [CrossRef]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc. Natl. Acad. Sci. USA 2017, 114, 6406–6411. [Google Scholar] [CrossRef]
- Zamecnik, J. The extracellular space and matrix of gliomas. Acta Neuropathol. 2005, 110, 435–442. [Google Scholar] [CrossRef]
- O’Hayre, M.; Vazquez-Prado, J.; Kufareva, I.; Stawiski, E.W.; Handel, T.M.; Seshagiri, S.; Gutkind, J.S. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 2013, 13, 412–424. [Google Scholar] [CrossRef]
- Diakowska, D. Cytokines association with clinical and pathological changes in esophageal squamous cell carcinoma. Dis. Markers 2013, 35, 883–893. [Google Scholar] [CrossRef]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef]
- Kumar, S.; O’Malley, J.; Chaudhary, A.K.; Inigo, J.R.; Yadav, N.; Kumar, R.; Chandra, D. Hsp60 and IL-8 axis promotes apoptosis resistance in cancer. Br. J. Cancer 2019, 121, 934–943. [Google Scholar]
- Li, A.; Dubey, S.; Varney, M.L.; Dave, B.J.; Singh, R.K. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J. Immunol. 2003, 170, 3369–3376. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Cherukumilli, M.; Mahmoudpour, S.H.; Brand, K.; Bandapalli, O.R. ShRNA-mediated knock-down of CXCL8 inhibits tumor growth in colorectal liver metastasis. Biochem. Biophys. Res. Commun. 2018, 500, 731–737. [Google Scholar] [CrossRef]
- Long, X.; Ye, Y.; Zhang, L.; Liu, P.; Yu, W.; Wei, F.; Ren, X.; Yu, J. IL-8, a novel messenger to cross-link inflammation and tumor EMT via autocrine and paracrine pathways (Review). Int. J. Oncol. 2016, 48, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; He, H.; Liu, H.; Li, R.; Chen, Y.; Qi, Y.; Jiang, Q.; Chen, L.; Zhang, P.; Zhang, H.; et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 2019, 68, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, J.; Li, Y.; Jiang, Y.; Ma, J.; Li, Q.; Pang, T. CXCL8 is associated with the recurrence of patients with acute myeloid leukemia and cell proliferation in leukemia cell lines. Biochem. Biophys. Res. Commun. 2018, 499, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yang, X.; Xu, C.; Sun, J.; Fang, Z.; Pan, H.; Han, W. Comprehensive analysis of the expression and prognostic value of CXC chemokines in colorectal cancer. Int. Immunopharmacol. 2020, 89, 107077. [Google Scholar] [CrossRef]
- Sun, F.; Wang, J.; Sun, Q.; Li, F.; Gao, H.; Xu, L.; Zhang, J.; Sun, X.; Tian, Y.; Zhao, Q.; et al. Interleukin-8 promotes integrin beta3 upregulation and cell invasion through PI3K/Akt pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 449. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, X.; Miao, W.; Wang, B.; Qiu, Y. CXCL8 promotes the invasion of human osteosarcoma cells by regulation of PI3K/Akt signaling pathway. APMIS 2017, 125, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Hosono, M.; Koma, Y.I.; Takase, N.; Urakawa, N.; Higashino, N.; Suemune, K.; Kodaira, H.; Nishio, M.; Shigeoka, M.; Kakeji, Y.; et al. CXCL8 derived from tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression by promoting migration and invasion of cancer cells. Oncotarget 2017, 8, 106071–106088. [Google Scholar] [CrossRef]
- Wu, J.; Gao, F.X.; Wang, C.; Qin, M.; Han, F.; Xu, T.; Hu, Z.; Long, Y.; He, X.M.; Deng, X.; et al. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 321. [Google Scholar] [CrossRef]
- Pawluczuk, E.; Lukaszewicz-Zajac, M.; Gryko, M.; Kulczynska-Przybik, A.; Mroczko, B. Serum CXCL8 and Its Specific Receptor (CXCR2) in Gastric Cancer. Cancers 2021, 13, 5186. [Google Scholar] [CrossRef]
- Litman-Zawadzka, A.; Łukaszewicz-Zając, M.; Gryko, M.; Kulczyńska-Przybik, A.; Mroczko, B. Serum chemokine CXCL-8 as a better biomarker for diagnosis and prediction of pancreatic cancer than its specific receptor CXCR-2, CRP and classical tumor markers (CA 19-9 and CEA). Pol. Arch. Intern. Med. 2018, 128, 524–531. [Google Scholar] [CrossRef]
- Paczek, S.; Lukaszewicz-Zajac, M.; Gryko, M.; Mroczko, P.; Kulczynska-Przybik, A.; Mroczko, B. CXCL-8 in Preoperative Colorectal Cancer Patients: Significance for Diagnosis and Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2040. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Xu, Y.; Ding, R.; Qiu, K.; Zhang, R.; Wang, H.; Huang, L.; Xie, X.; Yan, H.; Deng, Y.; et al. Extensive serum biomarker analysis in patients with non-small-cell lung carcinoma. Cytokine 2020, 126, 154868. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.S.; LaBaer, J. The sentinel within: Exploiting the immune system for cancer biomarkers. J. Proteome Res. 2005, 4, 1123–1133. [Google Scholar] [CrossRef] [PubMed]


| Variables | Verification Phase | Validation Phase | ||
|---|---|---|---|---|
| (n = 140) | (n = 280) | |||
| ESCC (n = 70) | NC (n = 70) | ESCC (n = 140) | NC (n = 140) | |
| Gender | ||||
| Male, n (%) | 40 (57.1) | 40 (57.1) | 99 (70.7) | 98 (70.0) |
| Female, n (%) Age, years | 30 (42.9) | 30 (42.9) | 41 (29.3) | 42 (30.0) |
| Mean age ± SD | 64.28 ± 8.23 | 64.74 ± 8.26 | 63.64 ± 8.64 | 64.31 ± 8.75 |
| Age range | 45–88 | 45–84 | 41–87 | 41–88 |
| Tumor site, n (%) | ||||
| Upper thorax | 1 (1.4) | 25 (17.9) | ||
| Middle thorax | 19 (27.2) | 72 (51.4) | ||
| Lower thorax | 8 (11.4) | 40 (28.6) | ||
| Unknown | 42 (60.0) | 3 (2.1) | ||
| Family tumor history, n (%) | ||||
| Yes | 12 (17.1) | 29 (20.7) | ||
| No | 56 (80.0) | 79 (56.4) | ||
| Unknown | 2 (2.9) | 32 (22.9) | ||
| Histological grade, n (%) | ||||
| High | 3 (4.3) | 3 (2.1) | ||
| Medium | 16 (22.8) | 46 (32.9) | ||
| Low | 8 (11.4) | 41 (29.3) | ||
| Unknown | 43 (61.5) | 50 (35.7) | ||
| TNM stage, n (%) | ||||
| I | 8 (11.4) | 45 (32.2) | ||
| II | 5 (7.1) | 31 (22.1) | ||
| III | 12 (17.1) | 30 (21.4) | ||
| IV | 6 (8.6) | 8 (5.7) | ||
| Unknown | 39 (55.7) | 26 (18.6) | ||
| Lymph node metastasis, n (%) | ||||
| Positive | 18 (25.8) | 54 (38.6) | ||
| Negative | 12 (17.1) | 71 (50.7) | ||
| Unknown | 40 (57.1) | 15 (10.7) | ||
| Distant metastasis, n (%) | ||||
| Yes | 6 (8.6) | 9 (6.4) | ||
| No | 25 (35.7) | 105 (75.0) | ||
| Unknown | 39 (55.7) | 26 (18.6) | ||
| Gene Symbol | Full Name | Degree | Function |
|---|---|---|---|
| CXCL8 | C-X-C motif chemokine ligand 8 | 86 | CXCL8 is a chemotactic factor and participates with inflammatory responses and neovascularization, and regulates immune response. |
| MMP9 | matrix metallopeptidase 9 | 82 | MMP9 is involved in the breakdown of extracellular matrix in normal physiological processes. |
| IL17A | interleukin 17A | 57 | IL17A mediated downstream pathways induce the production of inflammatory molecules, chemokines, and antimicrobial peptides. |
| SPP1 | secreted phosphoprotein 1 | 57 | The protein encoded by this gene is involved in the attachment of osteoclasts to the mineralized bone matrix. The encoded protein is secreted and binds hydroxyapatite with high affinity. |
| KNG1 | kininogen 1 | 55 | KNG1 is involved in signaling receptor binding and cysteine-type endopeptidase inhibitor activity. |
| CXCL1 | C-X-C motif chemokine ligand 1 | 53 | CXCL1 is associated with the growth and progression of certain tumors. |
| CXCL10 | C-X-C motif chemokine ligand 10 | 51 | Pro-inflammatory cytokine is involved in a wide variety of processes, such as chemotaxis and differentiation. |
| CRP | C-reactive protein | 50 | This gene is involved in complement activation and amplification via communication with complement initiation pattern recognition molecules. |
| CSF2 | colony stimulating factor 2 | 48 | CSF2 controls the production, differentiation, and function of granulocytes and macrophages. |
| CCL20 | C-C motif chemokine ligand 20 | 47 | CCL20 is involved in immunoregulatory and inflammatory processes. |
| Cohorts | AUC | 95%CI | Se (%) | Sp (%) | YI | +LR | −LR | PPV (%) | NPV (%) | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| Verification | 0.713 | 0.624–0.801 | 35.7 | 82.9 | 0.186 | 2.088 | 0.776 | 67.6 | 56.3 | 0.593 |
| Validation | 0.751 | 0.696–0.808 | 47.1 | 77.9 | 0.250 | 2.131 | 0.679 | 68.0 | 59.6 | 0.621 |
| Total | 0.739 | 0.692–0.787 | 44.3 | 81.4 | 0.257 | 2.420 | 0.684 | 70.5 | 59.4 | 0.628 |
| Variables | Number | Frequency (%) | p |
|---|---|---|---|
| Gender | |||
| Male | 139 | 41.7 | 0.296 |
| Female | 71 | 49.3 | |
| Age range (years) | |||
| <65 | 104 | 38.5 | 0.092 |
| ≥65 | 106 | 50.0 | |
| Family tumor history | |||
| Yes | 41 | 43.9 | 0.885 |
| No | 135 | 45.2 | |
| Tumor site | |||
| Upper and middle | 117 | 48.7 | 0.119 |
| Lower | 48 | 35.4 | |
| Differentiation | |||
| Moderate and high | 69 | 39.1 | 0.961 |
| Poor | 48 | 39.6 | |
| TNM stage | |||
| I–II | 89 | 38.2 | 0.234 |
| III–IV | 56 | 48.2 | |
| Lymphatic metastasis | |||
| Positive | 66 | 51.5 | 0.276 |
| Negative | 89 | 42.7 | |
| Distant metastasis | |||
| Yes | 15 | 33.3 | 0.436 |
| No | 130 | 43.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Sun, G.; Han, Z.; Wang, H.; Li, J.; Ye, H.; Song, C.; Zhang, J.; Wang, P. Anti-CXCL8 Autoantibody: A Potential Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Medicina 2022, 58, 1480. https://doi.org/10.3390/medicina58101480
Chen H, Sun G, Han Z, Wang H, Li J, Ye H, Song C, Zhang J, Wang P. Anti-CXCL8 Autoantibody: A Potential Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Medicina. 2022; 58(10):1480. https://doi.org/10.3390/medicina58101480
Chicago/Turabian StyleChen, Huili, Guiying Sun, Zhuo Han, Huimin Wang, Jiaxin Li, Hua Ye, Chunhua Song, Jianying Zhang, and Peng Wang. 2022. "Anti-CXCL8 Autoantibody: A Potential Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma" Medicina 58, no. 10: 1480. https://doi.org/10.3390/medicina58101480
APA StyleChen, H., Sun, G., Han, Z., Wang, H., Li, J., Ye, H., Song, C., Zhang, J., & Wang, P. (2022). Anti-CXCL8 Autoantibody: A Potential Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Medicina, 58(10), 1480. https://doi.org/10.3390/medicina58101480







