Automated Detection of Sudden Cardiac Death by Discrete Wavelet Transform of Electrocardiogram Signal
Abstract
:1. Introduction
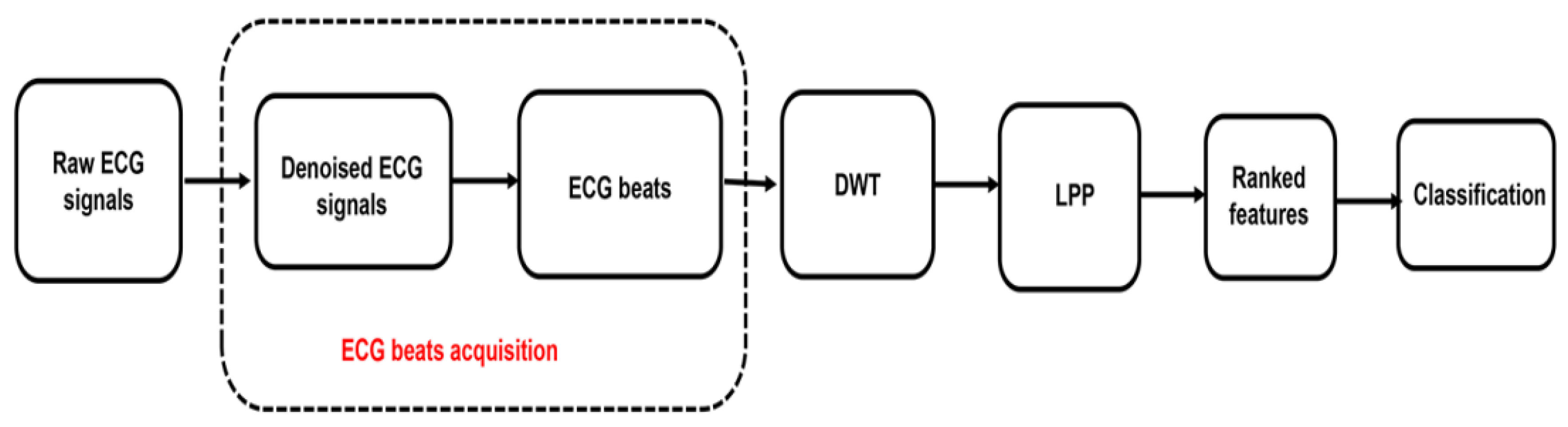
2. Materials and Methods
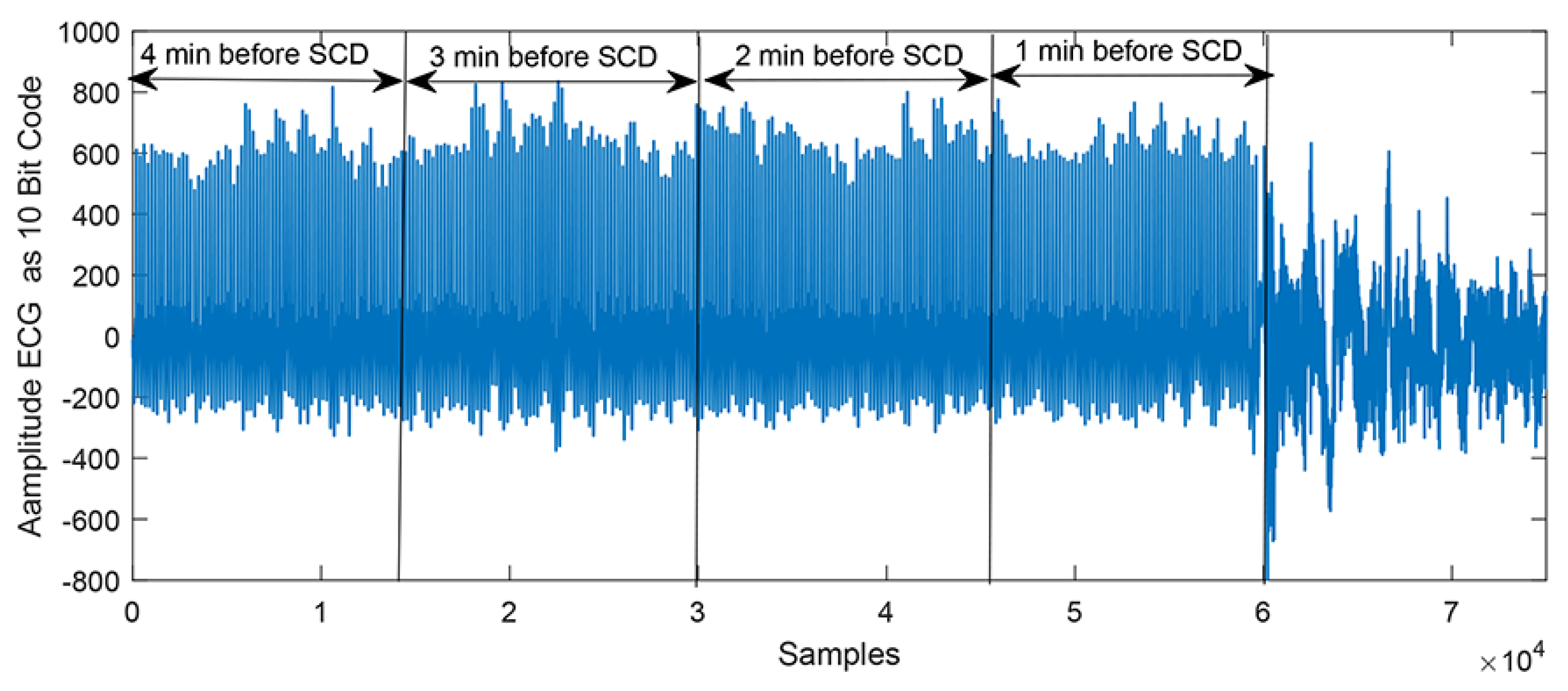
2.1. Data Acquisition
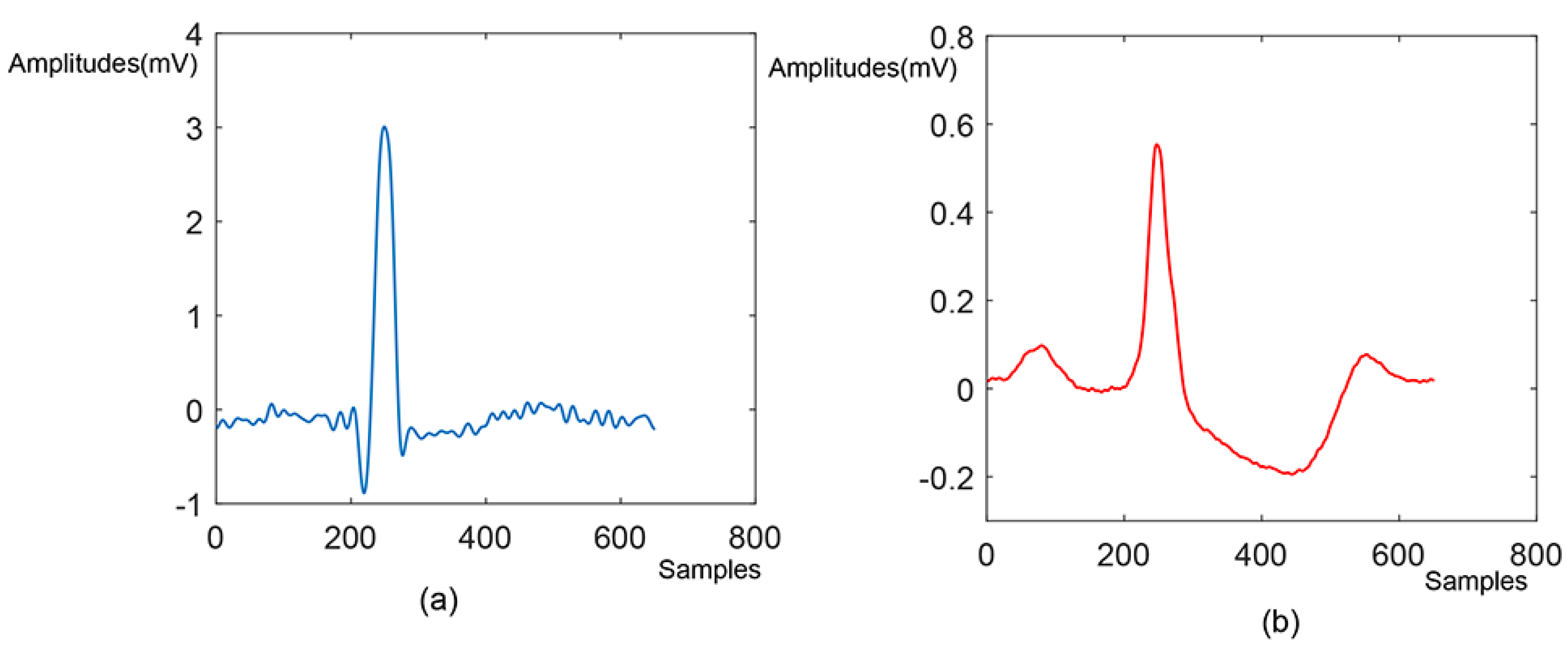
2.2. Pre-Processing and Beats Segmentation
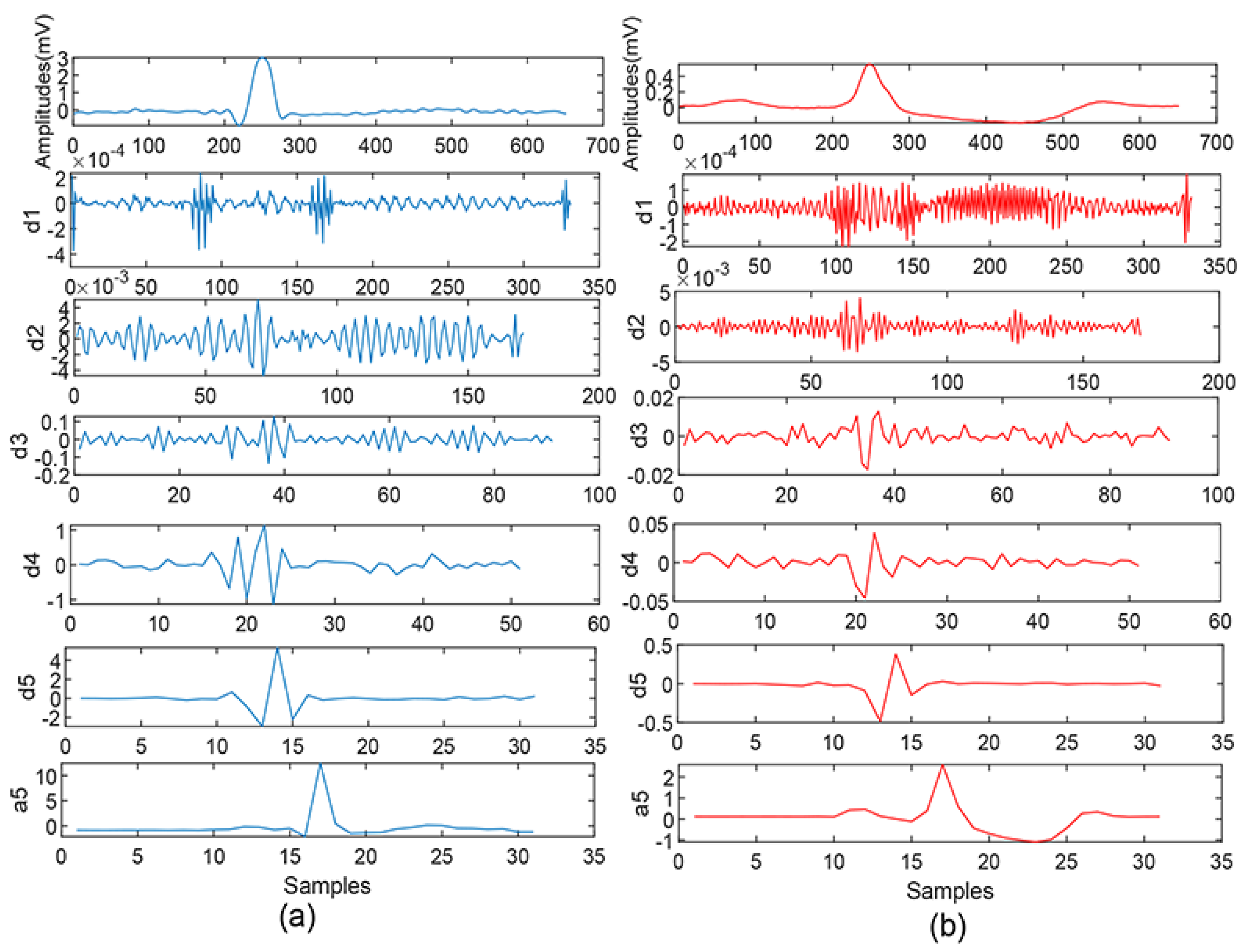
2.3. Feature Extraction-Discrete Wavelet Transformation
2.4. Features Reduction—Locality Preserving Projections (LPP)
2.5. Feature Ranking
2.6. Classification
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fujita, H.; Acharya, U.R.; Sudarshan, V.K.; Ghista, D.N.; Sree, S.V. Sudden cardiac death (SCD) prediction based on nonlinear heart rate variability features and SCD index. Appl. Soft Comput. 2016, 43, 510–519. [Google Scholar] [CrossRef]
- Ebrahimzadeh, E.; Pooyan, M.; Bijar, A. A Novel Approach to Predict Sudden Cardiac Death (SCD) Using Nonlinear and Time-Frequency Analyses from HRV Signals. PLoS ONE 2014, 9, e81896. [Google Scholar] [CrossRef]
- Nichols, M.; Townsend, N.; Scarborough, P.; Rayner, M. Cardiovascular disease in Europe: Epidemiological update. Eur. Heart J. 2013, 34, 3028–3034. [Google Scholar] [CrossRef]
- Pagidipati, N.J.; Gaziano, T.A. Estimating Deaths from Cardiovascular Disease: A Review of Global Methodologies of Mortality Measurement. Circulation 2013, 127, 749–756. [Google Scholar] [CrossRef]
- Passman, R.; Goldberger, J.J. Predicting the future: Risk stratification for sudden cardiac death in patients with left ventricular dysfunction. Circulation 2012, 125, 3031–3037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, T.W.; Shen, H.P.; Lin, C.; Ou, Y.L. Detection and Prediction of Sudden Cardiac Death (SCD) For Personal Healthcare. In Proceedings of the 29th Annual International Conference of the IEEE, Lyon, France, 26–29 March 2007; Volume 21, pp. 2575–2578. [Google Scholar]
- Kahney, E.W.; Ranjan, R.; Gleason, R.J.; Chen, X. Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harb. Symp. Quant. Biol. 2018, 82, 305–318. [Google Scholar] [CrossRef] [Green Version]
- Acharya, U.R.; Fujita, H.; Shu, L.O.; Hagiwara, Y.; Adam, M. Application of Deep Convolutional Neural Network for Automated Detection of Myocardial Infarction Using ECG Signals. Inf. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Adam, M.; Lih, O.S.; Tan, R.S. Automated characterization and classification of coronary artery disease and myocardial infarction by decomposition of ECG signals: A comparative study. Inf. Sci. 2017, 377, 17–29. [Google Scholar] [CrossRef]
- Huikuri, H.V.; Makikallio, T.H.; Raatikainen, M.J.; Perkiomaki, J. Prediction of sudden cardiac death: Appraisal of the studies and methods assessing the risk of sudden arrhythmic death. Circulation 2003, 108, 110–115. [Google Scholar] [CrossRef]
- VanHoogenhuyze, D.; Martin, G.; Weiss, J.; Schaad, J.; Singer, D. Spectrum of heart rate variability. Proc. Comput. Cardiol 1989. [Google Scholar]
- Ebrahimzadeh, E.; Pooyan, M. Early detection of sudden cardiac death by using classical linear techniques and time-frequency methods on electrocardiogram signals. Biomed. Sci. Eng. 2011, 11, 699–706. [Google Scholar] [CrossRef] [Green Version]
- Voss, A.; Kurths, J.; Kleiner, H.J.; Witt, A. The application of methods of non-linear dynamics for the improved and predictive recognition of patients threatened by sudden cardiac death. Cardiovasc. Res. 1996, 31, 419–433. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Vidya, K.S.; Ghista, D.N.; Lim, W.J.E.; Koh, J.E.W. Automated prediction of sudden cardiac death risk using kolmogorov complexity and recurrence quantification analysis features extracted from HRV signals. In Proceedings of the 2015 IEEE International Conference on Systems, Man, and Cybernetics, Hongkong, China, 9–12 October 2015. [Google Scholar]
- Shi, M.; Zhan, C.; He, H.; Jin, Y.; Wu, R.; Sun, Y.; Shen, B. Renyi distribution entropy analysis of short-term heart rate variability signals and its application in coronary artery disease detection. Front. Physiol. 2019, 10, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murukesan, L.; Murugappan, M.; Iqbal, M.; Saravanan, K. Machine Learning Approach for Sudden Cardiac Arrest Prediction Based on Optimal Heart Rate Variability Features. J. Med. Imaging Health Inform. 2014, 4, 521–532. [Google Scholar] [CrossRef]
- Mirhoseini, S.R.; Jahedmotlagh, M.R.; Pooyan, M. Improve Accuracy of Early Detection Sudden Cardiac Deaths (SCD) Using Decision Forest and SVM. In Proceedings of the International Conference on Robotics and Artificial Intelligence (ICRAI2016), Los Angeles, CA, USA, 20–22 April 2016. [Google Scholar]
- Ebrahimzadeh, E.; Manuchehri, M.S.; Amoozegar, S. A time local subset feature selection for prediction of sudden cardiac death from ECG signal. Med. Biol. Eng. Comput. 2018, 56, 1253–1270. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimzadeh, E.; Foroutan, A.; Shams, M.; Baradaran, R.; Rajabion, L.; Joulani, M.; Fayaz, F. An optimal strategy for prediction of sudden cardiac death through a pioneering feature-selection approach from HRV signal. Comput. Methods Programs Biomed. 2019, 169, 19–36. [Google Scholar] [CrossRef]
- Shi, M.; He, H.; Geng, W.; Wu, R.; Zhan, C.; Jin, Y.; Zhu, F.; Ren, S.; Shen, B. Early Detection of Sudden Cardiac Death by Using Ensemble Empirical Mode Decomposition-Based Entropy and Classical Linear Features From Heart Rate Variability Signals. Front. Physiol. 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Sree, V.S.; Eugene, L.W.J.; Ghista, D.N.; Tan, R.S. An integrated index for detection of Sudden Cardiac Death using Discrete Wavelet Transform and nonlinear features. Knowl.-Based Syst. 2015, 83, 149–158. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 2007, 3, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Chiradeja, P.; Ngaopitakkul, A. Classification of Lightning and Faults in Transmission Line Systems Using Discrete Wavelet Transform. Math. Probl. Eng. 2018, 2018, 1847968. [Google Scholar] [CrossRef]
- Yan, L.; Liu, Y.; Liu, Y. Application of Discrete Wavelet Transform in Shapelet-Based Classification. Math. Probl. Eng. 2020, 1, 6523872. [Google Scholar] [CrossRef]
- Liu, X.; Lin, S.; Fan, H. Face recognition algorithm based on Gabor wavelet and locality preserving projections. Mod. Phys. Lett. B 2017, 31, 1740041. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Wang, R.; Nie, F.; Wang, F.; Yu, Q.; Yang, X. An improved locality preserving projection with l(1)-norm minimization for dimensionality reduction. Neurocomputing 2018, 316, 322–331. [Google Scholar] [CrossRef]
- Kopra, A.R.; Lawandow, A.; Sabelli, H. Asymmetry, Symmetry and Beauty. Symmetry 2010, 2, 1591–1624. [Google Scholar]
- Desai, K.D.; Sankhe, M.S. A real-time fetal ECG feature extraction using multiscale discrete wavelet transform. In Proceedings of the International Conference on Biomedical Engineering & Informatics, Chongqing, China, 16–18 October 2012. [Google Scholar]
- Guzzetti, S.; Rovere, M.; Pinna, G.D.; Maestri, R.; Malliani, A. Different spectral components of 24 h heart rate variability are related to different modes of death in chronic heart failure. Eur. Heart J. 2005, 26, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, C.; Li, K. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med. Biol. Eng. Comput. 2015, 53, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Shen, L.; Sun, H.; Shen, B. Physiological Informatics: Collection and Analyses of Data from Wearable Sensors and Smartphone for Healthcare. Adv. Exp. Med. Biol. 2017, 1028, 17–37. [Google Scholar]
| Ref. | Data | Method | Classification | Cross Validation | Accuracy |
|---|---|---|---|---|---|
| Early SCD Detection Using HRV Signals | |||||
| [6] | No. of data: 23 SCD, 20 normal Source: the MIT/BIH SCD ans Normal Sinus Rhythm (NSR) databases Prediction time resolution: 2 min interval | Wavelet analysis and TF domain method | Artificial neural networks (ANN); Back propagation (BP) | Not mentioned | 87.5% (2 min before) |
| [12] | No. of data: 35 SCD, 35 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain, and nonlinear methods (poincaré plot (PP), Detrended fluctuation analysis (DFA)) | KNN; Multilayer perceptron (MLP) | Not mentioned | 91.23% (2 min before) |
| [2] | No. of data: 35 SCD, 35 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain, and nonlinear methods (PP, DFA) | KNN; Multilayer Perceptron Neural Network (PNN) | Leave one out cross-validation | 83.93% (4 min before) |
| [16] | No. of data: 23 SCD, 18 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain, and nonlinear methods (PP, DFA) | SVM, PNN | 10-fold cross-validation | 96.36% (2 min before) |
| [14] | No. of data: 40 SCD, 36 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Recurrence Quantification Analysis (RQA), and Kolmogorov, complexity parameters | SVM, KNN, PNN, DT | 10-fold cross-validation | 86.8% (4 min before) |
| [17] | No. of data: 19 SCD and18 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain, and nonlinear methods (PP) | SVM | 10-fold cross-validation | 83.24% (1 min before) |
| [1] | No. of data:20 SCD and18 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Wavelet transform, nonlinear methods (Renyi entropy (REn), fuzzy entropy (FuEn), Hjorth’s parameters, Tsallis entropy) | DT, KNN, SVM | 10-fold cross-validation | 94.7% (4 min before) |
| [18] | No. of data: 35 SCD, 35 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain and, nonlinear methods (PP, DFA) | MLP | Leave one out cross-validation | 95.23% (4 min before) 83.88% (12 min before) |
| [19] | No. of data: 35 SCD, 35 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | Linear (time, frequency domain), TF domain, and nonlinear methods (PP, DFA) | MLP, SVM, KNN | Not mentioned | 95.24% (4 min before) 84.28% (13 min before) |
| [20] | No. of data: 40 SCD, 36 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | EEMD, linear (time, frequency domain), TF domain, and nonlinear methods (REn, FuEn, dispersion entropy, Renyi distribution entropy (RdisEn), and improved multiscale permutation entropy | KNN | 10-fold cross-validation | 94.7% (4 min before) 96.1% (14 min before) |
| Early SCD detection using ECG signals | |||||
| [21] | No. of data: 40 SCD, 36 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | DWT, Nonlinear methods (Fractal Dimension (FD), DFA Hurst’s exponent (H), Sample Entropy, Approximate Entropy, and Correlation Dimension (CD)) | KNN, SVM, DT | 10-fold cross-validation | 92.11% (4 min before) |
| Current study | No. of data: 40 SCD, 36 normal Source: the MIT/BIH SCD ans NSR databases Prediction time resolution: 1 min interval | DWT and LPP | KNN, SVM, DT | 10-fold cross-validation | 98.1% (4 min before) 97.6% (14 min before) |
| Case | Number of Beats | |
|---|---|---|
| SCD | Normal | |
| 1st 1-min | 2975 | 2934 |
| 2nd 1-min | 2849 | 2938 |
| 3rd 1-min | 2925 | 3036 |
| 4th 1-min | 3112 | 3021 |
| 5th 1-min | 3013 | 3100 |
| 6th 1-min | 2923 | 3052 |
| 7th 1-min | 3001 | 3043 |
| 8th 1-min | 3038 | 2961 |
| 9th 1-min | 2908 | 3032 |
| 10th 1-min | 2904 | 3135 |
| 11th 1-min | 2907 | 3175 |
| 12th 1-min | 2758 | 3147 |
| 13th 1-min | 2865 | 3062 |
| 14th 1-min | 2861 | 3026 |
| Feature | Normal | SCD | p-Value | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
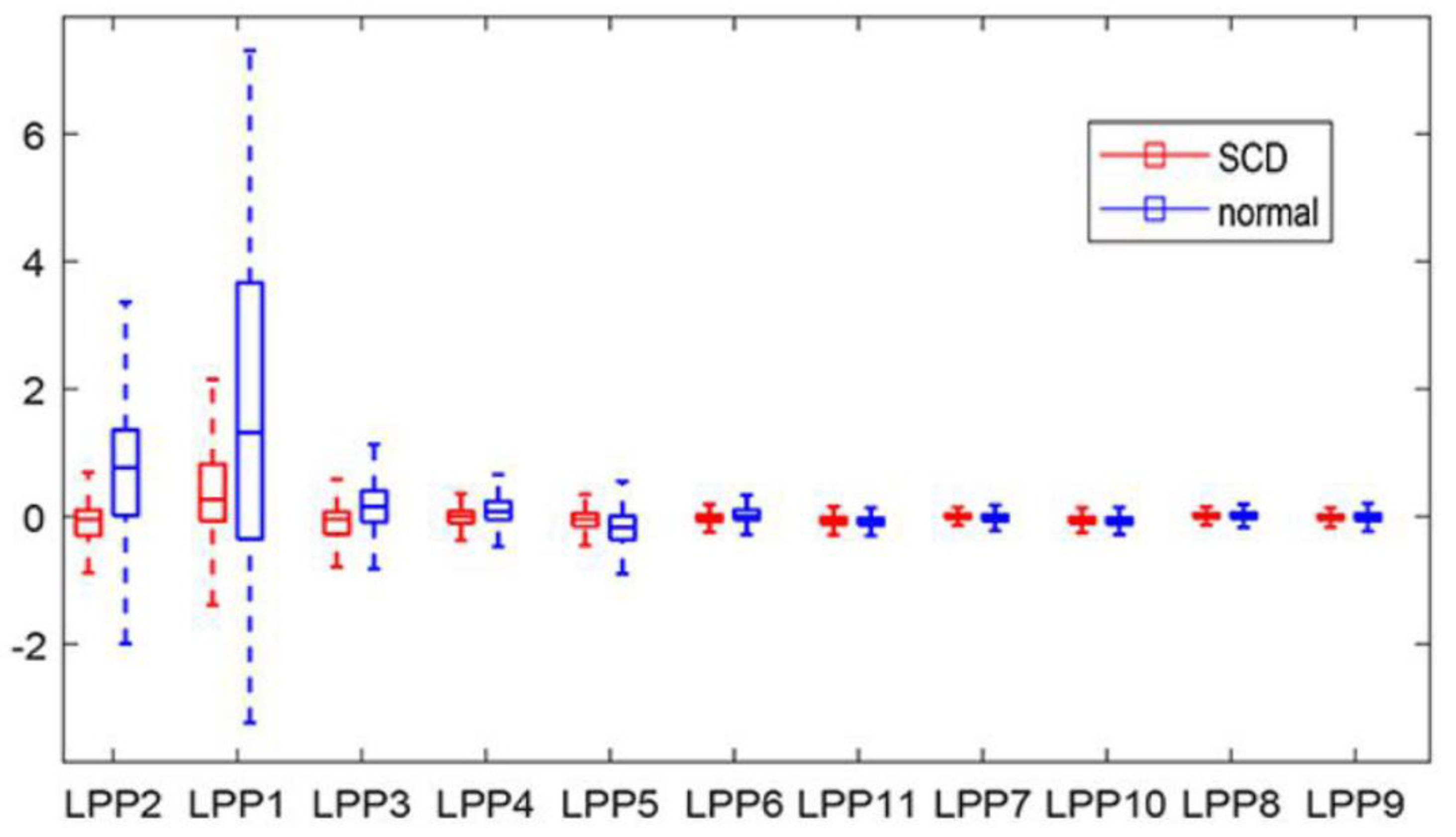
| LPP2 | −0.032 | 0.475 | 0.749 | 1.307 | 5.52 × 10−199 |
| LPP1 | 0.443 | 0.843 | 1.567 | 2.457 | 1.05 × 10−122 |
| LPP3 | −0.118 | 0.294 | 0.104 | 0.539 | 4.04 × 10−87 |
| LPP4 | −0.072 | 0.444 | 0.082 | 0.257 | 3.33 × 10−60 |
| LPP5 | −0.070 | 0.330 | −0.164 | 0.321 | 3.57 × 10−29 |
| LPP6 | −0.010 | 0.179 | 0.043 | 0.259 | 2.00 × 10−20 |
| LPP11 | −0.060 | 0.172 | −0.085 | 0.127 | 4.88 × 10−10 |
| LPP7 | 0.011 | 0.379 | −0.020 | 0.146 | 2.32 × 10−5 |
| LPP10 | −0.061 | 0.216 | −0.071 | 0.119 | 0.032 |
| LPP8 | 0.019 | 0.350 | 0.012 | 0.115 | 0.293 |
| LPP9 | −0.011 | 0.236 | −0.011 | 0.149 | 0.934 |
| Classifier | Features | TP | TN | FP | FN | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
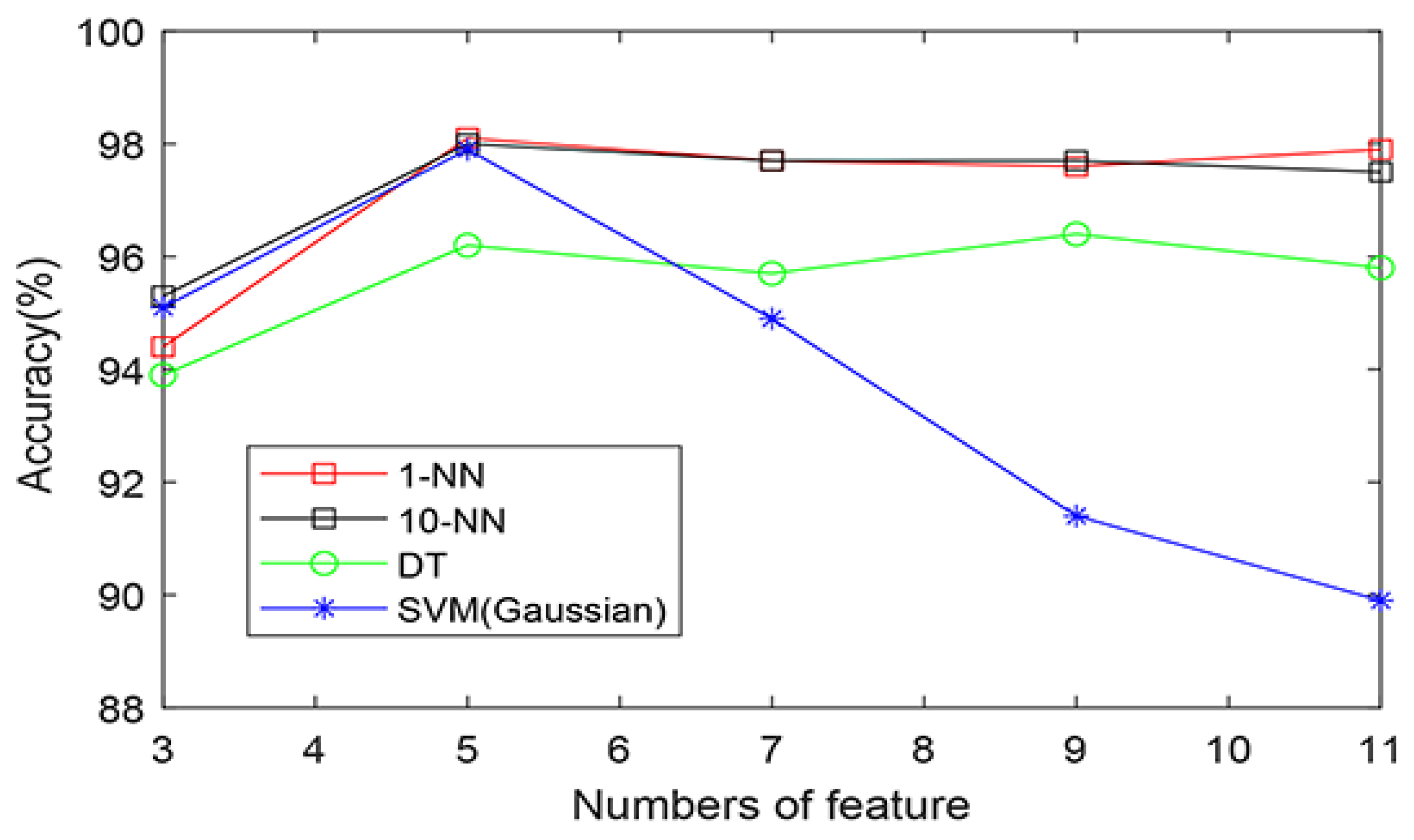
| KNN (1) | 5 | 3054 | 2962 | 58 | 59 | 98.1% | 98.1% | 98.0% |
| KNN (10) | 5 | 3050 | 2959 | 62 | 62 | 98.0% | 98.0% | 97.9% |
| SVM(Gaussian) | 5 | 3043 | 2960 | 69 | 61 | 97.9% | 97.8% | 98.0% |
| DT | 8 | 3012 | 2904 | 100 | 117 | 96.5% | 96.8% | 96.1% |
| Ranking Method | Features | TP | TN | FP | FN | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| t-test | 5 | 3054 | 2962 | 58 | 59 | 98.1% | 98.1% | 98.0% |
| Bhattacharyya | 5 | 3042 | 2838 | 70 | 83 | 97.5% | 97.8% | 97.2% |
| Wilcoxon | 8 | 3059 | 2945 | 53 | 76 | 97.9% | 98.3% | 97.5% |
| entropy | 6 | 3050 | 2941 | 62 | 80 | 97.7% | 98.0% | 97.4% |
| Wavelet Basis | Features | TP | TN | FP | FN | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| db1 | 7 | 3062 | 2944 | 50 | 77 | 97.9% | 98.4% | 97.5% |
| db2 | 12 | 3067 | 2942 | 45 | 79 | 98.0% | 98.6% | 97.4% |
| db3 | 12 | 3058 | 2954 | 54 | 69 | 98.0% | 98.3% | 97.7% |
| db4 | 9 | 3020 | 2911 | 92 | 110 | 96.7% | 97.0% | 96.4% |
| db5 | 7 | 3053 | 2940 | 59 | 81 | 97.7% | 98.1% | 97.3% |
| db6 | 5 | 3054 | 2962 | 58 | 59 | 98.1% | 98.1% | 98.0% |
| haar | 8 | 2976 | 2862 | 136 | 159 | 95.2% | 95.6% | 94.7% |
| coif1 | 7 | 3058 | 2954 | 54 | 69 | 98.0% | 98.3% | 97.7% |
| coif2 | 5 | 3046 | 2955 | 66 | 66 | 97.8% | 97.9% | 97.8% |
| coif3 | 10 | 3041 | 2920 | 71 | 101 | 97.2% | 97.7% | 96.7% |
| Case | Features | TP | TN | FP | FN | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| The 1st 1-min | 9 | 2875 | 2816 | 100 | 118 | 96.3% | 96.6% | 96.0% |
| The 2nd 1-min | 7 | 2771 | 2814 | 78 | 124 | 96.5% | 97.3% | 95.8% |
| The 3rd 1-min | 7 | 2848 | 2915 | 77 | 121 | 96.7% | 97.4% | 96.0% |
| The 4th 1-min | 5 | 3054 | 2962 | 58 | 59 | 98.1% | 98.1% | 98.0% |
| The 5th 1-min | 6 | 2957 | 3009 | 56 | 91 | 97.6% | 98.1% | 97.1% |
| The 6th 1-min | 4 | 2797 | 2876 | 126 | 176 | 94.9% | 95.7% | 94.2% |
| The 7th 1-min | 4 | 2914 | 2886 | 87 | 157 | 96.0% | 97.1% | 94.8% |
| The 8th 1-min | 6 | 2973 | 2869 | 65 | 92 | 97.4% | 97.9% | 96.9% |
| The 9th 1-min | 6 | 2842 | 2918 | 66 | 114 | 97.0% | 97.7% | 96.2% |
| The 10th 1-min | 6 | 2844 | 3033 | 60 | 102 | 97.3% | 97.9% | 96.7% |
| The 11th 1-min | 4 | 2828 | 3056 | 79 | 119 | 96.7% | 97.3% | 96.3% |
| The 12th 1-min | 6 | 2688 | 3072 | 70 | 75 | 97.5% | 97.5% | 97.6% |
| The 13th 1-min | 7 | 2811 | 2997 | 54 | 65 | 98.0% | 98.1% | 97.9% |
| The 14th 1-min | 5 | 2800 | 2944 | 61 | 82 | 97.6% | 97.9% | 97.3% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, M.; Yu, H.; Wang, H. Automated Detection of Sudden Cardiac Death by Discrete Wavelet Transform of Electrocardiogram Signal. Symmetry 2022, 14, 571. https://doi.org/10.3390/sym14030571
Shi M, Yu H, Wang H. Automated Detection of Sudden Cardiac Death by Discrete Wavelet Transform of Electrocardiogram Signal. Symmetry. 2022; 14(3):571. https://doi.org/10.3390/sym14030571
Chicago/Turabian StyleShi, Manhong, Hongjie Yu, and Hongjie Wang. 2022. "Automated Detection of Sudden Cardiac Death by Discrete Wavelet Transform of Electrocardiogram Signal" Symmetry 14, no. 3: 571. https://doi.org/10.3390/sym14030571
APA StyleShi, M., Yu, H., & Wang, H. (2022). Automated Detection of Sudden Cardiac Death by Discrete Wavelet Transform of Electrocardiogram Signal. Symmetry, 14(3), 571. https://doi.org/10.3390/sym14030571





