Abstract
COPD is a chronic lung disorder characterized by a progressive and irreversible airflow obstruction, and persistent pulmonary inflammation. It has become a global epidemic affecting 10% of the population, and is the third leading cause of death worldwide. Respiratory viruses are a primary cause of COPD exacerbations, often leading to secondary bacterial infections in the lower respiratory tract. COPD patients are more susceptible to viral infections and associated severe disease, leading to accelerated lung function deterioration, hospitalization, and an increased risk of mortality. The airway epithelium plays an essential role in maintaining immune homeostasis, and orchestrates the innate and adaptive responses of the lung against inhaled and pathogen insults. A healthy airway epithelium acts as the first line of host defense by maintaining barrier integrity and the mucociliary escalator, secreting an array of inflammatory mediators, and initiating an antiviral state through the interferon (IFN) response. The airway epithelium is a major site of viral infection, and the interaction between respiratory viruses and airway epithelial cells activates host defense mechanisms, resulting in rapid virus clearance. As such, the production of IFNs and the activation of IFN signaling cascades directly contributes to host defense against viral infections and subsequent innate and adaptive immunity. However, the COPD airway epithelium exhibits an altered antiviral response, leading to enhanced susceptibility to severe disease and impaired IFN signaling. Despite decades of research, there is no effective antiviral therapy for COPD patients. Herein, we review current insights into understanding the mechanisms of viral evasion and host IFN antiviral defense signaling impairment in COPD airway epithelium. Understanding how antiviral mechanisms operate in COPD exacerbations will facilitate the discovery of potential therapeutic interventions to reduce COPD hospitalization and disease severity.
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a common chronic inflammatory airways disease affecting 1.2 million people in the UK, and 10% of the global population aged over 45 []. It is characterized by progressive and irreversible airflow obstruction, and chronic persistent pulmonary inflammation [,,]. Respiratory virus infections are major drivers of acute COPD exacerbations, and are associated with COPD hospitalization, morbidity, and mortality [,]. In fact, COPD is the third leading cause of death globally, imposing a substantial social and economic burden [,,].
The airway epithelium plays an essential role in maintaining immune homeostasis, and modulating the innate and adaptive immune response in the lung against inhaled and pathogen insults []. As a primary site of respiratory viral infection, airway epithelial cells act as the first line of host defense by maintaining barrier integrity and the mucociliary escalator to prevent virus binding and entry into the host [,]. It also initiates the innate and adaptive immune response by secreting an array of inflammatory mediators, such as cytokines, chemokines, growth factors, lipid mediators, inflammasomes, and proteases [,,]. Upon viral invasion, airway epithelial cells rapidly recognize pathogens through pattern recognition receptors (PRRs) and intracellular viral sensors, and induce an antiviral state through the production of interferons (IFNs) [,,]. Consequently, IFNs activate a downstream signaling cascade through the expression of various interferon-stimulated genes (ISGs) [,,].
Substantial evidence suggests that the COPD airway epithelium is more susceptible to viral infection and associated severe diseases, leading to accelerated lung function deterioration, hospitalization, and an increased risk of mortality [,,]. The Global Initiative for Chronic Obstructive Lung Disease (GOLD) acknowledges that COPD patients are among the worst affected by COVID-19, also known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) []. In addition, impaired antiviral immunity has been implicated in COPD airway epithelial cells in response to a range of respiratory viruses, including SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) [,,,,]. However, much remains unknown about how respiratory viruses subvert or suppress the host antiviral response, leading to increased virulence of infection, and susceptibility to severe disease in COPD airways.
In this review, we initially discuss what is known about IFN antiviral defense signaling in airway epithelium, and COPD-associated impairment of IFN signaling. We also highlight current advances in understanding mechanisms of how respiratory viruses modulate or evade these antiviral defenses.
2. Virus Sensing Pathways in Airway Epithelium
The first phase of the antiviral response depends on the ability of host cells to rapidly sense the invasion of foreign pathogens, and promote the production of innate pro-inflammatory mediators, including IFNs, to impede virus-associated disease. Interaction with the airway epithelium represents the first encounter of virus with the human host, which initiates the innate immune response [,,]. As illustrated in Figure 1, upon virus entry, airway epithelial cells rapidly recognize the pathogenic viral genome via activation of a variety of innate immune signaling pathways which are mediated by PRRs. PRRs detect pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), mainly through the signaling of toll-like-receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) [,,,]. Consequently, PRR activation drives the transcription of IFN regulatory factors (IRFs) and NF-κB. IRFs further regulate transcriptional factors in the nucleus, which, in turn, stimulate the unique downstream signaling of IFNs and other pro-inflammatory cytokines to induce antiviral response [,,,].
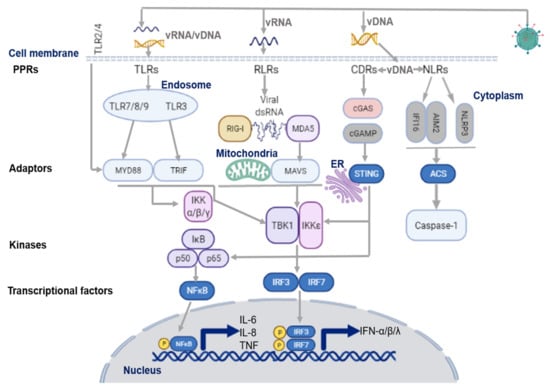
Figure 1.
Virus sensing pathways on airway epithelium. TLR7/8/9 and TLR3 in the endosome sense ssRNA, and dsRNA of the viral genome, respectively. They then activate their adaptor proteins, MyD88 for the former, and TRIF for the latter. The cell surface receptors, TLR2/4/6, also recognize respiratory viruses in the airway by modulating the adaptor protein, MyD88, to induce the antiviral response. DNA and RNA viruses release their genomes in the cytoplasm, where host innate sensors for viral RNA/DNA reside. Upon ss/dsRNA binding, RLRs interact with the adaptor protein, MAVS, on the mitochondrial outer membrane. CDRs (such as cGAS receptor) sense dsDNA and the RNA: DNA hybrids, and induce the synthesis of cGAMP, which then binds to the adaptor protein, STING. NLRs recognize DNA and RNA viruses via the NLRP3 inflammasome. NLRP3 activates and recruits ASC and procaspase-1 to form an inflammasome complex; IFI16 can recruit STING in response to cytoplasmic DNA through a molecular mechanism yet to be described. NLRs modulate the recruitment of their adaptor protein, ASC, to induce inflammation through the activation and secretion of pro-IL-1β and pro-IL-18 via caspase-1. The maturation of these cytokines further stimulates the production of IFNs and other cytokines. On the other hand, adaptor proteins, MyD88, STING, and MAVS, stimulate downstream signaling cascades that involve multiple kinases (TBK1, IKKs), and finally lead to IRF3/7 phosphorylation and nuclear translocation. The primary consequence of these virus-sensing pathways is the induction of type I/III IFN and pro-inflammatory cytokines and chemokines. ASC, adapter protein apoptosis-associated speck-like protein containing a caspase recruitment domain; CDR, cytosolic DNA receptors; cGAS, cyclic GMP-AMP synthase; cGAMP, 2′3′guanosine-adenosine monophosphate; IFI16, interferon-g inducible protein 16; IFN, interferon; IKK, IκB kinase; IRF3, interferon regulatory factor 3; MAVS, mitochondrial antiviral-signaling protein; MyD88, myeloid differentiation primary response 88; NLR, (NOD)-like receptor; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; RIG-I, retinoic acid inducible gene-I; RLR, RIG-1-like receptors; ss/dsRNA, single-stranded/double-stranded RNA; vRNA/DNA, viral RNA/DNA; STING, stimulator of interferon genes; TANK, TRAF-associated NF-κB activator; TBK1, TANK binding kinase 1; TLR, toll like receptor; TRIF, toll/IL-1R domain-containing adaptor-inducing IFN-β. Figure created with BioRender.com (accessed on 10 October 2021).
2.1. Virus Recognition by TLRs
As described elsewhere [], TLRs are type I integral membrane glycoproteins that recognize a variety of respiratory viruses. To date, the expression of 10 TLRs (TLR1-10) has been identified in human respiratory epithelial cells [,]. The interaction between TLRs and respiratory viruses trigger the signaling cascades of mitogen-activated protein (MAP) kinases and several transcription factors, including NF-κB, activating transcription factor 2 (ATF2), IRF-3, and IRF-7/9, leading to the secretion of type I (α and β) and type III (λ) IFNs, and pro-inflammatory cytokines (IL-1, IL-6, TNFα) and chemokines (IL-8, MCP-1) [,,]. TLRs contribute to both viral clearance and disease pathogenesis during infection in COPD [,,,].
TLRs are expressed either on the cell surface (TLR-1, -2, -4, -5, -6, and -10) or in the endosome compartment (TLR-3, -7, -8, and -9). Intracellular TLRs, in particular, TLR3 and TLR7, sense double (ds)- and single-stranded (ss) RNA viruses, including respiratory syncytial virus (RSV), rhinoviruses, influenza viruses, and SARS-CoV-2 [,,]. Apart from TLR3, which signals through TRIF, all other TLRs utilize their cytoplasmic toll-interleukin-1 receptor (TIR) domain to regulate cellular responses via their adaptor protein, MyD88, and kinases to initiate innate inflammatory responses [,,]. In addition, TLR activation upregulates signaling via the reactive oxygen species (ROS)-TNFα-converting enzyme (TACE)-epidermal growth factor receptor (EGFR) pathway, which results in IL-8 and vascular endothelial growth factor (VEGF) production []. The cell surface receptors (TLR-2, -4, and -6), however, might also be involved in the recognition of respiratory viruses by the airway epithelium [,]. The RSV F and G proteins have been shown to interact with TLR4, TLR2, and TLR6 to modulate the innate immune response [,,]. Most recently, TLR2 has been implicated in sensing the SARS-CoV-2 envelope protein to initiate virus-induced cytokine production, which is correlated to disease severity []. Interestingly, cigarette smoke enhanced TLR2 and TLR4 expression in the lung tissue of rodents and humans [,,,], suggesting a link to increased responses to COVID-19 in COPD airways. In addition, TLR2 and TLR4 may also contribute to COPD inflammation [].
2.2. Virus Recognition by RLRs
RIG-I (encoded by the DDX58 gene), melanoma differentiation-associated gene 5 (MDA5; encoded by the IFIH1 gene), and laboratory of genetics and physiology 2 (LGP2) are RLR family members present in the cytosol that detect intracellular dsRNA and ssRNA of a number of viruses [,,,]. RLRs bind RNA through a C-terminal helicase domain, which promotes a conformational change that exposes the N-terminal caspase activation and recruitment domain (CARD) [,,,,,,,]. In airway epithelial cells, induction of type I and type III interferon antiviral responses have been demonstrated for several respiratory viruses via the interaction of RIG-I/MDA5, CARD, and their adaptor proteins mitochondrial antiviral-signaling protein (MAVS), IFN-β promoter stimulator 1 (IPS-1), leading to activation of downstream NF-κB and IRF-3 pathways [,,,,].
On the other hand, the intracellular sensor cGAS (cyclic GMP-AMP (cGAMP) synthase) detects retroviral replication products, dsDNA and RNA/DNA hybrids, to induce the synthesis of cGAMP, which binds and activates STING (stimulator of interferon genes) to regulate type I/III IFN signaling [,,,,,,,,,]. Moreover, STING and MAVS also regulate downstream multiple kinase signaling cascades, leading to IRF-3 phosphorylation and NF-κB nuclear translocation, and the production of inflammatory cytokines/chemokines [,]. Lung tissue from COPD patients shows a constitutive decreased expression of IFN-β, IRF-7, RIG-I, and MDA5, suggesting that this deficiency contributes to increased susceptibility of the COPD epithelium to RNA virus infection []. Elevated levels of self-DNA have been linked to cigarette-smoke-induced inflammation in mice, and poor prognosis in COPD [,,]. This cell-free DNA may result from cigarette-smoke-induced oxidative stress, cellular senescence, and apoptosis- or necrosis-associated cell death [,]. Self-DNA is also recognized by cGAS-STING DNA virus sensors to regulate type I IFN-dependent lung inflammation in COPD [].
2.3. Virus Recognition by NLRs
NLRs represent another major group of cytoplasmic pathogen recognition receptors. The sensing of DNA and RNA viruses (e.g., RSV, influenza A virus (IAV), measles, encephalomyocarditis virus, hepatitis C virus) via NLRP3 (NLR family pyrin domain containing 3) activation represents a common pathway for viral detection by host cells [,,,,,]. NLRP3 modulates inflammasome-induced inflammation through the activation and secretion of pro-IL-1β and pro-IL-18. The maturation of these cytokines further stimulates the production of caspase-1, IFNs, and other cytokines [,,,,]. This is detrimental to the virus, leading to pyroptosis and elimination of virus-infected cells from the host [,]. NLRP3 also has a strong link to mitochondrial dysfunction [,]. Mitochondria contribute to antiviral responses in cells by facilitating RLR signaling []. Elevated NLRP3 has been reported in an in vivo mouse model of COPD exacerbation, and in the lung epithelium of COPD patients during exacerbation [,]. The NLRP3 inflammasome induces mitochondrial damage and mitophagy in COPD epithelium, resulting in marked impairment of MAVS-mediated antiviral immunity, thus regulating the production of antiviral type I and type III IFNs, and pro-inflammatory cytokines following viral infection [,,]. Some respiratory viruses, such as rhinoviruses, induce mitochondrial dysfunction to impair the efficiency of host antivirus defense [,]. Cigarette-smoke-associated mitochondrial dysfunction and the impaired antiviral response may contribute to the increased susceptibility to respiratory infections in COPD epithelium.
2.4. Viral Evasion during Virus Sensing
Respiratory viruses use different strategies to disguise themselves from host-sensing systems to avoid being recognized by the host as dangerous. In order to readily replicate in host cells, rhinoviruses and coronaviruses (CoV, including SARS-CoV-2) modify their recognizable viral nucleic acid products in the cytosol to form ‘replication organelles’ (RO), which can shield from host-sensing signaling cascades [,,,,,,]. RNA viruses, such as CoVs, rhinoviruses, and RSV, replicate in the cytosol of respiratory epithelial cells, and can form ROs []. In the cytosol, RSV induces the formation of ‘inclusion bodies’ that are found associated with cytosolic occluded structures []. Non-structural protein (nsp)-3 and nsp4 of CoVs, and the N and P proteins of RSV generate similar intracellular membrane structures inside membrane-bound vesicles or invaginations that are undetectable by host virus sensors, thus promoting productive virus replication machinery [,,].
It is still unclear whether the host innate defense system can also recognize and attack virus replication organelles to restrain virus infection. However, the activation of the type I IFN signaling cascade may induce effectors that compromise the integrity of ROs [,]. ROs may be recognized and targeted by GBPs (guanylate-binding proteins), including Mx GTPases, to initiate an antiviral response, and disrupt viral replication [,,,,].
Interestingly, unlike other RNA viruses, influenza virus circumvents RIG-I and MDA5 RNA sensors and TLRs by replicating in the nucleus rather than in the cytosol []. However, as mentioned above, the host can initiate countermeasures against influenza amplification by GBPs that are localized in the nucleus and the cytosol [,].
Some respiratory viruses also avoid host innate immune viral sensing by directly modifying their RNA by adding a cap or a cap mimic-structure to the 5′-end, which is identical to host mRNA [,,,,]. Rhinovirus can attach a cap-mimicking peptide, VPg, to its RNA 5′-end to protect it from recognition by innate RNA sensors [,,]. The RNA-dependent RNA polymerase (RdRp) complex of IAV cap-snatches host mRNA from short, 5′-capped transcripts produced by the DNA dependent RNA polymerase II (RNAPII) in the nucleus to prime transcription of viral mRNA []. RSV and CoVs utilize the enzymatic function in their polymerase complexes to add cap-structures to their own mRNAs to escape detection by host innate sensors [,]. CoVs use the 2′-O-methyltransferase activity of nsp16 to methylate their cap structures to avoid MDA5 and associated innate immune responses [].
Respiratory viruses also employ endoribonuclease activity to avoid virus sensing. CoVs and influenza encode endonuclease activity to destroy their own RNA, thus preventing viral recognition [,]. Likewise, some respiratory viruses induce host shut-off; a process in which viruses halt cellular protein expression, thereby suppressing innate immune responses, and facilitating viral replication and evasion [,,,]. The NS1 and PA-X proteins of influenza virus play important roles in host shut-off during influenza infection []. NS1 inhibits 3′-end cleavage and polyadenylation of cellular mRNAs, preventing host gene expression, including IFN and pro-inflammatory responses [,]. The NSP1 protein of CoVs, including SARS-CoV and Middle East respiratory syndrome-related coronavirus (MERS-CoV), mediates host shut-off by binding to cellular transcriptional and translational factors of host mRNA []. The 2A protease of rhinoviruses can also cleave host translation initiation factor elF4G to induce host shut-off during infection [].
Viral infection is associated with the formation of stress granules, an integral part of host stress responses, to store stalled untranslated mRNAs, and coordinate cellular processes during stress, including antiviral responses [,,]. Some RNA viruses, such as RSV, rhinoviruses, influenza viruses, and MERS-CoV, counteract the formation of stress granules to benefit their replication [,]. Stress granule factor proteins, such as G3BP1 and G3BP2, have been implicated in the formation of antiviral stress granule formation during RSV, rhinovirus, and SARS-CoV-2 infections [,]. Rhinoviruses use their 2A papain-like protease (PLpro) to cleave host proteins in the innate immune response to facilitate invasion []. A classic example of this is the RNA-sensing effector, MAVS, which is cleaved by both 2A and 3C proteases of rhinovirus to impede type I IFN signal transduction []. Some RNA viruses suppress the host innate immune response by manipulating the ubiquitin system and associated ISG15 protein [,,]. CoV PLpro interact with the deubiquitinating activity of ISG15 and E3 ligase RCHY1 to inhibit type I IFN secretion and apoptosis []. In addition, the NS1 protein of influenza and RSV targets TRIM25 to suppress RIG-I ubiquitination, and thus, benefit virus infection [,]. Influenza NS1 has been reported to induce the expression of the deubiquitinase A20 to downregulate RIG-I activation []. Influenza NS1 was also recently shown to affect the deubiquitinating activity of the MDM2 E3 ligase in a way that supports IAV infection [].
3. Type I and Type III Interferon Signaling Pathways in Airway Epithelium
Interferons are key defense molecules, which control and eliminate viral replication, thus playing a critical role in the host antiviral response. There are three distinct types of interferons, namely type I, type II, and type III interferons [,]. Though type II interferon (IFN-γ) is mainly expressed by immune cells, type I and type III interferons are expressed by airway epithelial cells, and will therefore be the focus of the remainder of this review.
3.1. Type I Interferon Signaling
Human type I interferons consist of 17 subtypes, including 13 isoforms of IFN-α, along with IFN-β, IFN-ε, IFN-κ, and IFN-ω [,,]. Among these, IFN-α and IFN-β are the most studied, classical, antiviral cytokines. Upon viral infection, however, airway epithelial cells mainly secrete IFN-β protein, and IFN-α is undetectable []. Type I IFN production is predominately initiated by IRF-3 and IRF-7 [,,]. It has been well documented that IFN-β is mainly produced from infected cells via IRF-3, and that IFN-β receptor binding induces IRF-7, leading to late-phase IFN-β and IFN-α production [,]. As described in Figure 2, type I interferons are translated and secreted from the infected cell to exert their antiviral effect through autocrine and paracrine pathways [,]. Type I IFN receptors are found on most cell types, including epithelial cells in the lung, and they signal through canonical and non-canonical pathways to mediate the expression of ISGs [,]. As outlined in Figure 2, they first bind to the extracellular part of type I IFN heterodimeric receptor complex IFN-α/β R1 and IFN-α/β R2 on the surface of surrounding cells. Receptor engagement subsequently activates IFN-α/β R1 and IFN-α/β R2, resulting in conformational changes in Tyk2 protein tyrosine kinase for the former, and Janus tyrosine kinase JAK1 for the latter [,]. JAKs mediate the phosphorylation and activation of the signal transducer and activator of transcription (STAT) family of proteins through the classical JAK–STAT-signaling pathways [,,,,,].
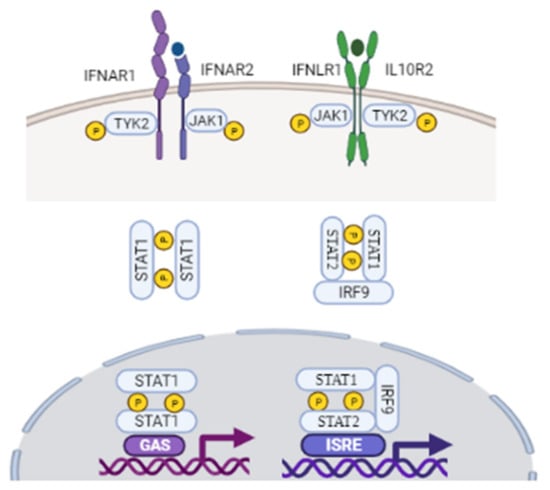
Figure 2.
Upon binding to their receptors, IFNs induce the activation of the JAK/STAT signaling pathway. Type I IFNs first bind to the extracellular part of type I IFN heterodimeric receptor complex IFN-α/β R1 and IFN-α/β R2. Receptor engagement subsequently activates IFN-alpha/beta R1 and IFN-α/β R2, resulting in conformational changes in their associated intracellular tyrosine residues Tyk2 and JAK1 protein tyrosine. Both type I and III IFNs use JAK1 for their signaling. Apart from IFNL-R1 receptor, type III IFNs also use IL-10 receptor beta (IL-10R2) receptor complex. Following JAK activation, STAT1/STAT2 are recruited and activated, which leads to their dimerization and binding to IRF9 and ISRE, forming the ISGF3 complex. Upon JAK activation, STAT1 homodimer complex are also formed, which translocate into the nucleus, and drives ISG production. STAT1 homodimers interact with GAS to induce a pro-inflammatory response. GAS, gamma activated sequences; IFN, interferon; IRF9, interferon regulatory factor 9; ISGF3, interferon stimulated gene factor 3; ISRE, interferon-stimulated regulatory element, JAK, Janus kinase; STAT, signal transducer and activator of transcription; TYK2, tyrosine Kinase 2. Figure created with BioRender.com (accessed on 10 October 2021).
Upon activation, STAT1 and STAT2, together with IRF-9, regulate the dimerization and the assembly of the IFN-stimulated gene factor 3 (ISGF3), which then translocates to the nucleus to induce transcription of ISGs. JAKs also mediate the phosphorylation of STAT1 and STAT3 homodimer complexes. STAT1 homodimers regulate signaling through gamma-activated sequences (GAS) to induce pro-inflammatory responses [,,,]. STAT3 homodimers indirectly suppress the transcription of inflammatory genes by activating genes associated with NF-κB gene inhibition [,], thus limiting the innate pro-inflammatory response. These signaling cascades comprise the canonical type I IFN pathway, which induces the downstream transcription of an array of ISGs to further limit viral infection []. The suppressor of cytokine signaling (SOCS) family of proteins inhibit the JAK-STAT signal transduction pathway by forming a negative feedback loop [,]. Type I IFNs also signal through the non-canonical pathway to initiate the transcription of ISGs. The main non-canonical pathways used by type I IFNs include the mitogen-activated protein kinase (MAPK) pathway and the phosphoinositide 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathways. Other non-canonical pathways include sirtuin (SIRT)-2 and the Schlafen (SLFN) family of proteins [,].
Many respiratory viruses, such as RSV and SARS-CoV-2, evade host antiviral responses by restraining the production of type I IFNs [,]. The NS1 and NS2 proteins of RSV inhibit the type I IFN host response by interrupting signal transduction and activation of the JAK-STAT pathway []. The NSP1, NSP3, ORF3b, ORF6, and N proteins of SARS-CoV are also type I IFNs antagonists [,]. Interestingly, ORF6 also suppresses the expression of mRNA export proteins’ Nup98-Rae1 complex by blocking STAT1 nuclear translocation []. PLpro inhibits IRF-3 phosphorylation to prevent its homodimerization and nuclear translocation, and antagonizes innate immunity by acting on downstream NF-κB and IFN-β pathways in both SARS-CoV- and SARS-CoV-2-infected cells [,]. Some viruses, such as RSV, rhinoviruses, and influenza viruses, induce the expression of SOCS genes to inhibit the antiviral effect of ISGs and IFN induction [,,]. Reduced expression of IFN-β and its transcription factor IRF-7 have been demonstrated in the COPD airway epithelium and macrophages compared with non-COPD cells [].
3.2. Type III Interferon Signaling
Type III IFNs (IFN-λ) are a relatively new class of IFNs, which include IFN-λ1/IL-29, IFN-λ2/IL-28A, IFN-λ3/IL-28B, and the most recently discovered, IFN-λ4 [,]. IFN-λ1 gene expression is regulated by both IRF-3 and IRF-7, whereas IFN-λ2/λ3 genes appear to be induced only by IRF-7 [,]. However, influenza virus- and rhinovirus-induced EGFR activation suppressed IRF-1-induced IFN-λ production, and increased viral infection []. Type III IFNs share similar antiviral pathways with those of type I IFNs, but expression of IFN-λ receptors is largely restricted to epithelial cells, suggesting a frontline role in the antiviral response. The IFN-λ receptor is composed of IL-10R2 and IFNLR1 [,,]. As IL-10R2 is ubiquitously expressed, the expression of IFNLR1 is restricted to certain cell types, including airway epithelial cells, suggesting that differential transcriptional regulation of the IFNLR1 gene is associated with the cell-type specificity of IFN-λ.
Despite differential receptor constitution, type III IFNs also act through the canonical signaling pathways, and are recognized by the same PRRs as those used by type I IFNs (Figure 2). Upon virus binding, receptor-ligand complexes trigger JAK1 and Tyk2 phosphorylation and activation, resulting in phosphorylation of STAT1:STAT2 heterodimers. These heterodimers signal through ISGF3, as described in type I IFN pathways, to promote the transcription of downstream ISGs, and induce an antiviral state [,,,].
3.3. ISGs Induce an Antiviral State in Airway Epithelium
IFNs induce a myriad of ISGs, which introduce a second round of autocrine and paracrine signaling in cells. This has the effect of establishing an antiviral state in the host cells to attenuate viral-associated pathogenesis in the infected cells before trigging the adaptive immune response, leading to viral clearance [,,,,]. ISGs interact with transcription factors and other signaling mediators to amplify the innate immune response. ISGs also interact with antiviral proteins to disturb virus replication by degrading viral RNA or blocking host cell translation [,]. Consequently, ISGs efficiently prevent further spread of the infection in bystander cells by restricting viral transcription and replication [,]. ISGs regulate the production of the pro-apoptotic p53 protein and several antiviral proteins, including the GTPase myxovirus resistance 1 (MX1), IFN-inducible double-stranded RNA-dependent protein kinase (PKR), 2′-5′-oligoadenylate synthetase (OAS), IFN-induced transmembrane proteins (IFITMs), apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC1), the tripartite motif-containing (TRIM) family of molecules, and viperin (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible) to subvert viral replication [,]. RSV, rhinoviruses, and influenza viruses induce the expression of viperin in airway epithelium to fight against respiratory viral invasion [,,].
Accumulated evidence suggests that many viruses escape host antiviral defenses by preventing ISG secretion, and thus, perturb IFN-α/β and associated signaling pathways. The influenza virus NS1 protein has been shown to inhibit PKR activation, resulting in increased viral replication in the lung []. Impaired IFN-λ and IFN-β have been demonstrated in airway epithelial cells of active smokers compared with that of non-smokers []. Airway epithelium in asthma and COPD patients have been reported to have a deficient or delayed IFN-β and IFN-λ response to rhinovirus infection, as well as the secretion of downstream ISGs, including PKR, OAS1, and viperin [,]. Elevated IL-17A in the lung tissue of patients with severe COPD correlated with lung function decline, and IL-17A attenuated virus-induced IFN-λ expression by enhancing SOCS1 and SOCS3 expression to inhibit autocrine signaling loops in human airway epithelial cells []. Downregulated MxA, OAS1, and viperin have also been shown in the sputum of severe COPD patients []. This defective IFN response in the respiratory tract may contribute to the increased susceptibility to infection resulting in impaired apoptosis and increased virus load in infected cells. Singanayagam and colleagues found that ISG-induced CXCL10 downregulation in immune cells from the lungs of COPD patients was associated with impaired IFN-α, IFN-β, and IFN-λ secretion following rhinovirus infection compared with cells from healthy subjects []. Impaired IFN-β and IFN-λ have also been demonstrated in IAV infected primary bronchial epithelial cells from COPD patients [,].
4. SARS-CoV-2 Associated IFN Signaling and Implications for COPD
As a global pandemic, SARS-CoV-2 has had a hugely detrimental impact worldwide. SARS-CoV-2 is a non-segmented, positive sense, single-strand RNA virus, which causes both upper and lower respiratory tract infections [,]. The majority of patients exhibit mild flu-like upper respiratory symptoms; however, a subset of patients develop severe life-threatening acute respiratory distress syndrome (ARDS) [,,]. Upon infection, individuals with COPD and current smokers are among the high-risk conditions associated with the development of SARS-CoV-2-associated disease and mortality [,,,]. The infection is initiated by the attachment of the virus (spike) S protein to the host cellular entry receptor angiotensin converting enzyme 2 (ACE2), and other virus entry co-factors, such as transmembrane serine protease 2 (TMPRSS2). Elevated expression of ACE2 has been found in airway epithelium of COPD and smokers [,,]. ACE2 in COPD has also been shown to inversely correlate with lung function []. Upregulated ACE and TMPRSS2 expression has been demonstrated on non-ciliated goblet cells (primary target of SARS-CoV-2 infection) in COPD airway epithelium []. Increased replication has also been demonstrated in well-differentiated COPD bronchial epithelial cells compared with cells from heathy subjects, suggesting that COPD-associated goblet cell hyperplasia may contribute to the increased susceptibility to SARS-CoV-2-associated severe disease []. Most recently, ACE2 has been identified as an ISG, suggesting that SARS-CoV-2 may exploit IFN-driven ACE2 upregulation to enhance infection. []. Another study found that SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and virus-induced hyperinflammation, indicating a role of TLR4 activation in SARS-CoV-2 pathogenesis and associated innate immune response []. Upregulation of TLR4 has also been implicated in COPD inflammation and exacerbations, and may be a therapeutic target in SARS-CoV-2-induced severe disease [].
SARS-CoV-2 perturbs host antiviral innate immune responses through multiple elaborate viral inhibitory mechanisms to evade the host immune response, as well as facilitating viral transmission and adaptation to its human host. These strategies include compromising PRR sensing through RIG-I and TLR signaling, antagonizing IFN production, and inhibiting IFN signaling [,,,].
SARS-CoV-2 infection also induces overt, but delayed, type-I IFN response by inhibiting IFN signaling during the early phase of infection [,,]. Xia and colleagues have demonstrated that the nsp1, nsp6, nsp13, ORF3a, M, and ORF7b proteins of SARS-CoV-2 block STAT1 phosphorylation; whereas nsp6, nsp13, ORF7a, and ORF7b suppress STAT2 phosphorylation, and ORF6 inhibits nuclear translocation of STAT1 []. SARS-CoV-2 nsp1, nsp3, nsp12, nsp13, nsp14, orf3, orf6, and M protein have also been shown to inhibit Sendai-virus-induced IFN-β promoter activation, and ORF6 inhibits both type I IFN production and downstream signaling [,].
The SARS-CoV-2 PLpro protein exits its deubiquitinating and deISGylating activity by directly removing ubiquitin-like ISG15 modifications from IRF3 and MDA5, and directly cleaving IRF. The PLpro protein can interact with key regulators in the STING, NF-κB, MAPK, and TGF-β pathways, and thereby suppress the innate immune response [,,,,]. Therefore, PLpro may also be an important therapeutic target for the treatment of SARS-CoV-2.
In summary, SARS-CoV-2 utilizes mechanisms to circumvent host antiviral immunity, thus facilitating virus replication and spread. Direct IFN-β treatment has shown to effectively block SARS-CoV-2 replication in the early stage of infection []. Impaired IFN responses have also been demonstrated in stable COPD airway epithelium, and during COPD exacerbations. Taken together, impaired IFN responses may contribute to the increased susceptibility to SARS-CoV-2-associated severe disease and SARS-CoV-2-induced exacerbations in COPD airway epithelium.
5. Therapeutic Implications of IFN Signalling Cascades in COPD Exacerbations: Current Development and Future Perspectives
TLR activation initiates host self-defense mechanisms to fight bacteria or viral pathogens. However, dysregulated TLR2 and TLR4 expression and signaling activation have been demonstrated in cigarette-smoke-induced airway inflammation, and during COPD exacerbations [,]. The activation of these receptors initiates downstream MyD88- and NF-κB-dependent signaling cascades, and the associated inflammatory response in the airways [,,]. Inhibition of TLR2 and TLR4 signaling pathways may represent an effective strategy in correcting the excessive inflammatory response in COPD. In response to TLR3 and TLR7/8 stimulation, lung tissue explants from COPD patients display elevated proinflammatory response compared with healthy smokers [].
In theory, inhibitors or antagonists of TLR signaling that directly block TLR receptor- ligands binding or TLR signaling transduction pathways may represent potential therapeutic targets for treating COPD inflammation. However, this approach is still in its infancy, and to date there are no clinical studies of TLR antagonists in COPD. A number of TLR blockers are currently in Phase II clinical trials for the treatment of other inflammatory conditions, and may offer a promising future perspective for this approach [,]
The targeting of mediators in the IFN signaling or even augmentation of IFN signaling may also protect COPD patients from viral-induced exacerbations, thus improving quality of life. In a randomized clinic trial, orally inhaled IFN-β therapy in corticosteroid-resistant asthmatic patients resulted in improved lung function, and a reduced requirement for oral corticosteroids [] However, there are side effects associated with IFN-β administration which may hamper the clinical application of this approach []. In a recent study, Watson et al. reported that exogenous IFN-β treatment of influenza-infected COPD macrophages and airway epithelium had no effect on virus shedding, nor IFN and ISG production []. However, giving IFN-β as a prophylactic before infection significantly improved antiviral efficiency in both COPD macrophages and epithelium, indicating that the timing of IFN admission may be critical in clinical trial design. Data from a Phase II randomized trial of on-demand inhaled interferon beta-1a in severe asthmatics subjects with upper respiratory tract infection indicated that inhaled interferon beta-1a treatment did not reduce the severe exacerbation rate, but improved lung function [].
In conclusion, mediators that regulate IFN antiviral response, and the complex network of IFN signaling transduction, may play an important role in host defense against viral-induced COPD exacerbation. Elucidating the role of IFNs signaling cascades in the pathogenesis of COPD may provide a platform to understand the disease mechanism in this condition, leading to new routes to combat virus-induced exacerbations in COPD.
Author Contributions
H.G.-P. wrote the manuscript. D.L., C.C.T., J.C.K., S.W. critically revised the draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Mater Hospital Young Philanthropist (YP) trustees (to J.C.K.), Pfizer UK and Chiesi Farmaceutici (to C.C.T and J.C.K.), and NI Chest Heart and Stroke funding (to JK/CT/HGP).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Barnes, P.J. Oxidative stress-based therapeutics in COPD. Redox Biol. 2020, 33, 101544. [Google Scholar] [CrossRef] [PubMed]
- Linden, D.; Guo-Parke, H.; Coyle, P.V.; Fairley, D.; McAuley, D.F.; Taggart, C.C.; Kidney, J. Respiratory viral infection: A potential “missing link” in the pathogenesis of COPD. Eur. Respir. Rev. 2019, 28, 180063. [Google Scholar] [CrossRef] [Green Version]
- Guo-Parke, H.; Linden, D.; Weldon, S.; Kidney, J.C.; Taggart, C.C. Mechanisms of Virus-Induced Airway Immunity Dysfunction in the Pathogenesis of COPD Disease, Progression, and Exacerbation. Front. Immunol. 2020, 11, 1205. [Google Scholar] [CrossRef] [PubMed]
- Vareille, M.; Kieninger, E.; Edwards, M.R.; Regamey, N. The airway epithelium: Soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011, 24, 210–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, R.N.; Brighton, L.E.; Mueller, L.; Xiang, Z.; Rager, J.E.; Fry, R.C.; Peden, D.B.; Jaspers, I. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. J. Allergy Clin. Immunol. 2012, 130, 958–967.e14. [Google Scholar] [CrossRef] [Green Version]
- Rovina, N.; Koutsoukou, A.; Koulouris, N.G. Inflammation and immune response in COPD: Where do we stand? Mediators Inflamm. 2013, 2013, 413735. [Google Scholar] [CrossRef] [Green Version]
- Alexander-Brett, J.; Holtzman, M.J. Chapter 53—Virus Infection of Airway Epithelial Cells, in Mucosal Immunology, 4th ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 1013–1021. [Google Scholar]
- The Global Initiative for Chronic Obstructive Lung Disease GOLD. 2021. Available online: https://goldcopd.org/2021-gold-reports (accessed on 10 October 2021).
- Singanayagam, A.; Loo, S.L.; Calderazzo, M.; Finney, L.J.; Torralbo, M.B.T.; Bakhsoliani, E.; Girkin, J.; Veerati, P.; Pathinayake, P.S.; Nichol, K.S.; et al. Antiviral immunity is impaired in COPD patients with frequent exacerbations. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 317, L893–L903. [Google Scholar] [CrossRef]
- Veerati, P.C.; Troy, N.M.; Reid, A.T.; Li, N.F.; Nichol, K.S.; Kaur, P.; Maltby, S.; Wark, P.A.B.; Knight, D.A.; Bosco, A.; et al. Airway Epithelial Cell Immunity Is Delayed During Rhinovirus Infection in Asthma and COPD. Front. Immunol. 2020, 11, 974. [Google Scholar] [CrossRef]
- Hsu, A.C.; Starkey, M.R.; Hanish, I.; Parsons, K.; Haw, T.J.; Howland, L.J.; Barr, I.; Mahony, J.B.; Foster, P.S.; Knight, D.A.; et al. Targeting PI3K-p110α Suppresses Influenza Virus Infection in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2015, 191, 1012–1023. [Google Scholar] [CrossRef]
- Hsu, A.C.; Dua, K.; Starkey, M.R.; Haw, T.J.; Nair, P.M.; Nichol, K.; Zammit, N.; Grey, S.T.; Baines, K.J.; Foster, P.S.; et al. MicroRNA-125a and -b inhibit A20 and MAVS to promote inflammation and impair antiviral response in COPD. JCI Insight. 2017, 2, e90443. [Google Scholar] [CrossRef] [Green Version]
- Finney, L.J.; Glanville, N.; Farne, H.; Aniscenko, J.; Fenwick, P.; Kemp, S.V.; Trujillo-Torralbo, M.B.; Loo, S.L.; Calderazzo, M.A.; Wedzicha, J.A.; et al. Inhaled corticosteroids downregulate the SARS-CoV-2 receptor ACE2 in COPD through suppression of type I interferon. J. Allergy Clin. Immunol. 2021, 147, 510–519.e5. [Google Scholar] [CrossRef]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNAsensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Sousa, C.R.E. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Taniguchi, T. IRFs: Master regulators of signaling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006, 6, 644–658. [Google Scholar] [CrossRef]
- Sartorius, R.; Trovato, M.; Manco, R.; D’Apice, L.; De Berardinis, P. Exploiting viral sensing mediated by Toll-like receptors to design innovative vaccines. NPJ Vaccines 2021, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, I.; Ye, F.; McNally, B.; Willette, M.; Flano, E. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J. Virol. 2013, 87, 3261–3270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovach, M.A.; Standiford, T.J. Toll like receptors in diseases of the lung. Int. Immunopharmacol. 2011, 11, 1399–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sioud, M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol. Med. 2006, 12, 167–176. [Google Scholar] [CrossRef]
- Schulz, O.; Diebold, S.S.; Chen, M.; Näslund, T.I.; Nolte, M.A.; Alexopoulou, L.; Azuma, Y.T.; Flavell, R.A.; Liljeström, P.; Sousa, C.R.E. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 2005, 433, 887–892. [Google Scholar] [CrossRef]
- Volkova, M.; Zhang, Y.; Shaw, A.C.; Lee, P.J. The role of Toll-like receptors in age-associated lung diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2012, 67, 247–253. [Google Scholar] [CrossRef]
- Pace, E.; Giarratano, A.; Ferraro, M.; Bruno, A.; Siena, L.; Mangione, S.; Johnson, M.; Gjomarkaj, M. TLR4 upregulation underpins airway neutrophilia in smokers with chronic obstructive pulmonary disease and acute respiratory failure. Hum. Immunol. 2011, 72, 54–62. [Google Scholar] [CrossRef]
- Wang, R.; Ahmed, J.; Wang, G.; Hassan, I.; Strulovici-Barel, Y.; Salit, J.; Mezey, J.G.; Crysta, R.G. Airway epithelial expression of TLR5 is downregulated in healthy smokers and smokers with chronic obstructive pulmonary disease. J. Immunol. 2012, 189, 2217–2225. [Google Scholar] [CrossRef] [Green Version]
- Haw, T.J.; Starkey, M.R.; Pavlidis, S.; Fricker, M.; Arthurs, A.L.; Nair, P.M.; Liu, G.; Hanish, I.; Kim, R.Y.; Foster, P.S.; et al. Toll-like receptor 2 and 4 have opposing roles in the pathogenesis of cigarette smoke-induced chronic obstructive pulmonary disease. Am. J. Physiol. Lung. Cell Mol. Physiol. 2018, 314, L298–L317. [Google Scholar] [CrossRef]
- Onofrio, L.; Caraglia, M.; Facchini, G.; Margherita, V.; Placido, S.; Buonerba, C. Toll-like receptors and COVID-19: A two-faced story with an exciting ending. Future Sci. OA 2020, 6, FSO605. [Google Scholar] [CrossRef]
- Koff, J.L.; Shao, M.X.; Ueki, I.F.; Nadel, J.A. Multiple TLRs activate EGFR via a signaling cascade to produce innate immune responses in airway epithelium. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 294, L1068–L1075. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Karki, R.; William, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, H.K. Innate immune recognition of respiratory syncytial virus infection. BMB Rep. 2014, 47, 184–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murawski, M.R.; Bowen, G.N.; Cerny, A.M.; Anderson, L.J.; Haynes, L.M.; Tripp, R.A.; Kurt-Jones, E.A.; Finberg, R.W. Respiratory syncytial virus activates innate immunity through Toll-like receptor 2. J. Virol. 2009, 83, 1492–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshaghdali, K.; Saeed, M.; Kamal, M.A.; Saeed, A. Interaction of ectodomain of Respiratory Syncytial Virus G protein with TLR2/TLR6 heterodimer: An in vitro and in silico approach to decipher the role of RSV G protein in pro-inflammatory response against the virus. Curr. Pharm. Des. 2021, 27, 4464–4476. [Google Scholar] [CrossRef]
- Pace, E.; Ferraro, M.; Siena, L.; Melis, M.; Montalbano, A.M.; Johnson, M.; Bonsignore, M.R.; Bonsignore, G.; Gjomarkaj, M. Cigarette smoke increases Toll-like receptor 4 and modifies lipopolysaccharide-mediated responses in airway epithelial cells. Immunology 2008, 124, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Bracke, K.R.; Vermaelen, K.Y.; Demedts, I.K.; Joos, G.F.; Pauwels, R.A.; Brusselle, G.G. Murine TLR4 is implicated in cigarette smoke-induced pulmonary inflammation. Int. Arch. Allergy Immunol. 2006, 141, 354–368. [Google Scholar] [CrossRef]
- Simpson, J.L.; McDonald, V.M.; Baines, K.J.; Oreo, K.M.; Wang, F.; Hansbro, P.M.; Gibson, P.G. Influence of age, past smoking, and disease severity on TLR2, neutrophilic inflammation, and MMP-9 levels in COPD. Mediat. Inflamm. 2013, 2013, 462934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidletskaya, K.; Vitkina, T.; Denisenko, Y. The Role of Toll-Like Receptors 2 and 4 in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstr. Pulmon. Dis. 2020, 15, 1481–1493. [Google Scholar] [CrossRef]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell Mol. Immunol. 2021, 18, 539–555. [Google Scholar] [CrossRef]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Valero, J.; Olloquequi, J.; Montes, J.F.; Rodríguez, E.; Martín-Satué, M.; Texidó, L.; Ferrer Sancho, J. Deficient pulmonary IFN-β expression in COPD patients. PLoS ONE 2019, 14, e0217803. [Google Scholar]
- Nascimento, M.; Gombault, A.; Lacerda-Queiroz, N.; Panek, C.; Savigny, F.; Sbeity, M.; Bourinet, M.; Le Bert, M.; Riteau, N.; Ryffel, B.; et al. Self-DNA release and STING-dependent sensing drives inflammation to cigarette smoke in mice. Sci. Rep. 2019, 9, 14848. [Google Scholar] [CrossRef]
- Avriel, A.; Rozenberg, D.; Raviv, Y.; Heimer, D.; Bar-Shai, A.; Gavish, R.; Sheynin, J.; Douvdevani, A. Prognostic utility of admission cell-free DNA levels in patients with chronic obstructive pulmonary disease exacerbations. Int. J. Chron. Obstr. Pulmon. Dis. 2016, 11, 3153–3161. [Google Scholar] [CrossRef] [Green Version]
- Ma, R.; Ortiz Serrano, T.P.; Davis, J.; Prigge, A.D.; Ridge, K.M. The cGAS-STING pathway: The role of self-DNA sensing in inflammatory lung disease. FASEB J. 2020, 34, 13156–13170. [Google Scholar] [CrossRef]
- Jacobs, S.R.; Damania, B. NLRs, inflammasomes, and viral infection. J. Leukoc. Biol. 2012, 92, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Shrivastava, G.; León-Juárez, M.; García-Cordero, J.; Meza-Sánchez, D.E.; Cedillo-Barrón, L. Inflammasomes and its importance in viral infections. Immunol. Res. 2016, 64, 1101–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segovia, J.; Sabbah, A.; Mgbemena, V.; Tsai, S.Y.; Chang, T.H.; Berton, M.T.; Morris, I.R.; Allen, I.C.; Ting, J.P.; Bose, S. TLR2/MyD88/NF-κB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS ONE 2012, 7, e29695. [Google Scholar] [CrossRef] [PubMed]
- Allen, I.C.; Scull, M.A.; Moore, C.B.; Holl, E.K.; McElvania-TeKippe, E.; Taxman, D.J.; Guthrie, E.H.; Pickles, R.J.; Ting, J.P. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 2009, 30, 556–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komune, N.; Ichinohe, T.; Ito, M.; Yanagi, Y. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1β secretion. J Virol. 2011, 85, 13019–13026. [Google Scholar] [CrossRef] [Green Version]
- Rajan, J.V.; Rodriguez, D.; Miao, E.A.; Aderem, A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J Virol. 2011, 85, 4167–4172. [Google Scholar] [CrossRef] [Green Version]
- Burdette, D.; Haskett, A.; Presser, L.; McRae, S.; Iqbal, J.; Waris, G. Hepatitis C virus activates interleukin-1β via caspase-1-inflammasome complex. J. Gen. Virol. 2012, 93 Pt 2, 235–246. [Google Scholar] [CrossRef]
- Holley, C.L.; Schroder, K. The rOX-stars of inflammation: Links between the inflammasome and mitochondrial meltdown. Clin. Transl. Immunol. 2020, 9, e01109. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.P.; Amar, S.; Gehlot, P.; Patra, S.K.; Kanwar, N.; Kanwal, A. Mitochondrial Modulations, Autophagy Pathways Shifts in Viral Infections: Consequences of COVID-19. Int. J. Mol. Sci. 2021, 22, 8180. [Google Scholar] [CrossRef]
- Nachmias, N.; Langier, S.; Brzezinski, R.Y.; Siterman, M.; Stark, M.; Etkin, S.; Avriel, A.; Schwarz, Y.; Shenhar-Tsarfaty, S.; Bar-Shai, A. NLRP3 inflammasome activity is upregulated in an in-vitro model of COPD exacerbation. PLoS ONE 2019, 14, e0214622. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Lv, C.; Wang, S.; Ying, H.; Weng, Y.; Yu, W. NLRP3 Inflammasome Involves in the Acute Exacerbation of Patients with Chronic Obstructive Pulmonary Disease. Inflammation 2018, 41, 1321–1333. [Google Scholar] [CrossRef]
- Yoon, C.M.; Nam, M.; Oh, Y.M.; Dela Cruz, C.S.; Kang, M.J. Mitochondrial Regulation of Inflammasome Activation in Chronic Obstructive Pulmonary Disease. J. Innate Immun. 2016, 8, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Zhang, M.Y.; Qu, Y.Q. The Underlying Role of Mitophagy in Different Regulatory Mechanisms of Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstr. Pulmon. Dis. 2020, 15, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome—A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michi, A.N.; Yipp, B.G.; Dufour, A.; Lopes, F.; Proud, D. PGC-1α mediates a metabolic host defense response in human airway epithelium during rhinovirus infections. Nat. Commun. 2021, 12, 3669. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Worm, S.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Santerre, M.; Arjona, S.P.; Allen, C.N.; Shcherbik, N.; Sawaya, B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021, 268, 2013–2022. [Google Scholar] [CrossRef]
- Roulin, P.S.; Lötzerich, M.; Torta, F.; Tanner, L.B.; van Kuppeveld, F.J.; Wenk, M.R.; Greber, U.F. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe 2014, 16, 677–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lifland, A.W.; Jung, J.; Alonas, E.; Zurla, C.; Crowe, J.E., Jr.; Santangelo, P.J. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 2012, 86, 8245–8258. [Google Scholar] [CrossRef] [Green Version]
- Oudshoorn, D.; Rijs, K.; Limpens, R.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8, e01658-17. [Google Scholar] [CrossRef] [Green Version]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef]
- Scutigliani, E.M.; Kikkert, M. Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Factor Rev. 2017, 37, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Praefcke, G.J.K. Regulation of innate immune functions by guanylate-binding proteins. Int. J. Med. Microbiol. 2018, 308, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Graf, L.; Chen, T.; Liao, Q.; Bai, T.; Petric, P.P.; Zhu, W.; Yang, L.; Dong, J.; Lu, J.; et al. Rare variant MX1 alleles increase human susceptibility to zoonotic H7N9 influenza virus. Science 2021, 373, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551–566. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, P.B. Metastable biomolecular condensates of interferon-inducible antiviral Mx-family GTPases: A paradigm shift in the last three years. J. Biosci. 2021, 46, 72. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lu, Y.; Thulasi Raman, S.N.; Xu, F.; Wu, Q.; Li, Z.; Brownlie, R.; Liu, Q.; Zhou, Y. Nuclear-resident RIG-I senses viral replication inducing antiviral immunity. Nat. Commun. 2018, 9, 3199. [Google Scholar] [CrossRef]
- Nordmann, A.; Wixler, L.; Boergeling, Y.; Wixler, V.; Ludwig, S. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J. 2012, 26, 1290–1300. [Google Scholar] [CrossRef]
- Flanegan, J.B.; Petterson, R.F.; Ambros, V.; Hewlett, N.J.; Baltimore, D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl. Acad. Sci. USA 1977, 74, 961–965. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.F.; Nomoto, A.; Detjen, B.M.; Wimmer, E. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 1977, 74, 59–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barral, P.M.; Sarkar, D.; Fisher, P.B.; Racaniello, V.R. RIG-I is cleaved during picornavirus infection. Virology 2009, 391, 171–176. [Google Scholar] [CrossRef] [Green Version]
- De Vlugt, C.; Sikora, D.; Pelchat, M. Insight into Influenza: A Virus Cap-Snatching. Viruses 2018, 10, 641. [Google Scholar] [CrossRef] [Green Version]
- Barik, S. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 1993, 74, 485–490. [Google Scholar] [CrossRef]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindler, E.; Gil-Cruz, C.; Spanier, J.; Li, Y.; Wilhelm, J.; Rabouw, H.H.; Züst, R.; Hwang, M.; V’kovski, P.; Stalder, H.; et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017, 13, e1006195. [Google Scholar] [CrossRef]
- Yuan, P.; Bartlam, M.; Lou, Z.; Chen, S.; Zhou, J.; He, X.; Lv, Z.; Ge, R.; Li, X.; Deng, T.; et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 2009, 458, 909–913. [Google Scholar] [CrossRef]
- Nogales, A.; Villamayor, L.; Utrilla-Trigo, S.; Ortego, J.; Martinez-Sobrido, L.; De Diego, M.L. Natural Selection of H5N1 Avian Influenza A Viruses with Increased PA-X and NS1 Shutoff Activity. Viruses 2021, 13, 1760. [Google Scholar] [CrossRef]
- Kong, J.Q.; Shen, J.H.; Huang, Y.; Ruan, R.Y.; Xiang, B.; Zheng, X.D.; Cheng, K.D.; Wang, W. Development of a yeast two-hybrid screen for selection of A/H1N1 influenza NS1 non-structural protein and human CPSF30 protein interaction inhibitors. Yao Xue Xue Bao 2010, 45, 388–394. [Google Scholar] [PubMed]
- Terada, Y.; Kawachi, K.; Matsuura, Y.; Kamitani, W. MERS coronavirus nsp1 participates in an efficient propagation through a specific interaction with viral RNA. Virology 2017, 511, 95–105. [Google Scholar] [CrossRef]
- Petersen, J.F.; Cherney, M.M.; Liebig, H.D.; Skern, T.; Kuechler, E.; James, M.N. The structure of the 2A proteinase from a common cold virus: A proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 1999, 18, 5463–5475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zheng, Y.; Huang, J.; Li, J. Mitophagy in Antiviral Immunity. Front. Cell Dev. Biol. 2021, 9, 723108. [Google Scholar] [CrossRef] [PubMed]
- Eiermann, N.; Haneke, K.; Sun, Z.; Stoecklin, G.; Ruggieri, A. Dance with the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses 2020, 12, 984. [Google Scholar] [CrossRef] [PubMed]
- Montero, H.; Trujillo-Alonso, V. Stress granules in the viral replication cycle. Viruses 2011, 3, 2328–2338. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, M.E.; Lifland, A.W.; Utley, T.J.; Santangelo, P.; Crowe, J.E., Jr. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 2010, 84, 12274–12284. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.; Langereis, M.A.; van Kuppeveld, F.J. Induction and suppression of innate antiviral responses by picornaviruses. Cytokine Growth Factor Rev. 2014, 25, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.; Prasad, K.; AlAsmari, A.F.; Alharbi, M.; Rashid, S.; Kumar, V. Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation. Int. J. Biol. Macromol. 2021, 10, 636–648. [Google Scholar] [CrossRef]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Heaton, S.M.; Borg, N.A.; Dixit, V.M. Ubiquitin in the activation and attenuation of innate antiviral immunity. J. Exp. Med. 2016, 213, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ma-Lauer, Y.; Carbajo-Lozoya, J.; Hein, M.Y.; Müller, M.A.; Deng, W.; Lei, J.; Meyer, B.; Kusov, Y.; von Brunn, B.; Bairad, D.R.; et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. USA 2016, 113, E5192–E5201. [Google Scholar] [CrossRef]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Ban, J.; Lee, N.R.; Lee, N.J.; Lee, J.K.; Quan, F.S.; Inn, K.S. Human Respiratory Syncytial Virus NS 1 Targets TRIM25 to Suppress RIG-I Ubiquitination and Subsequent RIG-I-Mediated Antiviral Signaling. Viruses 2018, 10, 716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzorno, A.; Dubois, J.; Machado, D.; Cartet, G.; Traversier, A.; Julien, T.; Lina, B.; Bourdon, J.C.; Rosa-Calatrava, M.; Terrier, O. Influenza A viruses alter the stability and antiviral contribution of host E3-ubiquitin ligase Mdm2 during the time-course of infection. Sci. Rep. 2018, 8, 3746. [Google Scholar] [CrossRef] [PubMed]
- Walker, F.C.; Sridhar, P.R.; Baldridge, M.T. Differential roles of interferons in innate responses to mucosal viral infections. Trends Immunol. 2021, 42, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Mazewski, C.; Perez, R.E.; Fish, E.N.; Platanias, L.C. Type I Interferon (IFN)-Regulated Activation of Canonical and Non-Canonical Signaling Pathways. Front. Immunol. 2020, 11, 606456. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 2019, 21, 1445. [Google Scholar] [CrossRef] [Green Version]
- Khaitov, M.R.; Laza-Stanca, V.; Edwards, M.R.; Walton, R.P.; Rohde, G.; Contoli, M.; Papi, A.; Stanciu, L.A.; Kotenko, S.V.; Johnston, S.L. Respiratory virus induction of alpha-, beta- and lambda-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy 2009, 64, 375–386. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Jefferies, C.A. Regulating IRFs in IFN Driven Disease. Front. Immunol. 2019, 10, 325. [Google Scholar] [CrossRef] [Green Version]
- Bouwman, W.; Verhaegh, W.; Holtzer, L.; van de Stolpe, A. Measurement of Cellular Immune Response to Viral Infection and Vaccination. Front. Immunol. 2020, 11, 575074. [Google Scholar] [CrossRef]
- Moore, E.C.; Barber, J.; Trip, R.A. Respiratory syncytial virus (RSV) attachment and nonstructural proteins modify the type I interferon response associated with suppressor of cytokine signaling (SOCS) proteins and IFN-stimulated gene-15 (ISG15). Virol. J. 2008, 5, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, D.K.; Forero, A. Mechanisms of Antiviral Immune Evasion of SARS-CoV-2. J. Mol. Biol. 2021, 22, 167265. [Google Scholar] [CrossRef] [PubMed]
- Spann, K.M.; Tran, K.C.; Collins, P.L. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 2005, 79, 5353–5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokugamage, K.G.; Hage, A.; de Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94, e01410-20. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Devaraj, S.G.; Wang, N.; Chen, Z.; Chen, Z.; Tseng, M.; Barretto, N.; Lin, R.; Peters, C.J.; Tseng, C.T.; Baker, S.C.; et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007, 282, 32208–32221. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Gielen, V.; Sykes, A.; Zhu, J.; Chan, B.; Macintyre, J.; Regamey, N.; Kieninger, E.; Gupta, A.; Shoemark, A.; Bossley, C.; et al. Increased nuclear suppressor of cytokine signaling 1 in asthmatic bronchial epithelium suppresses rhinovirus induction of innate interferons. J. Allergy Clin. Immunol. 2015, 136, 177–188.e11. [Google Scholar] [CrossRef] [Green Version]
- Pauli, E.K.; Schmolke, M.; Wolff, T.; Viemann, D.; Roth, J.; Bode, J.G.; Ludwig, S. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog. 2008, 4, e1000196. [Google Scholar] [CrossRef]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Ueki, I.F.; Min-Oo, G.; Kalinowski, A.; Ballon-Landa, E.; Lanier, L.L.; Nadel, J.A.; Koff, J.L. Respiratory virus-induced EGFR activation suppresses IRF1-dependent interferon λ and antiviral defense in airway epithelium. J. Exp. Med. 2013, 210, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Ye, L.; Schnepf, D.; Staeheli, P. Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019, 19, 614–625. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- McGillivary, G.; Jordan, Z.B.; Peeples, M.E.; Bakaletz, L.O. Replication of respiratory syncytial virus is inhibited by the host defense molecule viperin. J. Innate Immun. 2013, 5, 60–71. [Google Scholar] [CrossRef]
- Proud, D.; Turner, R.B.; Winther, B.; Wiehler, S.; Tiesman, J.P.; Reichling, T.D.; Juhlin, K.D.; Fulmer, A.W.; Ho, B.Y.; Walanski, A.A.; et al. Gene expression profiles during in vivo human rhinovirus infection: Insights into the host response. Am. J. Respir. Crit. Care Med. 2008, 178, 962–968. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.S.; Olfat, F.; Phoon, M.C.; Hsu, J.P.; Howe, J.L.C.; Seet, J.E.; Chin, K.C.; Chow, V.T.K. In vivo and in vitro studies on the antiviral activities of viperin against influenza H1N1 virus infection. J. Gen. Virol. 2012, 93, 1269–1277. [Google Scholar] [CrossRef]
- Bergmann, M.; Garcia-Sastre, A.; Carnero, E.; Pehamberger, H.; Wolff, K.; Palese, P.; Muster, T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 2000, 74, 6203–6206. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zhang, W.; Booth, J.L.; Hutchings, D.C.; Wang, X.; White, V.L.; Youness, H.; Cross, C.D.; Zou, M.H.; Burian, D.; et al. Human primary airway epithelial cells isolated from active smokers have epigenetically impaired antiviral responses. Respir. Res. 2016, 17, 111. [Google Scholar] [CrossRef] [Green Version]
- Niwa, M.; Fujisawa, T.; Mori, K.; Yamanaka, K.; Yasui, H.; Suzuki, Y.; Karayama, M.; Hozumi, H.; Furuhashi, K.; Enomoto, N.; et al. IL-17A Attenuates IFN-λ Expression by Inducing Suppressor of Cytokine Signaling Expression in Airway Epithelium. J. Immunol. 2018, 201, 2392–2402. [Google Scholar] [CrossRef] [Green Version]
- Hilzendeger, C.; da Silva, J.; Henket, M.; Schleich, F.; Corhay, J.L.; Kebadze, T.; Edwards, M.R.; Mallia, P.; Johnston, S.L.; Louis, R. Reduced sputum expression of interferon-stimulated genes in severe COPD. Int. J. Chron. Obstr. Pulmon. Dis. 2016, 11, 1485–1494. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course 881 and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, 882 retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.M.; Niikura, M.; Yang, C.W.T.; Sin, D.D. COVID-19 and COPD. Eur. Respir. J. 2020, 56, 2002108. [Google Scholar] [CrossRef] [PubMed]
- Sin, D.D. COVID-19 in COPD: A growing concern. E Clin. Med. 2020, 26, 100546. [Google Scholar] [CrossRef]
- Leung, J.M.; Yang, C.X.; Tam, A.; Shaipanich, T.; Hackett, T.L.; Singhera, G.K.; Dorscheid, D.R.; Sin, D.D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: Implications for COVID-19. Eur. Respir. J. 2020, 55, 2000688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, A.; Öberg, L.; Angermann, B.; Spalluto, C.M.; Hühn, M.; Burke, H.; Cellura, D.; Freeman, A.; Muthas, D.; Etal, D.; et al. Dysregulation of COVID-19 related gene expression in the COPD lung. Respir. Res. 2021, 22, 164. [Google Scholar] [CrossRef] [PubMed]
- Osan, J.K.; Talukdar, S.N.; Feldmann, F.; DeMontigny, B.A.; Jerome, K.; Bailey, K.L.; Feldmann, H.; Mehedi, M. Goblet Cell Hyperplasia Increases SARS-CoV-2 Infection in COPD. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ziegler, C.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef] [PubMed]
- Aboudounya, M.M.; Heads, R.J. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediat. Inflamm. 2021. [Google Scholar] [CrossRef] [PubMed]
- Taefehshokr, N.; Taefehshokr, S.; Hemmat, N.; Heit, B. COVID-19: Perspectives on Innate Immune Evasion. Front. Immunol. 2020, 11, 580641. [Google Scholar] [CrossRef]
- Kasuga, Y.; Zhu, B.; Jang, K.J.; Yoo, J.S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.; Chan, J.F.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef]
- Freitas, B.T.; Durie, I.A.; Murray, J.; Longo, J.E.; Miller, H.C.; Crich, D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020, 6, 2099–2109. [Google Scholar] [CrossRef]
- Liu, G.; Lee, J.H.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; van Gent, M.; Riedl, W.; Davis-Gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef]
- Moustaqil, M.; Ollivier, E.; Chiu, H.-P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infec. 2021, 10, 178–195. [Google Scholar] [CrossRef]
- Anwar, M.A.; Shah, M.; Kim, J.; Choi, S. Recent clinical trends in toll-like receptor targeting therapeutics. Med. Res. Rev. 2019, 39, 1053–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomerenke, A.; Lea, S.R.; Herrick, S.; Lindsay, M.A.; Singh, D. Characterization of TLR-induced inflammatory responses in COPD and control lung tissue explants. Int. J. Chron. Obstr. Pulmon. Dis. 2016, 11, 2409–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djukanović, R.; Harrison, T.; Johnston, S.L.; Gabbay, F.; Wark, P.; Thomson, N.C.; Niven, R.; Singh, D.; Reddel, H.K.; Davies, D.E.; et al. The effect of inhaled IFN-β on worsening of asthma symptoms caused by viral infections. A randomized trial. Am. J. Respir. Crit. Care Med. 2014, 190, 145–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolb-Mäurer, A.; Goebeler, M.; Mäurer, M. Cutaneous Adverse Events Associated with Interferon-β Treatment of Multiple Sclerosis. Int. J. Mol. Sci. 2015, 16, 14951–14960. [Google Scholar] [CrossRef] [Green Version]
- Watson, A.; Spalluto, C.M.; McCrae, C.; Cellura, D.; Burke, H.; Cunoosamy, D.; Freeman, A.; Hicks, A.; Hühn, M.; Ostridge, K.; et al. Dynamics of IFN-β Responses during Respiratory Viral Infection. Insights for Therapeutic Strategies. Am. J. Respir. Crit. Care Med. 2020, 201, 83–94. [Google Scholar] [CrossRef]
- McCrae, C.; Olsson, M.; Gustafson, P.; Malmgren, A.; Aurell, M.; Fagerås, M.; Da Silva, C.A.; Cavallin, A.; Paraskos, J.; Karlsson, K.; et al. INEXAS: A Phase 2 Randomized Trial of On-demand Inhaled Interferon Beta-1a in Severe Asthmatics. Clin. Exp. Allergy 2021, 51, 273–283. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).