Long Non-Coding RNA Expression in Laser Micro-Dissected Luminal A and Triple Negative Breast Cancer Tissue Samples—A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients’ Characteristics and Tumor Samples
2.2. Laser Capture Microdissection (LCM)
2.3. RNA Extraction
2.4. LncRNA Profiling in FFPE Tissues
2.5. LncRNA Validation
2.6. Cell Lines
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Ethics Approval
Consent to Participate and for Publication
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network., Genome sequencing centres: Washington University in St Louis., Koboldt, D.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70.
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blows, F.M.; Driver, K.E.; Schmidt, M.K.; Broeks, A.; van Leeuwen, F.E.; Wesseling, J.; Cheang, M.C.; Gelmon, K.; Nielsen, T.O.; Blomqvist, C.; et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010, 7, e1000279. [Google Scholar] [CrossRef] [PubMed]
- Almarzooq, R.; Alrayes, A.; Alaradi, H.; Abdulla, H. Molecular subtypes of breast cancer. Bahrain Med Bull. 2018, 40, 222–225. [Google Scholar]
- Klopfleisch, R.; Weiss, A.T.; Gruber, A.D. Excavation of aburied treasure DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol. Histopathol. 2011, 26, 797–810. [Google Scholar]
- Mathias, C.; Zambalde, E.P.; Rask, P.; Gradia, D.F.; de Oliveira, J.C. Long non-coding RNAs differential expression in breast cancer subtypes: What do we know? Clin. Genet. 2019, 95, 558–568. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grander, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [Green Version]
- Fu, P.F.; Zheng, X.; Fan, X.; Lin, A. Role of cytoplasmic lncRNAs in regulating cancer signaling pathways. J. Zhejiang Univ. Sci. B 2019, 20, 1–8. [Google Scholar] [CrossRef]
- Chen, C.; Li, Z.; Yang, Y.; Xiang, T.; Song, W.; Liu, S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol. Ther. 2015, 16, 856–865. [Google Scholar] [CrossRef] [Green Version]
- Augoff, K.; Mccue, B.; Plow, E.F.; Sossey-Alaoui, K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 2012, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Merry, C.R.; McMahon, S.; Forrest, M.E.; Bartels, C.F.; Saiakhova, A.; Bartel, C.A.; Scacheri, P.C.; Thompson, C.L.; Jackson, M.W.; Harris, L.N.; et al. Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer. Oncotarget 2016, 7, 53230–53244. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Wulfkuhle, J.D.; Calvert, V.S.; VanMeter, A.; Zhou, W.; Coukos, G.; Geho, D.H.; Petricoin, E.F., III; Liotta, L.A. Laser-capture microdissection. Nat. Protoc. 2006, 1, 586–603. [Google Scholar] [CrossRef] [PubMed]
- Nariţa, D.; Anghel, A.; Seclaman, E.; Ilina, R.; Cireap, N.; Ursoniu, S. Molecular profiling of ADAM12 gene in breast cancers. Rom. J. Morphol. Embryol. 2010, 51, 669–676. [Google Scholar]
- Seclaman, E.; Narita, D.; Anghel, A.; Cireap, N.; Ilina, R.; Sirbu, I.O.; Marian, C. MicroRNA Expression in Laser Micro-dissected Breast Cancer Tissue Samples—A Pilot Study. Pathol. Oncol. Res. 2019, 25, 233–239. [Google Scholar] [CrossRef]
- Mihala, A.; Alexa, A.A.; Samoilă, C.; Dema, A.; Vizitiu, A.C.; Anghel, A.; Tămaş, L.; Marian, C.V.; Sîrbu, I.O. A pilot study on the expression of microRNAs resident on chromosome 21 in laser microdissected FFPE prostate adenocarcinoma samples. Rom. J. Morphol. Embryol. 2015, 56, 1063–1068. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget 2017, 8, 44917–44943. [Google Scholar] [CrossRef] [Green Version]
- Amaral, P.P.; Neyt, C.; Wilkins, S.J.; Askarian-Amiri, M.E.; Sunkin, S.M.; Perkins, A.C.; Mattick, J.S. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 2009, 15, 2013–2027. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Wang, G.; Zhou, C.F.; Zhang, H.B.; Sun, H.; Zhang, W.; Zhou, H.-H.; Liu, R.; Zhu, Y.-S. LncRNA Profile Study Reveals a Three-LncRNA Signature Associated with the Pathological Complete Response Following Neoadjuvant Chemotherapy in Breast Cancer. Front. Pharmacol. 2019, 10, 574. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, J.; Wang, Z.; Wang, P.; Gao, X.; Wang, Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed. Pharmacother. 2018, 104, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res. Treat. 2014, 145, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.Y.; Siu, M.T.; Ho, C.W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Zhou, Y.; Sun, A.J.; Xue, J.L. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J. Cell Physiol. 2018, 233, 8558–8566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, W.B.; Wang, Z.W.; Wang, X.H. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1020–1026. [Google Scholar]
- Pickard, M.R.; Williams, G.T. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget 2016, 7, 10104–10116. [Google Scholar] [CrossRef] [Green Version]
- Idogawa, M.; Ohashi, T.; Sasaki, Y.; Nakase, H.; Tokino, T. Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int. J. Cancer 2017, 140, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, S.; Li, Z.; Long, X.; Guo, Z.; Zhang, G.; Zu, J.; Chen, Y.; Wen, L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int. J. Biol. Macromol. 2017, 105, 346–353. [Google Scholar] [CrossRef]
- Li, X.; Deng, X.; Pang, X. LncRNA NEAT1 Silenced miR-133b Promotes Migration and Invasion of Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3616. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Rao, Y.; Dong, W. Quantitative analysis of lncRNA from human FF and FFPE brain speimens. FASEB J. 2018, 32, 525.1. [Google Scholar]
- Lv, Y.; Li, S.; Li, Z.; Tao, R.; Shao, Y.; Chen, Y. Quantitative analysis of noncoding RNA from paired fresh and formalin-fixed paraffin-embedded brain tissues. Int. J. Legal Med. 2020, 134, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Iddawela, M.; Rueda, O.M.; Klarqvist, M.; Graf, S.; Earl, H.M.; Caldas, C. Reliable gene expression profiling of formalin-fixed paraffin-embedded breast cancer tissue (FFPE) using cDNA-mediated annealing, extension, selection, and ligation whole-genome (DASL WG) assay. BMC Med. Genom. 2016, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Lehmann, B.D.; Shyr, Y.; Guo, Y. The Utilization of Formalin Fixed-Paraffin-Embedded Specimens in High Throughput Genomic Studies. Int. J. Genom. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, B.; Sheng, Q.; Seitz, R.S.; Lawrence, K.D.; Morris, S.W.; Thomas, L.R.; Hout, D.R.; Schweitzer, B.L.; Guo, Y.; Pietenpol, J.A.; et al. Comparison of triple-negative breast cancer molecular subtyping using RNA from matched fresh-frozen versus formalin-fixed paraffin-embedded tissue. BMC Cancer 2017, 17, 1–14. [Google Scholar]
| Characteristics | TNBC N (%) | Luminal A N (%) | |
|---|---|---|---|
| Age | >50 | 7 (77.78) | 8 (88.89) |
| <50 | 2 (22.22) | 1 (11.11) | |
| BMI | Normal | 5 (55.56) | 5 (55.56) |
| Obese | 4 (44.44) | 4 (44.44) | |
| Stage | I and II | 5 (55.56) | 4 (44.44) |
| III | 4 (44.44) | 5 (55.56) | |
| Tumor size | <5 cm | 5 (55.56) | 6 (66.67) |
| >5 cm | 4 (44.44) | 3 (33.33) | |
| Lymph node involvement | Yes | 5 (55.56) | 4 (44.44) * |
| No | 4 (44.44) | 3 (33.33) * | |
| HER2 | Positive | 5 (55.56) | 5 (55.56) |
| Negative | 4 (44.44) | 4 (44.44) | |
| Ki-67 | Positive | 9 (100.00) | 9 (100.00) |
| Negative | 0 (0.00) | 0 (0.00) | |
| ER | Positive | 0 (0.00) | 9 (100.00) |
| Negative | 9 (100.00) | 0 (0.00) | |
| PR | Positive | 0 (0.00) | 9 (100.00) |
| Negative | 9 (100.00) | 0 (0.00) |
| LncRNA Name | Fold Change | p Value | LncRNA Name | Fold Change | p Value |
|---|---|---|---|---|---|
| BCYRN1 | 0.967 | 0.457 | MEG3 | 5.394 | 0.605 |
| BDNF-AS | 0.894 | 0.469 | NEAT1 | 5.818 | 0.548 |
| DISC2 | 0.624 | 0.325 | OTX2-AS1 | 205.222 | 0.029 |
| EGOT | 1.634 | 0.860 | PANDAR | 1.101 | 0.236 |
| FTX | 13.518 | 0.456 | PTENP1-AS | 0.989 | 0.708 |
| GACAT1 | 1.347 | 0.728 | SNHG16 | 1.115 | 0.614 |
| GAS5 | 1.734 | 0.914 | SOX2-OT | 0.020 | 0.042 |
| H19 | 1.163 | 0.764 | ST7-AS2 | 2.206 | 0.956 |
| HEIH | 1.388 | 0.702 | TERC | 1.652 | 0.812 |
| HOTAIR | 5.771 | 0.543 | TMEM161B-AS1 | 0.909 | 0.474 |
| IPW | 1.163 | 0.656 | TRERNA1 | 0.444 | 0.372 |
| JPX | 1.014 | 0.512 | TUG1 | 1.453 | 0.814 |
| KCNQ1OT1 | 4.028 | 0.690 | UCA1 | 0.386 | 0.601 |
| FALEC | 1.204 | 0.633 | XIST | 0.769 | 0.316 |
| LINC-ROR | 0.624 | 0.174 | ZFAS1 | 1.680 | 0.868 |
| MALAT1 | 1.763 | 0.936 |
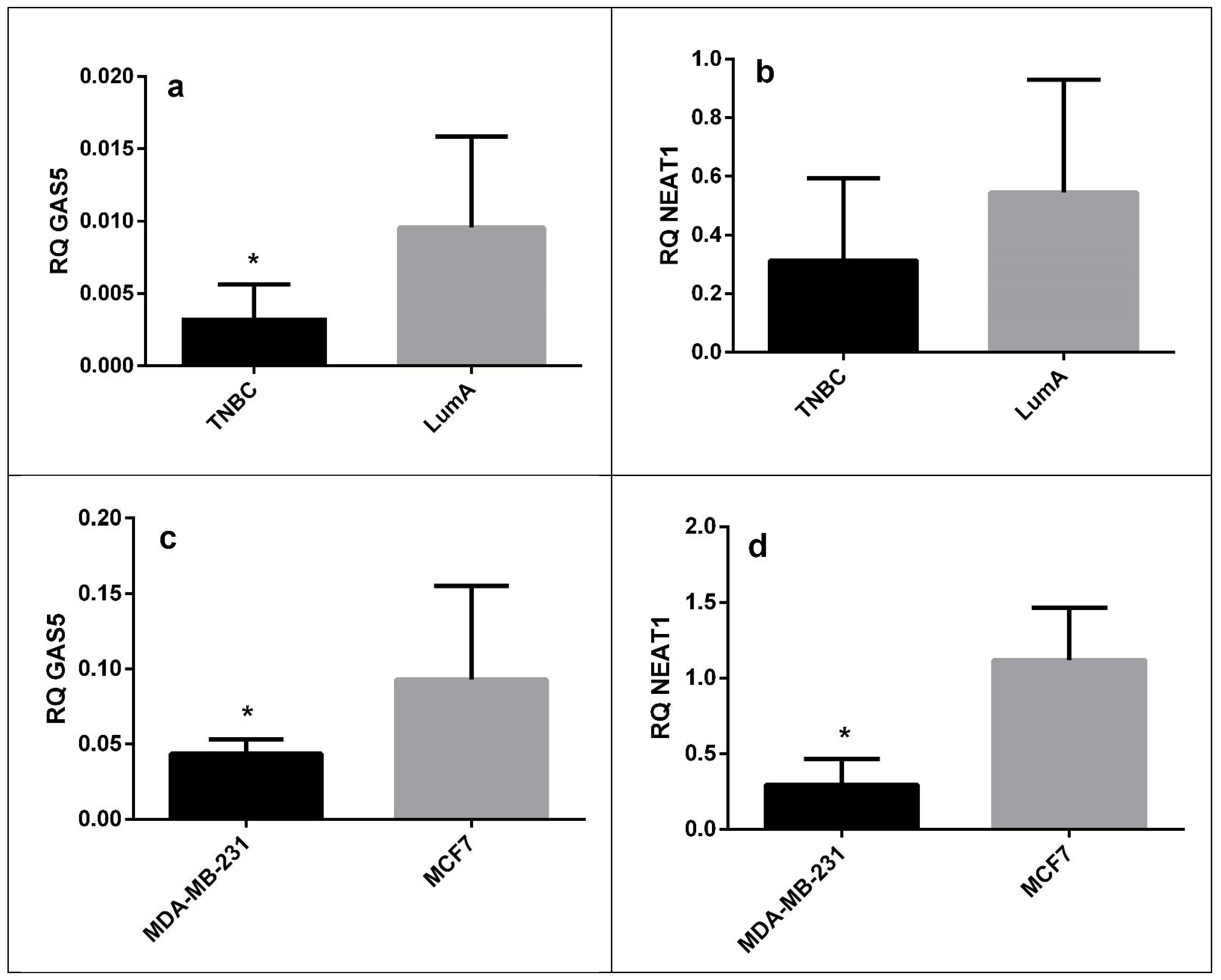
| LncRNA | Sample Type | Fold Change | p Value |
|---|---|---|---|
| GAS5 | FFPE tissue | 0.33 | 0.022 |
| Cell lines | 0.52 | 0.035 | |
| NEAT1 | FFPE tissue | 0.66 | 0.27 |
| Cell lines | 0.23 | 0.035 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcu, A.; Nitusca, D.; Vaduva, A.; Baderca, F.; Cireap, N.; Coricovac, D.; Dehelean, C.A.; Seclaman, E.; Ilina, R.; Marian, C. Long Non-Coding RNA Expression in Laser Micro-Dissected Luminal A and Triple Negative Breast Cancer Tissue Samples—A Pilot Study. Medicina 2021, 57, 371. https://doi.org/10.3390/medicina57040371
Marcu A, Nitusca D, Vaduva A, Baderca F, Cireap N, Coricovac D, Dehelean CA, Seclaman E, Ilina R, Marian C. Long Non-Coding RNA Expression in Laser Micro-Dissected Luminal A and Triple Negative Breast Cancer Tissue Samples—A Pilot Study. Medicina. 2021; 57(4):371. https://doi.org/10.3390/medicina57040371
Chicago/Turabian StyleMarcu, Anca, Diana Nitusca, Adrian Vaduva, Flavia Baderca, Natalia Cireap, Dorina Coricovac, Cristina Adriana Dehelean, Edward Seclaman, Razvan Ilina, and Catalin Marian. 2021. "Long Non-Coding RNA Expression in Laser Micro-Dissected Luminal A and Triple Negative Breast Cancer Tissue Samples—A Pilot Study" Medicina 57, no. 4: 371. https://doi.org/10.3390/medicina57040371
APA StyleMarcu, A., Nitusca, D., Vaduva, A., Baderca, F., Cireap, N., Coricovac, D., Dehelean, C. A., Seclaman, E., Ilina, R., & Marian, C. (2021). Long Non-Coding RNA Expression in Laser Micro-Dissected Luminal A and Triple Negative Breast Cancer Tissue Samples—A Pilot Study. Medicina, 57(4), 371. https://doi.org/10.3390/medicina57040371








