MAP3Kε1/2 Interact with MOB1A/1B and Play Important Roles in Control of Pollen Germination through Crosstalk with JA Signaling in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. MAP3Kε1/2 Were Expressed Ubiquitously in Different Tissues
2.2. The map3kε1 map3kε2 Double Mutant Enhanced Pollen Germination
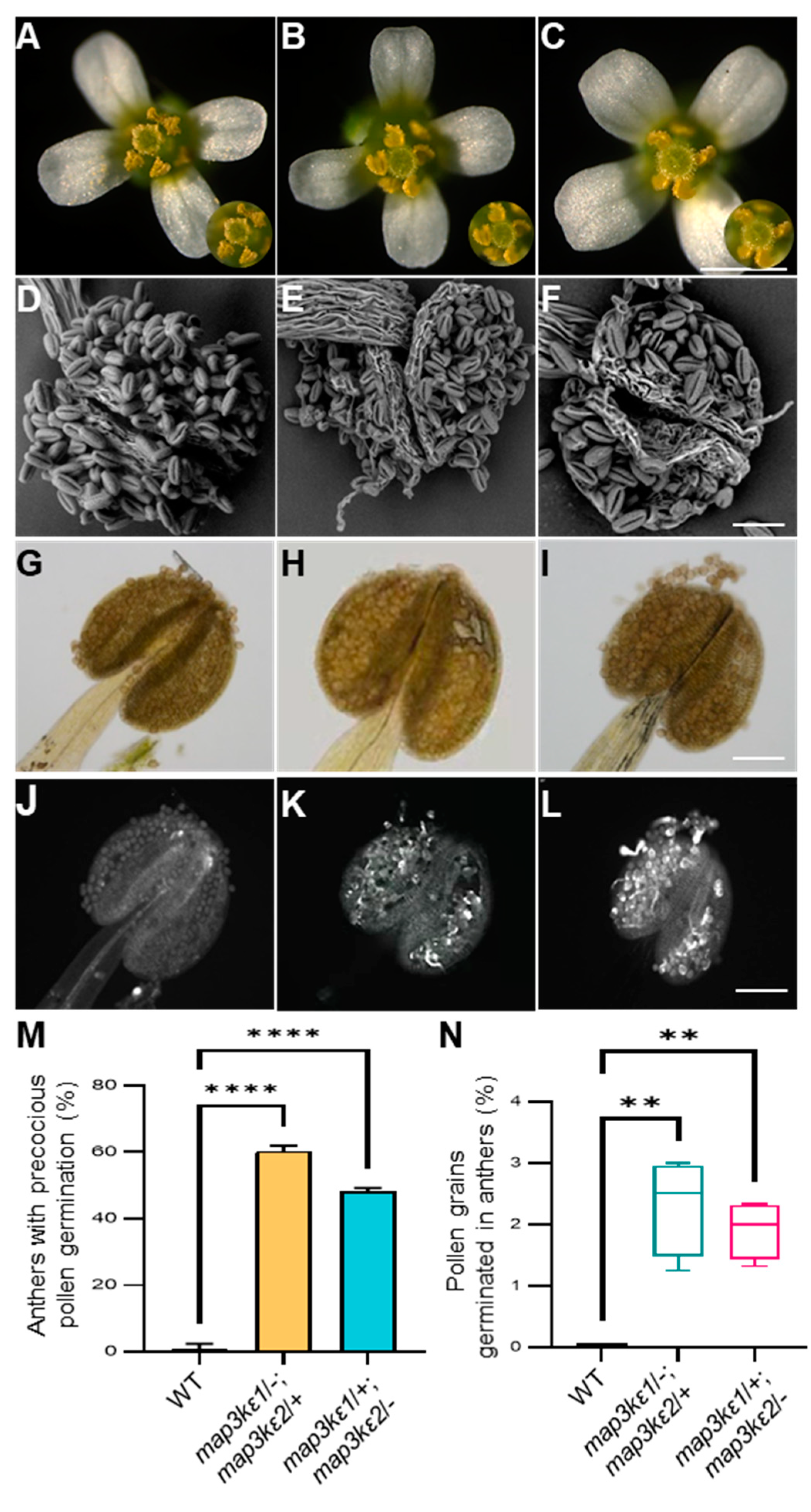
2.3. The map3kε1 map3kε2 Double Mutant Pollen Grains Precociously Germinated in the Dehiscent Anthers
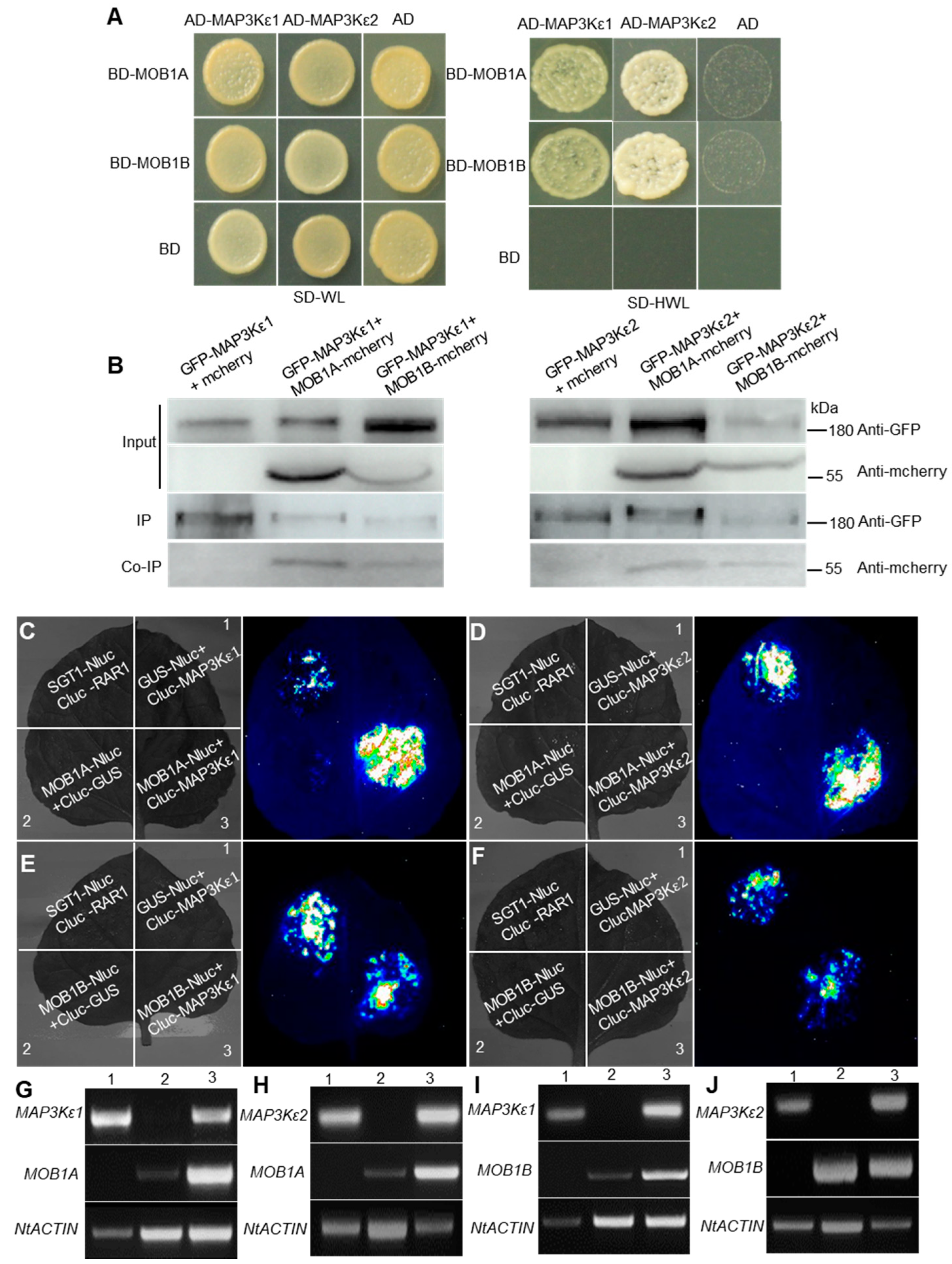
2.4. MAP3Kε1/2 Interacted with MOB1A/1B
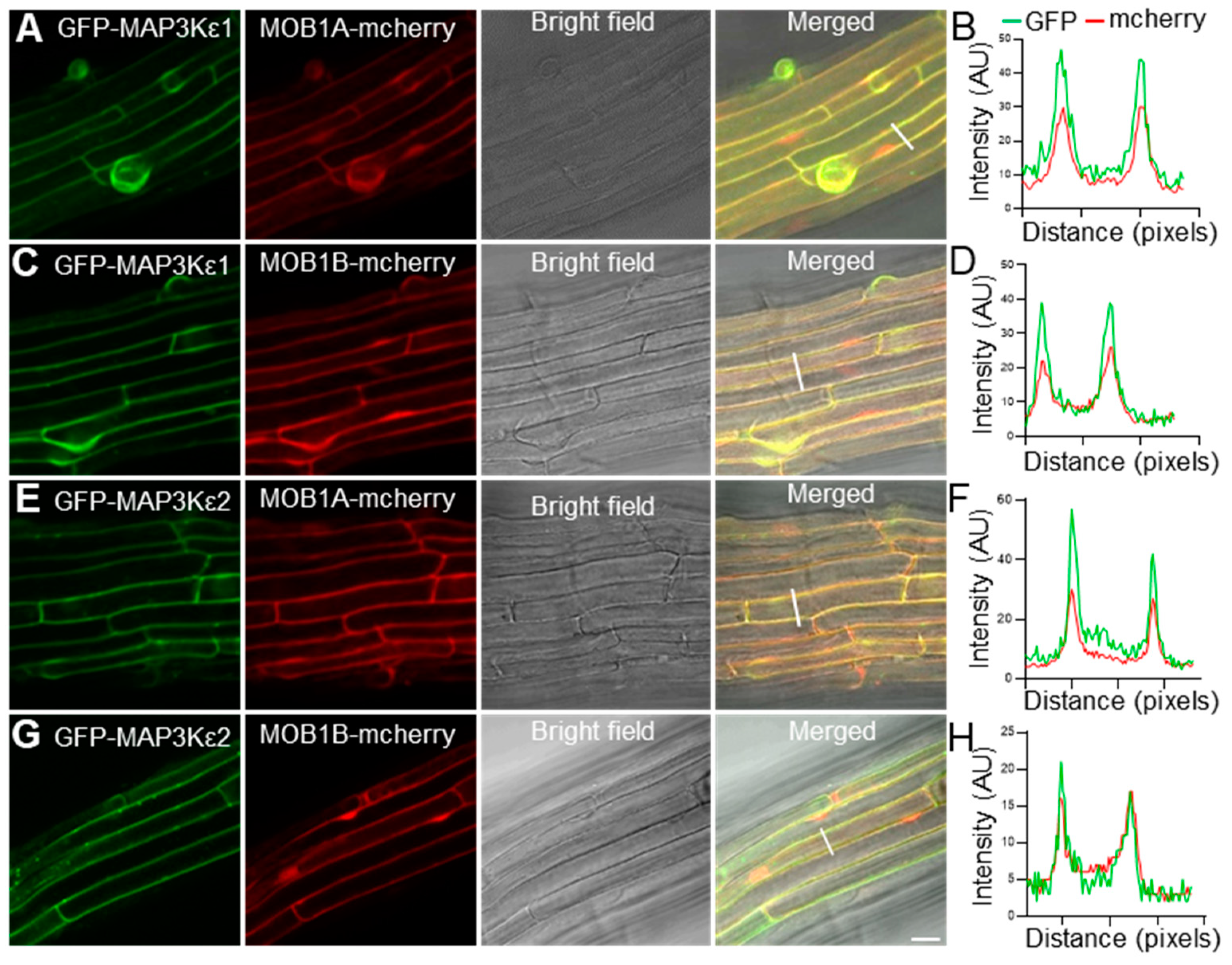
2.5. MAP3Kε1/2 Were Co-Localized with MOB1A/1B in the Cytoplasm and Plasma Membrane
2.6. JA Enhanced Pollen Germination
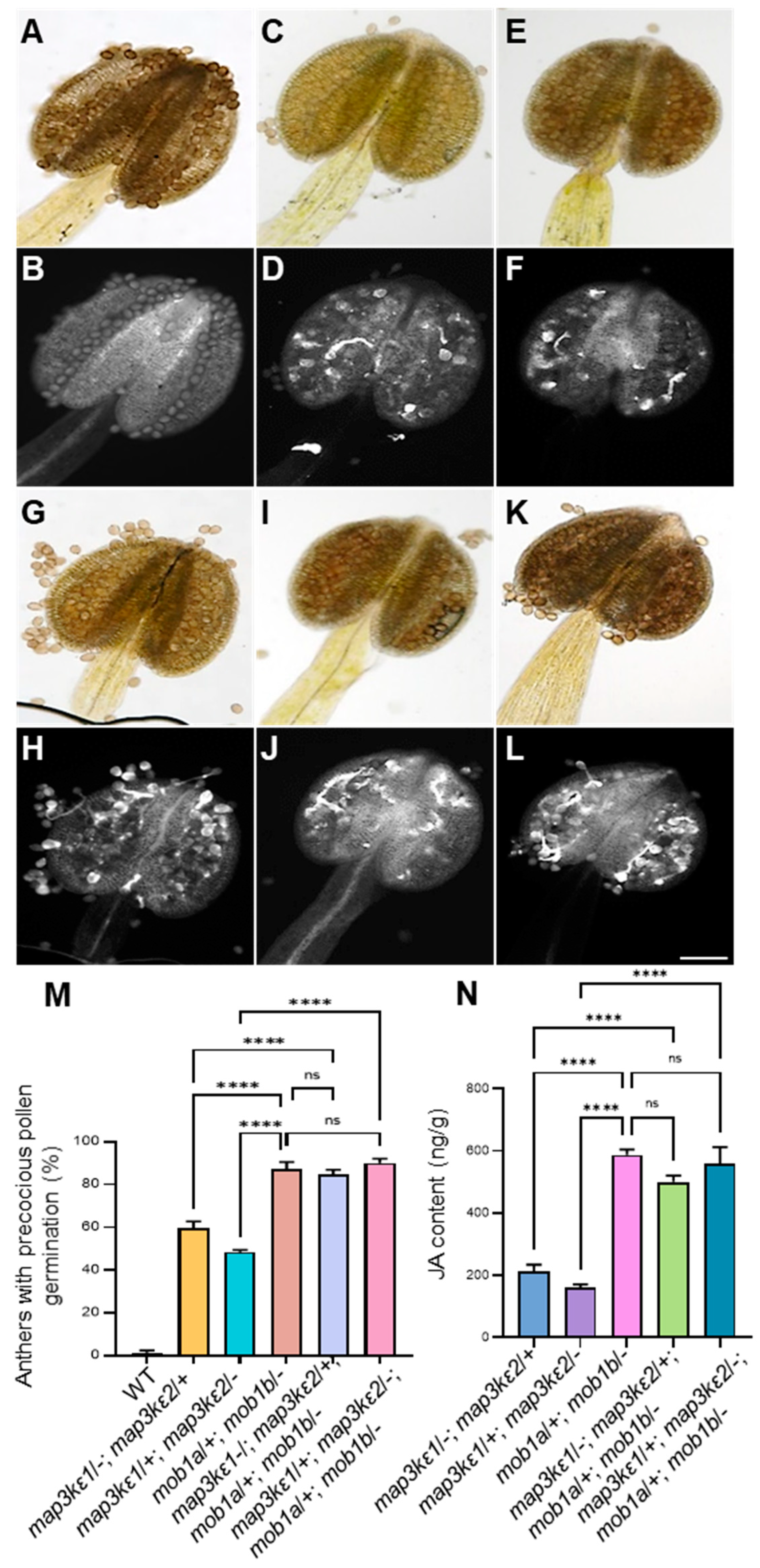
2.7. Genetic Interaction between MAP3Kε1/2 and MOB1A/1B
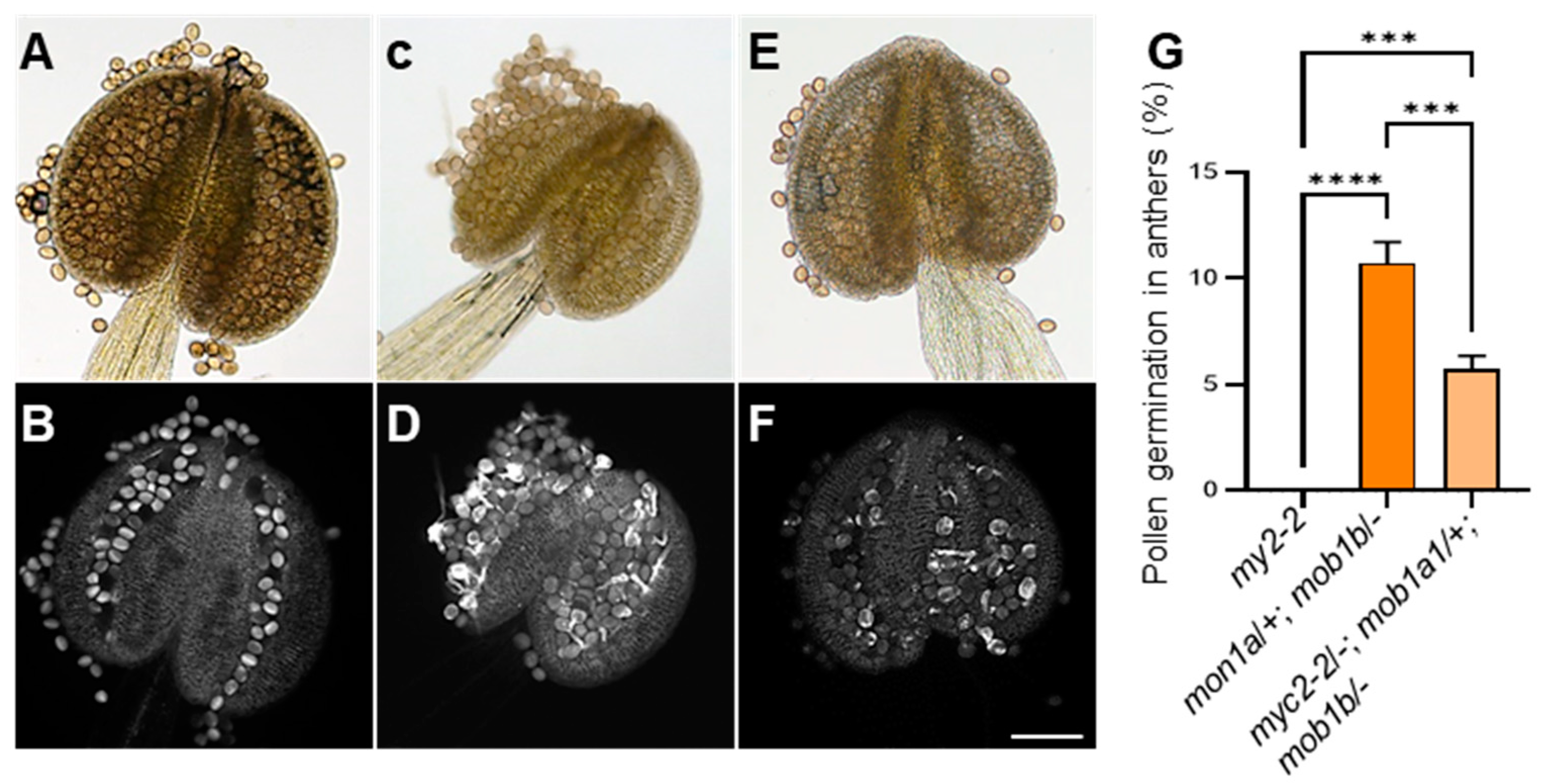
2.8. Genetic Interaction between MYC2 and MOB1A/1B
3. Discussion
3.1. MAP3Kε1 and MAP3Kε2 Functioned Redundantly and Involved in Control of Pollen Germination before Pollen Grains Landed on the Stigmas
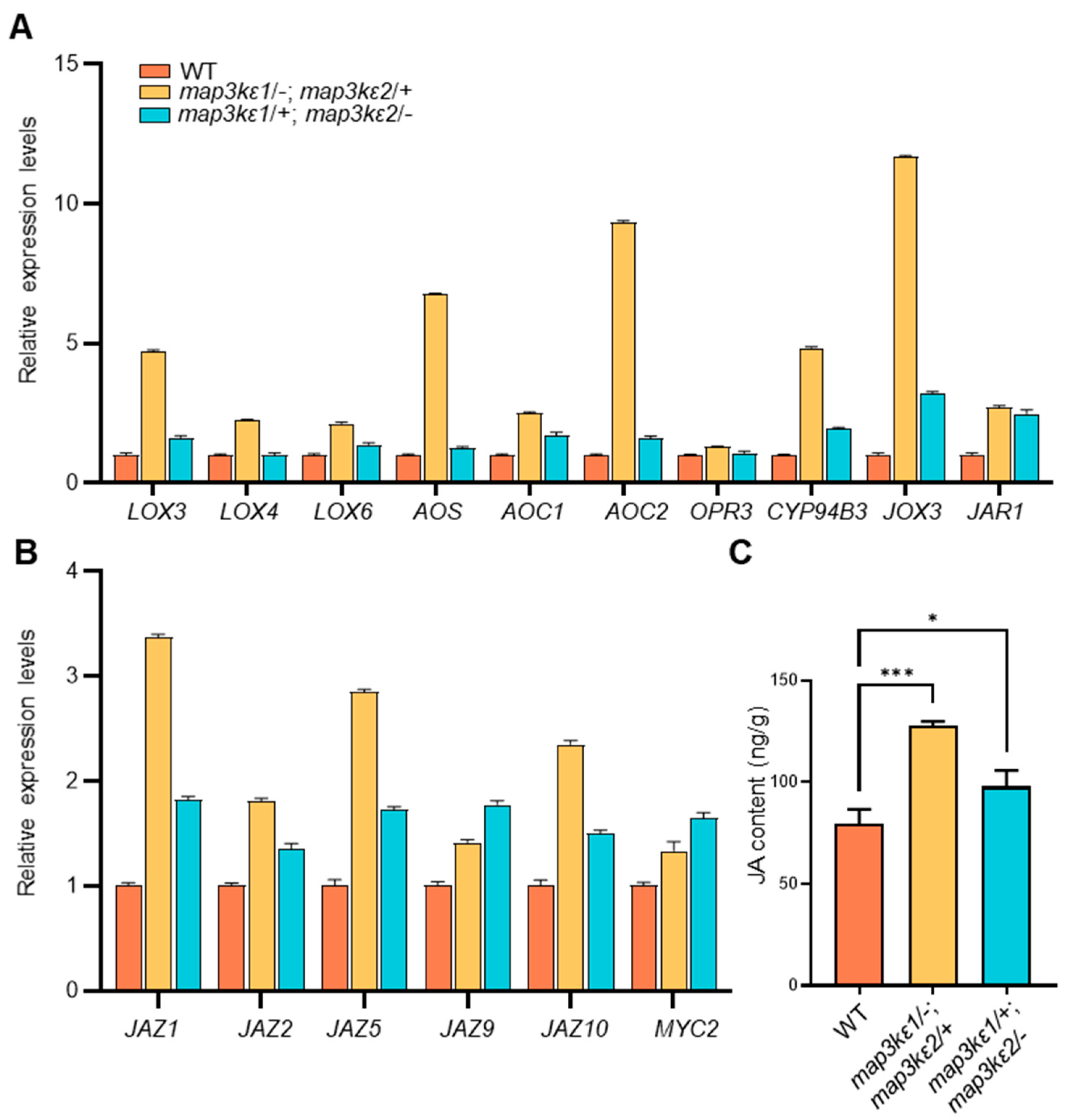
3.2. MAP3Kε1/2 Controlled Pollen Germination Possibly by Crosstalk with JA Signaling
3.3. Callose May Be Involved in Regulation of Pollen Germination
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RT-PCR and qRT-PCR Analysis
4.3. Molecular Cloning and Transformation
4.4. Phenotypic Analyses
4.5. Yeast Two-Hybrid Assays
4.6. Firefly Luciferase Complementation Imaging Assay
4.7. Co-Immunoprecipitation Assay
4.8. Subcellular Localization
4.9. Measurement of JA and Callose
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheung, A.Y.; Boavida, L.C.; Aggarwal, M.; Wu, H.M.; Feijo, J.A. The pollen tube journey in the pistil and imaging the in vivo process by two-photon microscopy. J. Exp. Bot. 2010, 61, 1907–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamamura, Y.; Nagahara, S.; Higashiyama, T. Double fertilization on the move. Curr. Opin. Plant Biol. 2012, 15, 70–77. [Google Scholar] [CrossRef]
- Malho, R.; Liu, Q.; Monteiro, D.; Rato, C.; Camacho, L.; Dinis, A. Signalling pathways in pollen germination and tube growth. Protoplasma 2006, 228, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Wengier, D.; Shuai, B.; Gui, C.P.; Muschietti, J.; McCormick, S.; Tang, W.H. The pollen receptor kinase LePRK2 mediates growth-promoting signals and positively regulates pollen germination and tube growth. Plant Physiol. 2008, 148, 1368–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirova, J.; Sedlarova, M.; Piterkova, J.; Luhova, L.; Petrivalsky, M. The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 2011, 181, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Liu, C.Z.; Li, D.D.; Zhao, T.T.; Li, F.; Jia, X.N.; Zhao, X.Y.; Zhang, X.S. The Arabidopsis KINβγ subunit of the SnRK1 complex regulates pollen hydration on the stigma by mediating the level of reactive oxygen species in pollen. PLoS Genet. 2016, 12, e1006228. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Jia, P.F.; Yang, W.C.; Li, H.J. Plasma membrane H(+)-ATPases-mediated cytosolic proton gradient regulates pollen tube growth. J. Integr. Plant Biol. 2020, 62, 1817–1822. [Google Scholar] [CrossRef]
- Johnson, S.A.; McCormick, S. Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic Mutant. Plant Physiol. 2001, 126, 685–695. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Kim, S.T.; Lord, E.M. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005, 138, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Xie, B.; Wang, X.; Hong, Z. Precocious pollen germination in Arabidopsis plants with altered callose deposition during microsporogenesis. Planta 2010, 231, 809–823. [Google Scholar] [CrossRef]
- Coimbra, S.; Costa, M.; Mendes, M.A.; Pereira, A.M.; Pinto, J.; Pereira, L.G. Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex. Plant Reprod. 2010, 23, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Cao, J.; Huang, L.; Yu, Y.; Lin, S. FLA14 is required for pollen development and preventing premature pollen germination under high humidity in Arabidopsis. BMC Plant Biol. 2021, 21, 254. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.N.; Takahashi, H.; Takahara, K.; Kawai-Yamada, M.; Kitazaki, K.; Shoji, K.; Goto, F.; Yoshihara, T.; Uchimiya, H. NAD+ accumulation during pollen maturation in Arabidopsis regulating onset of germination. Mol. Plant 2013, 6, 216–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhang, Q.; Zhao, K.; Luo, Q.; Bao, S.; Liu, H.; Men, S. The Arabidopsis GPR1 gene negatively affects pollen germination, pollen tube growth, and gametophyte senescence. Int. J. Mol. Sci. 2017, 18, 1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. S1), S153–S164. [Google Scholar] [CrossRef] [Green Version]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- McConn, M.; Browse, J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [Green Version]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [Green Version]
- von Malek, B.; van der Graaff, E.; Schneitz, K.; Keller, B. The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 2002, 216, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cetinbas-Genc, A.; Vardar, F. Effect of methyl jasmonate on in-vitro pollen germination and tube elongation of Pinus nigra. Protoplasma 2020, 257, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Li, X.R.; Li, H.J.; Yuan, L.; Liu, M.; Shi, D.Q.; Liu, J.; Yang, W.C. Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell 2014, 26, 619–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, Y.; Guo, L.; Cai, Q.; Ma, F.; Zhu, Q.Y.; Zhang, Q. Sodmergen, Arabidopsis JINGUBANG is a negative regulator of pollen germination that prevents pollination in moist environments. Plant Cell 2016, 28, 2131–2146. [Google Scholar] [CrossRef] [Green Version]
- Group, M.; Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Jouannic, S.; Champion, A.; Segui-Simarro, J.M.; Salimova, E.; Picaud, A.; Tregear, J.; Testillano, P.; Risueno, M.C.; Simanis, V.; Kreis, M.; et al. The protein kinases AtMAP3Kepsilon1 and BnMAP3Kepsilon1 are functional homologues of S. pombe cdc7p and may be involved in cell division. Plant J. 2001, 26, 637–649. [Google Scholar] [CrossRef]
- Simanis, V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003, 116 Pt 21, 4263–4275. [Google Scholar] [CrossRef] [Green Version]
- Bettignies, G.; Johnston, L.H. The mitotic exit network. Curr. Biol. 2003, 13, R301. [Google Scholar] [CrossRef] [Green Version]
- Maerz, S.; Seiler, S. Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr. Opin. Microbiol. 2010, 13, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Hergovich, A.; Hemmings, B.A. Hippo signalling in the G2/M cell cycle phase: Lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 2012, 23, 794–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, J.; Cui, X.; Yuan, X.; Yu, X.; Sun, J.; Gong, Q. The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 2016, 67, 1461–1475. [Google Scholar] [CrossRef] [Green Version]
- Galla, G.; Zenoni, S.; Marconi, G.; Marino, G.; Botton, A.; Pinosa, F.; Citterio, S.; Ruperti, B.; Palme, K.; Albertini, E.; et al. Sporophytic and gametophytic functions of the cell cycle-associated Mob1 gene in Arabidopsis thaliana L. Gene 2011, 484, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pinosa, F.; Begheldo, M.; Pasternak, T.; Zermiani, M.; Paponov, I.A.; Dovzhenko, A.; Barcaccia, G.; Ruperti, B.; Palme, K. The Arabidopsis thaliana Mob1A gene is required for organ growth and correct tissue patterning of the root tip. Ann. Bot. 2013, 112, 1803–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.; Guo, Z.; Song, L.; Wang, Y.; Cheng, Y. NCP1/AtMOB1A plays key roles in auxin-mediated Arabidopsis development. PLoS Genet. 2016, 12, e1005923. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Yue, X.; Cui, X.; Song, L.; Cheng, Y. AtMOB1 genes regulate jasmonate accumulation and plant development. Plant Physiol. 2020, 182, 1481–1493. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.M.; Liang, Y.; Mei, J.; Liao, H.Z.; Wang, P.; Hu, K.; Chen, L.Q.; Zhang, X.Q.; Ye, D. The Arabidopsis AGC kinases NDR2/4/5 interact with MOB1A/1B and play important roles in pollen development and germination. Plant J. 2021, 105, 1035–1052. [Google Scholar] [CrossRef]
- Chaiwongsar, S.; Otegui, M.S.; Jester, P.J.; Monson, S.S.; Krysan, P.J. The protein kinase genes MAP3K epsilon 1 and MAP3K epsilon 2 are required for pollen viability in Arabidopsis thaliana. Plant J. 2006, 48, 193–205. [Google Scholar] [CrossRef]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signaling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef]
- Wang, Z.; Gou, X. Receptor-like protein kinases function upstream of MAPKs in regulating plant development. Int. J. Mol. Sci. 2020, 21, 7638. [Google Scholar] [CrossRef] [PubMed]
- Bush, S.M.; Krysan, P.J. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 2007, 58, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Bruffett, K.; Lee, J.; Hause, G.; Walker, J.C.; Zhang, S. Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 2008, 20, 602–613. [Google Scholar] [CrossRef] [Green Version]
- Hord, C.L.; Sun, Y.J.; Pillitteri, L.J.; Torii, K.U.; Wang, H.; Zhang, S.; Ma, H. Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 2008, 1, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, H.; He, Y.; Liu, Y.; Walker, J.C.; Torii, K.U.; Zhang, S. A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 2012, 24, 4948–4960. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Lu, J.; Xu, J.; McClure, B.; Zhang, S. Two mitogen-activated protein kinases, MPK3 and MPK6, are required for funicular guidance of pollen tubes in Arabidopsis. Plant Physiol. 2014, 165, 528–533. [Google Scholar] [CrossRef] [Green Version]
- Lukowitz, W.; Roeder, A.; Parmenter, D.; Somerville, C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 2004, 116, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Su, S.H.; Krysan, P.J. A double-mutant collection targeting MAP kinase related genes in Arabidopsis for studying genetic interactions. Plant J. 2016, 88, 867–878. [Google Scholar] [CrossRef]
- Zeng, Q.; Chen, J.G.; Ellis, B.E. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011, 67, 895–906. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Ranf, S.; Pancholi, S.S.; Jayanty, S.; Walla, M.D.; Miller, W.; Howe, G.A.; Lincoln, D.E.; Stratmann, J.W. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 2007, 104, 12205–12210. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Yuan, B.; Liu, H.; Li, X.; Xu, C.; Wang, S. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 2010, 64, 86–99. [Google Scholar] [CrossRef] [PubMed]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, Y.J.; Kim, M.H.; Kwak, J.M. MAPK cascades in guard cell signal transduction. Front. Plant Sci. 2016, 7, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.Y.; Oh, S.I.; Ok, S.H.; Cho, S.K.; Shin, J.S. Identification of putative MAPK kinases in Oryza minuta and O. sativa responsive to biotic stresses. Mol. Cells 2007, 23, 108–114. [Google Scholar]
- Wilson, Z.A.; Song, J.; Taylor, B.; Yang, C. The final split: The regulation of anther dehiscence. J. Exp. Bot. 2011, 62, 1633–1649. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Han, X.; Lu, T.G. Callose synthesis during reproductive development in monocotyledonous and dicotyledonous plants. Plant Signal. Behav. 2016, 11, e1062196. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Chai, M.; Yang, J.; Ning, G.; Wang, G.; Ma, H. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiol. 2014, 164, 1893–1904. [Google Scholar] [CrossRef] [Green Version]
- Enns, L.C.; Kanaoka, M.M.; Torii, K.U.; Comai, L.; Okada, K.; Cleland, R.E. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 2005, 58, 333–349. [Google Scholar] [CrossRef]
- Garcia-Andrade, J.; Ramirez, V.; Flors, V.; Vera, P. Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J. 2011, 67, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Scalschi, L.; Sanmartin, M.; Camanes, G.; Troncho, P.; Sanchez-Serrano, J.J.; Garcia-Agustin, P.; Vicedo, B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015, 81, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Sanmartin, N.; Pastor, V.; Pastor-Fernandez, J.; Flors, V.; Pozo, M.J.; Sanchez-Bel, P. Role and mechanisms of callose priming in mycorrhiza-induced resistance. J. Exp. Bot. 2020, 71, 2769–2781. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.Z.; Zhu, M.M.; Cui, H.H.; Du, X.Y.; Tang, Y.; Chen, L.Q.; Ye, D.; Zhang, X.Q. MARIS plays important roles in Arabidopsis pollen tube and root hair growth. J. Integr. Plant Biol. 2016, 58, 927–940. [Google Scholar] [CrossRef]
- Cui, H.H.; Liao, H.Z.; Tang, Y.; Du, X.Y.; Chen, L.Q.; Ye, D.; Zhang, X.Q. ABORTED GAMETOPHYTE 1 is required for gametogenesis in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 1003–1016. [Google Scholar] [CrossRef]
- Zhu, L.; Chu, L.C.; Liang, Y.; Zhang, X.Q.; Chen, L.Q.; Ye, D. The Arabidopsis CrRLK1L protein kinases BUPS1 and BUPS2 are required for normal growth of pollen tubes in the pistil. Plant J. 2018, 95, 474–486. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Tan, Z.M.; Zhu, L.; Niu, Q.K.; Zhou, J.J.; Li, M.; Chen, L.Q.; Zhang, X.Q.; Ye, D. MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLoS Genet. 2013, 9, e1003933. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, S.L.; Xie, L.F.; Puah, C.S.; Zhang, X.Q.; Yang, W.C.; Sundaresan, V.; Ye, D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.L.; Liu, L.; Niu, Q.K.; Xia, C.; Yang, K.Z.; Li, R.; Chen, L.Q.; Zhang, X.Q.; Zhou, Y.; Ye, D. Male gametophyte defective 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J. 2011, 65, 647–660. [Google Scholar] [CrossRef]
- Yang, K.Z.; Xia, C.; Liu, X.L.; Dou, X.Y.; Wang, W.; Chen, L.Q.; Zhang, X.Q.; Xie, L.F.; He, L.; Ma, X.; et al. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009, 57, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.Y.; Yang, K.Z.; Ma, Z.X.; Chen, L.Q.; Zhang, X.Q.; Bai, J.R.; Ye, D. AtTMEM18 plays important roles in pollen tube and vegetative growth in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 679–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, D.J.; Cao, X.; Wang, W.; Tan, X.Y.; Zhang, X.Q.; Chen, L.Q.; Ye, D. GNOM-LIKE 2, encoding an adenosine diphosphate-ribosylation factor-guanine nucleotide exchange factor protein homologous to GNOM and GNL1, is essential for pollen germination in Arabidopsis. J. Integr. Plant Biol. 2009, 51, 762–773. [Google Scholar] [CrossRef]
- Tang, Y.; Yin, Z.; Zeng, Y.; Zhang, Q.; Chen, L.; He, Y.; Lu, P.; Ye, D.; Zhang, X. MTOPVIB interacts with AtPRD1 and plays important roles in formation of meiotic DNA double-strand breaks in Arabidopsis. Sci. Rep. 2017, 7, 10007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like Kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380.e6. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Chu, J.; Sun, X.; Wang, J.; Yan, C. Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 2012, 28, 1081–1087. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.L.; Blancaflor, E.B.; Kochian, L.V.; Gilroy, S. Spatial coordination of aluminium uptake, production of reactive oxygen species, callose production and wall rigidification in maize roots. Plant Cell Environ. 2006, 29, 1309–1318. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, J.; Zhou, P.; Zeng, Y.; Sun, B.; Chen, L.; Ye, D.; Zhang, X. MAP3Kε1/2 Interact with MOB1A/1B and Play Important Roles in Control of Pollen Germination through Crosstalk with JA Signaling in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 2683. https://doi.org/10.3390/ijms23052683
Mei J, Zhou P, Zeng Y, Sun B, Chen L, Ye D, Zhang X. MAP3Kε1/2 Interact with MOB1A/1B and Play Important Roles in Control of Pollen Germination through Crosstalk with JA Signaling in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(5):2683. https://doi.org/10.3390/ijms23052683
Chicago/Turabian StyleMei, Juan, Pengmin Zhou, Yuejuan Zeng, Binyang Sun, Liqun Chen, De Ye, and Xueqin Zhang. 2022. "MAP3Kε1/2 Interact with MOB1A/1B and Play Important Roles in Control of Pollen Germination through Crosstalk with JA Signaling in Arabidopsis" International Journal of Molecular Sciences 23, no. 5: 2683. https://doi.org/10.3390/ijms23052683
APA StyleMei, J., Zhou, P., Zeng, Y., Sun, B., Chen, L., Ye, D., & Zhang, X. (2022). MAP3Kε1/2 Interact with MOB1A/1B and Play Important Roles in Control of Pollen Germination through Crosstalk with JA Signaling in Arabidopsis. International Journal of Molecular Sciences, 23(5), 2683. https://doi.org/10.3390/ijms23052683




