Abstract
Background: Endometriosis diagnosis constitutes a considerable economic burden for the healthcare system with diagnostic tools often inconclusive with insufficient accuracy. We sought to analyze the human miRNAome to define a saliva-based diagnostic miRNA signature for endometriosis. Methods: We performed a prospective ENDO-miRNA study involving 200 saliva samples obtained from 200 women with chronic pelvic pain suggestive of endometriosis collected between January and June 2021. The study consisted of two parts: (i) identification of a biomarker based on genome-wide miRNA expression profiling by small RNA sequencing using next-generation sequencing (NGS) and (ii) development of a saliva-based miRNA diagnostic signature according to expression and accuracy profiling using a Random Forest algorithm. Results: Among the 200 patients, 76.5% (n = 153) were diagnosed with endometriosis and 23.5% (n = 47) without (controls). Small RNA-seq of 200 saliva samples yielded ~4642 M raw sequencing reads (from ~13.7 M to ~39.3 M reads/sample). Quantification of the filtered reads and identification of known miRNAs yielded ~190 M sequences that were mapped to 2561 known miRNAs. Of the 2561 known miRNAs, the feature selection with Random Forest algorithm generated after internally cross validation a saliva signature of endometriosis composed of 109 miRNAs. The respective sensitivity, specificity, and AUC for the diagnostic miRNA signature were 96.7%, 100%, and 98.3%. Conclusions: The ENDO-miRNA study is the first prospective study to report a saliva-based diagnostic miRNA signature for endometriosis. This could contribute to improving early diagnosis by means of a non-invasive tool easily available in any healthcare system.
1. Introduction
Endometriosis, defined by the presence of endometrium-like tissue outside the uterus, affects 2–10% of the female population, i.e., around 190 million women worldwide. It is a heterogenous disease with a poorly understood natural history and as such poses many challenges [1,2]. The first is timely diagnosis mainly because the symptoms of endometriosis are non-specific, and clinical examination is often either negative or results in a wrong diagnosis [3,4]. The second challenge is that complementary explorations, especially biomarkers [5,6] and imaging examinations [5,6,7,8], are often inconclusive and fail to diagnose early-stage endometriosis with sufficient accuracy or are of limited relevance for the severe forms. Consequently, therapeutic and follow-up strategies are compromised, and there is a high rate of conventional treatment failure [2,4]. Finally, endometriosis constitutes a considerable economic burden for the healthcare system linked not only to direct costs but also to indirect costs from school and work absenteeism. Overall, the annual cost of endometriosis was estimated at around 10,000 euros per patient in 2012, which is equivalent to that of diabetes in France in 2017 [9,10,11,12,13].
During the last decade, new diagnostic tools have been investigated to detect this debilitating disorder as early as possible [5,14,15,16,17,18]. Among these, microRNA (miRNA) analysis is emerging as a promising option supported by a growing body of evidence from studies in cancer and degenerative disorders [19,20,21,22,23]. Human miRNAs are single stranded, highly conserved, non-coding RNAs composed of 21–25 nucleotides. Partial binding to their complementary messenger RNA (mRNA) can regulate gene degradation and translation. It is estimated that about 60% of genes are regulated by miRNAs [24]. From a biological point of view, miRNAs are mainly transcribed from genes in intronic regions of coding or non-coding transcripts [24,25,26]. miRNAs are transcribed in the nucleus under hundreds of duplex nucleotide-long primary miRNAs (pri-miRNA) subsequently cleaved to generate precursor miRNAs (pre-miRNA). These pre-miRNAs are then transported from the nucleus to the cytoplasm, where the duplexes are cleaved to form mature miRNAs, which are incorporated into RNA silencing complexes (RISC) that regulate post-translational modifications by binding to the target mRNA [24,25,26]. Finally, the miRNAs are released from the cells into the circulation using various carriers, such as Argonaute, nucleophosmin 1, high-density lipoproteins, or extracellular vesicles (exosomes), which confer remarkable stability against endogenous RNAses. The miRNAs can then be detected in human fluids [24,25,26].
In the specific setting of endometriosis, several authors have evaluated the relevance of a blood-based miRNA signature, but the results are discordant because of methodological and control group issues [19,20,22,23,27,28]. Indeed, in previous studies, the control groups was composed of asymptomatic patients and/or patients undergoing a tubal ligation and/or pelvic inflammatory disease and/or gynecologic disorders but without knowledge of symptoms [19,20,22,23].
Similarly, saliva miRNAome analysis has been investigated by several teams exploring biomarkers for numerous benign and malignant disorders but never in the context of endometriosis [29,30,31,32].
Therefore, the aim of the prospective ENDO-miRNA study was to analyze the human miRNome to differentiate patients with and without endometriosis, to define a saliva-based diagnostic miRNA signature for endometriosis with an internal cross validation.
2. Material and Methods
2.1. Ethics Statement
The data and saliva used for analysis were collected from the prospective ENDO-miRNA study (ClinicalTrials.gov Identifier: NCT04728152) under the Research Protocol ID RCB: 2020-A03297-32. Informed consent was obtained from all the participants. The study and data analysis followed the Standards for Reporting of Diagnostic Accuracy (STARD) reporting guidelines [33] (Annex S1). The study consisted of two parts: (i) identification of a biomarker based on genome-wide miRNA expression profiling by small RNA sequencing using next-generation sequencing (NGS) and (ii) development of a saliva-based miRNA diagnostic signature according to expression and accuracy profiling using an ML algorithm [17,21,34,35,36,37,38,39,40,41,42].
2.2. Study Population
The prospective ENDO-miRNA study included 200 saliva samples obtained from women with chronic pelvic pain suggestive of endometriosis. All the saliva samples were collected between January and June 2021. All the patients underwent either a laparoscopic procedure (therapeutic or diagnostic laparoscopy) and/or MRI imaging [5,6,7,8]. The laparoscopic procedures were systematically videoed and then analyzed by two operators (C.T., Y.D.), blinded to the symptoms and imaging findings, to confirm the presence or absence of endometriosis. For the patients who underwent laparoscopy, diagnosis was confirmed by histology. For the patients diagnosed with endometriosis without laparoscopic evaluation, all had MRI with features of deep endometriosis with colorectal involvement and/or endometriomas confirmed by two expert radiologists. The miRNAs were analyzed blinded to the surgical and imaging findings. Following exploration by laparoscopy or MRI, the women were classified into two groups: an endometriosis group and a control group of women with various benign pathologies other than endometriosis or with symptoms suggestive of endometriosis but without clinical or MRI features and no endometriosis lesions found during laparoscopic inspection (complex patient). The study flow chart is reported in Figure 1. The patients with endometriosis were stratified according to the revised American Society of Reproductive Medicine (rASRM) classification [43].
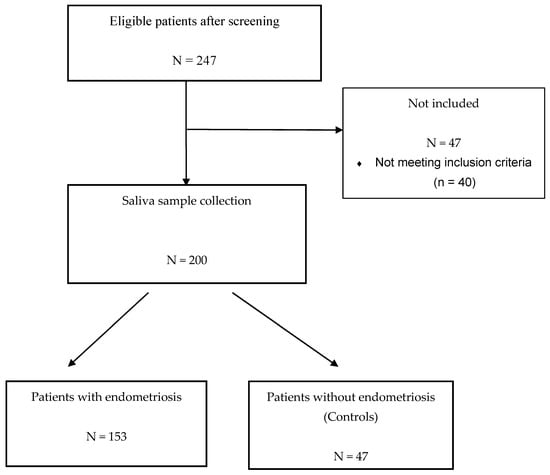
Figure 1.
Flow chart of ENDO-miRNA study.
2.3. Saliva Sample Collection
The saliva samples (2 mL) were collected in an all-in-one system including a nucleic acid stabilizing solution for the collection, stabilization, and transportation (RE-100, DNA Genotek Inc., 2 Beaverbrook Road, Ottawa, ON, Canada) using an at-home kit (https://www.dnagenotek.com/us/products/collection-infectious-disease/OME-505.html, accessed on 1 December 2021). All the samples were stored at room temperature prior to shipping.
2.4. RNA Sample Extraction, Preparation and Quality Control
The RNA was isolated from each saliva sample using the miRNeasy Kit (Qiagen, Inc., Germantown, MD, USA) according to the manufacturer’s instructions [29,31,44,45]. RNA quality was assessed using the Agilent Technologies Tapestation 2200. RNA-sequencing libraries were prepared using the QIAseq miRNA Library Kit (Qiagen) according to the manufacturer’s instructions. Samples were indexed in batches of 96, with a targeted sequencing depth of 17 million reads per sample. Sequencing was performed using 100 base single-end reads, using an Novaseq6000 sequencer (Illumina, San Diego, CA, USA) [46,47].
3. Bioinformatics
3.1. Raw Data Preprocessing (Raw, Filtered, Aligned Reads) and Quality
Sequencing reads were processed using the data processing pipeline. FastQ files were trimmed to remove adapter sequences using Cutadapt version v.1.18 and were aligned using Bowtie version 1.1.1 with the following transcriptome databases: the human reference genome available from NCBI (https://www.ncbi.nlm.nih.gov/genome/guide/human/, accessed on 1 December 2021) and miRBase21 (miRNAs) using the MirDeep2 v0.1.0 package. The raw sequencing data quality was assessed using FastQC software v0.11.7 [40,44,46,48,49,50].
3.2. Differential Expression Analysis of miRNAs
miRNA expression was quantified by miRDeep2 v0.1.0 [51]. Differential expression tests were then conducted in DESeq2 for miRNAs with read counts in ≥1 of the samples. DESeq2 V1.20 integrates methodological advances with several novel features to facilitate a more quantitative analysis of comparative RNA-seq data using shrinkage estimators for dispersion and fold change [52]. The resulting matrix was filtered for expressed miRNAs [53]. The miRNAs were considered as differentially expressed if the absolute value of log2 fold change was >1.5 (upregulated) and <0.5 (downregulated). The p-value adjusted for multiple testing was <0.05 [52]. The Annex S2 summarized and adapted the miRNA-nome Sequencing Analysis Pipeline used from the methods of Potla et al. [44].
4. Statistical Analysis
4.1. Development and Validation of the Diagnostic Model
Random Forest (RF) was considered to design the saliva signature [34,35,36,37,54,55,56]. Random Forest (RF) classifier is an ensemble method that trains several Decision Tree (DT) in parallel with bootstrapping followed by aggregation, jointly referred to as bagging. Bootstrapping indicates that several individual DTs are trained in parallel on various subsets of a training dataset using different subsets of available features. Bootstrapping ensures that each individual DT in the RF is unique, which reduces the overall variance of the RF classifier. For the final decision, RF classifier aggregates the decisions of individual DTs and consequently exhibits good generalization [54]. F1-score, sensitivity, specificity, and the ROC AUC were calculated to assess and compare the diagnostic performance of the diagnostic signature [57,58].
4.2. Validation of the Signature Accuracy
The accuracy and reproducibility of the signature were tested on 10 data sets randomly [41,59,60] composed of the same proportion of control and endometriosis patients. Each data set was randomly generated to conserve the initial ratio of endometriosis and control patient’s profile. Analysis was performed using Python (Python Software Foundation) with XGBoost 1.3.3, scikit-learn 0.19.1, and scipy 1.1 packages.
4.3. Other Statistical Analyses
Statistical analysis was based on the chi-square test as appropriate for categorical variables. Values of p < 0.05 were considered to denote significant differences. Data were managed with an Excel database (Microsoft, Redmond, WA, USA) and analyzed using R 2.15 software, available online (http://cran.r-project.org/, accessed on 1 December 2021).
5. Results
5.1. Description of the ENDO-miRNA Cohort
The clinical characteristics of the patients in the endometriosis and control groups are presented in Table 1. Among the 200 patients, 76.5% (n = 153) were diagnosed with endometriosis and 23.5% (n = 47) without (controls). In the control group, 51% (24) of the women had no abnormality and were defined as discordant or complex patients (Table 1). There were no significant differences in terms of age or BMI between the groups. The mean (±SD) time from symptom onset to diagnosis for endometriosis patients was 14.8 years (±17.88). In both groups, the patients had pain symptoms suggestive of endometriosis. Comparatively, for patients with and without endometriosis using Visual Analogical Scale (VAS), the dysmenorrhea/of dysmenorrhea (mean ± SD) were 6 ± 3.4 versus 5 ± 3.2, p < 0.001; dyspareunia was 5.28 ± 3.95 verus 4.95 ± 3.52, p < 0.001; and urinary pain during menstruation (mean ± SD) were 4.35 ±3.36 versus 2.84 ±2.76, p < 0.001. For the endometriosis patients, 52% (80) had rASRM stage I–II, and 48% (73) had stage III–IV.

Table 1.
Demographic Characteristics of the population.
5.2. Global Overview of miRNA Transcriptome
Small RNA-seq of 200 saliva samples yielded ~4.642 M raw sequencing reads (from ~13.7 M to ~39.3 M reads/sample). Pre-filtering and filtering steps retained 70% (~3.205 M) of initial raw reads. The majority of the filtered reads were of short read length. Quantification of the filtered reads and identification of known miRNAs yielded ~190 M sequences that were mapped to 2561 known miRNAs from miRBase v21. The number of expressed miRNAs ranged from 1250 (outlier) to 2561 per sample. The distribution of expressed miRNAs in the 200 saliva samples and the overall composition of processed reads is shown in Figure 2.
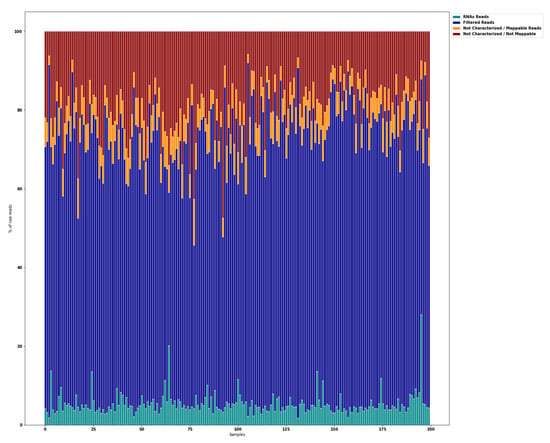
Figure 2.
Overall composition of processed reads. RNA reads, miRNAs + piRNAs + rRNAs + tRNAs + mRNAs + others; Filtered Reads, reads with no adapters + reads with low quality bases + reads too short; Not Characterized/Mappable reads, mapped reads to GRCh38 that could not be characterized as a particular type; Not Characterized/Not Mappable reads, reads that could not be mapped.
5.3. Feature Selection of miRNAs Relevant for a Diagnosis of Endometriosis
The expression and accuracy of the miRNA profiles were used to identify miRNAs related to endometriosis. Out of the 2561 known miRNAs, the feature selection method generated a subset of 109 miRNAs. The respective correlation and accuracy to diagnose endometriosis according to the F1-score, sensitivity, specificity, and AUC ranged from 18.9–87.7%, 11.6–99.4%, 8.5–97.9%, and 36.9–69.2%, respectively. Among the 109 miRNAs selected, 79% (n = 86) and 21% (n = 23) had an AUC value of <60 and ≥60% for correlation and accuracy, respectively; 83% (n = 91) and 17% (n = 18) had an F1-score ranging between 0–79%, and ≥80%, respectively; 83% (n = 91) and 17% (n = 18) had a sensitivity ranging between 0–79%, and ≥80%, respectively; and 83% (n = 91) and 17% (n = 18) had a specificity ranging between 0–79%, and ≥80%, respectively. Finally, 0% (n = 0) were identified as being downregulated and 6.4% (n = 7) as being upregulated. Annex S3 summarizes the diagnostic accuracy of the 109 miRNAs selected in the signature.
5.4. Saliva-Based Diagnostic Signature for Endometriosis
The overall performance of the diagnostic signature composed of 109 mi RNAs (Random Forest model) against the 10 randomized datasets is reported in Table 2. The sensitivity, specificity, and AUC ranges from 80% to 96.8%, 80% to 100%, and 79.9% to 98.4%, respectively. The signature, after internal cross validation on 10 different data sets, obtained its higher accuracy with a respective sensitivity, specificity, and AUC of 96.7%, 100% and 98.3% (Table 2).

Table 2.
Random Forest accuracies for endometriosis diagnosis.
5.5. Relation between Pathophysiology of Endometriosis and miRNA Expression
Among the 109 miRNAs composing the endometriosis diagnostic signature, 77% (84) have been reported to be associated with pathophysiologic pathways for benign and malignant disorders (Annex S3). Only miR-34c-5p and miR-19b-1-5p have previously been reported in the field of endometriosis. Among the 109 mi RNA of the signature, 29 (27%) are associated with the main signaling pathways of endometriosis: PI3K/Akt, PTEN, Wnt/β-catenin, HIF1α/NF κB, and YAP/TAZ/EGFR (Annex S3).
6. Discussion
To the best of our knowledge, the ENDO-miRNA study is the first prospective study to report a saliva-based diagnostic miRNA signature for endometriosis. This could contribute to improving early diagnosis by means of a non-invasive tool easily available in any healthcare system. Its value lies in the combination of the intrinsic quality of miRNA to condense endometriosis phenotypes (and its heterogeneity) and the modeling power of AI. Its reproducibility is based on our bioinformatics approach of miRNA-sequencing analysis and a statistical approach designed to overcome the complexity and heterogeneity of endometriosis.
We hypothesized that a saliva-based miRNA signature for endometriosis would be a low-cost and scalable method allowing samples to be collected anywhere by anyone. The tool would thus be available for underprivileged populations unlike methods based on blood samples, which are blood-volume and temperature dependent, imposing complex logistics of collecting peripheral blood and transporting it to a laboratory for analysis.
Saliva is an increasingly attractive body fluid in the search for disease biomarkers [29,30,31,32,45,61]. miRNAs exhibit remarkable stability in severe conditions, such as extended storage [24,29,32,62]. Zheng et al. demonstrated that saliva is not affected by coagulation that could induce a release of miRNAs. This is especially crucial as many studies evaluating miRNA expression in endometriosis have been performed on serum [23,61]. Zhang et al. first developed a technique to stabilize the saliva and process RNA analysis. The average range of RNA content in 1 L of bodily fluids is as low as 0.01 mg in urine to as high as 11.2 mg in saliva [63]. Moreover, blood, leukocytes, and saliva have lower standard deviations in their RNA content (<50% on average) compared with serum and urine. The average concentration of the isolated RNAs from all bodily fluids can be classified into high (>20 ng/μL) for blood, leukocytes, saliva, and cell-free saliva and low (<10 ng/μL) for plasma, serum, urine, and cell-free urine. Finally, more than 90% of miRNAs in saliva are shared with blood, leukocytes, and plasma, which further supports its stability and reproducibility [63].
In the specific setting of endometriosis, and despite the various endometriosis phenotypes, we were able to build an endometriosis diagnostic signature. The most accurate signature in our model provides a sensitivity, specificity and AUC of 96.7%, 100% and 98.3%, respectively. These values testify to the high accuracy of the signature, supporting its clinical value, and raise the issue to revise the current diagnostic strategy for exploring patients with symptoms suggestive of endometriosis, based on a diagnostic laparoscopy.
Multiple diagnostic biomarkers have been suggested as screening and triage tests to diagnose endometriosis [14,15], but none of them are of sufficient accuracy, i.e., a sensitivity of 0.94 and a specificity of 0.79 [5,14,15]. In accordance with the 2011 Biomarkers Definitions Working Group, a biomarker is “a characteristic that can be objectively measured and evaluated as an indicator of normal biological or pathogenic processes, or as an indicator of pharmacologic response to therapeutic interventions” [64]. The present study was able to quantify and analyze the miRNAome for (i) discordant/complex patients (women with chronic pelvic pain suggestive of endometriosis and both negative clinical examination and imaging findings), (ii) women with early-stage (stage I–II rASRM) and advanced-stage (stage III–IV rASRM) endometriosis, and (iii) women with other gynecological disorders sharing symptoms of endometriosis. To subscribe to biomarker criteria, we hypothesized the relevance of the exhaustive evaluation of all miRNAs associated with endometriosis for 200 saliva samples to unearth the complexity of the disease and its heterogeneity. To our knowledge, this is the first exhaustive sequencing of the human saliva miRNAome in the specific context of endometriosis, and we show that 97.3% of all miRNAs are detectable in the saliva with a homogeneous stability of reads. Our analysis resulted in the selection of a set of 109 miRNAs robustly tested.
Several studies have reported aberrant expression of miRNAs in affected tissues or peripheral blood samples of patients with endometriosis [20,22,23,62,63,64]. Several miRNAs have been shown to be dysregulated during the pathogenic process of endometriosis [20,22,23]. Diagnostic power of several miRNAs has been assessed in endometriosis [19,20,22,23,65]. For example, Maged et al. have shown that serum miR-122 and miR-199a had a sensitivity of 95.6 and 100.0% and a specificity of 91.4 and 100%, respectively, for diagnosis of disease status in women. Thus, these miRNAs are putative serum biomarkers for endometriosis [66]. To date, Moustafa et al. [65] is the only team to have attempted to build a blood-based miRNA diagnostic signature for endometriosis composed of six miRNAs based on Random Forest analysis. In agreement with previous studies [40,41,42], it would appear illusory that so few miRNAs could reflect the diversity of a multifactorial disorder such as endometriosis, which involves multiple and poorly known signaling pathways. Therefore, we hypothesized the value of (i) analyzing a specific selection of miRNAs, which resulted in a selection of 109; (ii) reducing the number of features to improve the final accuracy; and finally, (iii) using Random Forest model with high accuracy, which supports the value of AI technology. Such an approach has previously been validated in a study showing that a 100-miRNA blood signature was sufficiently stable to provide almost the same classification accuracy across different types of cancers and platforms [40,41]. Previous studies have demonstrated that saliva miRNA expression analysis can differentiate Crohn’s disease from ulcerative colitis and are of value in head and neck, pancreatic-biliary tract, and oral cancers but failed to report a true diagnostic signature [29,31]. Nevertheless, Rapado-González et al. reported a salivary miRNA signature composed of 22 miRNAs in colorectal cancer patients vs. healthy individuals in the discovery phase [30]. Moreover, Cheng et al. demonstrated the relevance of miRNA saliva expression to diagnose Yang and Yin Deficiency with a panel of 81 and 96 miRNAs, respectively [67].
From a pathophysiologic point of view, after systematic review, among the 109 miRNAs of our endometriosis signature, only four miRNAs (miR-34c-5p, miR-19b-1-5p, miR-149-5p, and miR-378a-3p) have previously been reported in patients with endometriosis; 25 have not been reported previously, implying that further studies should be conducted to confirm their involvement in the pathophysiology of endometriosis; and the remaining 80 are known to be involved in various signaling pathways, such as PI3K/Akt, PTEN, Wnt/β-catenin, HIF1α/NF κB, and YAP/TAZ/EGFR, with potential therapeutic implications.
Some limitations of the present study deserve to be discussed. First, although 97.3% of the human miRNAome were detectable and analyzed in the saliva samples, we cannot rule out that the remaining 2.7% of miRNAs are not involved in endometriosis. Second, as for blood sampling, a potential bias could be linked to the use of hormonal treatment: some patients in both the endometriosis and control group had undergone prior hormonal treatment that might have affected miRNA expression. However, previous studies [19,65] have reported that no significant miRNA changes are observed either during the menstrual cycle or in response to sex steroid hormone therapies [19]. Finally, as previously mentioned for the blood-based miRNA signature, another potential bias could be related to the inclusion of patients with deep endometriosis and/or endometrioma without laparoscopic control in the endometriosis group. However, previous studies have demonstrated the high accuracy of MRI to diagnose endometrioma and deep endometriosis with colorectal involvement, reaching the criteria for a replacement and SnNout triage test [5,7]. Although, our prospective study is the largest available on miRNA and saliva (n = 200) [61], the sample size, especially concerning the control group (n = 47), and the internal cross validation warrants an external validation. Our signature exhibits higher accuracy in patients over 18 years old, and our population did not include adolescents. Therefore, it is not possible to extrapolate our results in this specific population, impending further studies.
7. Conclusions and Perspectives
Despite some limits of the current prospective study, our data support the use of a saliva-based diagnostic miRNA signature for endometriosis in the diagnosis care pathways after an external validation to confirm these results. Saliva sampling is a cheap and non-invasive process and can be repeated multiple times, thus potentially improving both the diagnostic and therapeutic management of patients through early identification and for all populations. Finally, beyond the context of endometriosis, our methodology could be used as a blueprint to investigate other pathologies, both benign and malignant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm11030612/s1. Annex S1: STARD 2015: Stands for “Standards for Reporting Diagnostic accuracy studies; Annex S2: MIRNome Sequencing Analysis Pipeline adaptation from Potla et al. [43]; Annex S3: mi RNA signature accuracy, pathophysiology and signaling pathways.
Author Contributions
S.B., Y.D., S.S. and E.D. conceived and designed the study; S.B., Y.D., C.T., A.P. and E.D. included patients and performed the surgical procedures; all authors analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Part of this work was funded by a grant from the Conseil Régional d’Ile de France (grant number EX024087) and from Ziwig Health.
Institutional Review Board Statement
The experimental protocol was approved by le comité de protection des personnes (C.P.P.) Sud-Ouest et Outre-Mer 1 (CPP 1-20-095 ID 10476).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available at sofiane.bendifallah@aphp.fr.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Giudice, L.C. Clinical Practice. Endometriosis. N. Engl. J. Med. 2010, 362, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- van der Zanden, M.; Teunissen, D.A.M.; van der Woord, I.W.; Braat, D.D.M.; Nelen, W.L.D.M.; Nap, A.W. Barriers and Facilitators to the Timely Diagnosis of Endometriosis in Primary Care in the Netherlands. Fam. Pract. 2020, 37, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Duffy, J.M.N.; Adamson, G.D.; Benson, E.; Bhattacharya, S.; Bhattacharya, S.; Bofill, M.; Brian, K.; Collura, B.; Curtis, C.; Evers, J.L.H.; et al. Top 10 Priorities for Future Infertility Research: An International Consensus Development Study. Hum. Reprod. Oxf. Engl. 2020, 35, 2715–2724. [Google Scholar] [CrossRef]
- Nisenblat, V.; Bossuyt, P.M.; Farquhar, C.; Johnson, N.; Hull, M.L. Imaging Modalities for the Non-Invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016. [Google Scholar] [CrossRef]
- Bazot, M.; Lafont, C.; Rouzier, R.; Roseau, G.; Thomassin-Naggara, I.; Daraï, E. Diagnostic Accuracy of Physical Examination, Transvaginal Sonography, Rectal Endoscopic Sonography, and Magnetic Resonance Imaging to Diagnose Deep Infiltrating Endometriosis. Fertil. Steril. 2009, 92, 1825–1833. [Google Scholar] [CrossRef]
- Bazot, M.; Darai, E.; Hourani, R.; Thomassin, I.; Cortez, A.; Uzan, S.; Buy, J.-N. Deep Pelvic Endometriosis: MR Imaging for Diagnosis and Prediction of Extension of Disease. Radiology 2004, 232, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Lorusso, F.; Scioscia, M.; Rubini, D.; Stabile Ianora, A.A.; Scardigno, D.; Leuci, C.; De Ceglie, M.; Sardaro, A.; Lucarelli, N.; Scardapane, A. Magnetic Resonance Imaging for Deep Infiltrating Endometriosis: Current Concepts, Imaging Technique and Key Findings. Insights Imaging 2021, 12, 105. [Google Scholar] [CrossRef]
- Soliman, A.M.; Rahal, Y.; Robert, C.; Defoy, I.; Nisbet, P.; Leyland, N.; Singh, S. Impact of Endometriosis on Fatigue and Productivity Impairment in a Cross-Sectional Survey of Canadian Women. JOGC 2021, 43, 10–18. [Google Scholar] [CrossRef]
- Surrey, E.; Soliman, A.M.; Trenz, H.; Blauer-Peterson, C.; Sluis, A. Impact of Endometriosis Diagnostic Delays on Healthcare Resource Utilization and Costs. Adv. Ther. 2020, 37, 1087–1099. [Google Scholar] [CrossRef] [Green Version]
- As-Sanie, S.; Soliman, A.M.; Evans, K.; Erpelding, N.; Lanier, R.; Katz, N.P. Healthcare Utilization and Cost Burden among Women with Endometriosis by Opioid Prescription Status in the First Year after Diagnosis: A Retrospective Claims Database Analysis. J. Med. Econ. 2020, 23, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Shi, B.; Huang, X.; Duan, J.; Qiu, Y.; Ha, C.; Huang, R.; Xiao, D.; Liu, J.; Xuan, J. Cost-Effectiveness Analysis of Dienogest Compared With Gonadotropin-Releasing Hormone Agonist After Conservative Surgery for Endometriosis in China. Clin. Ther. 2021, 43, 1276–1284.e1. [Google Scholar] [CrossRef] [PubMed]
- Simoens, S.; Dunselman, G.; Dirksen, C.; Hummelshoj, L.; Bokor, A.; Brandes, I.; Brodszky, V.; Canis, M.; Colombo, G.L.; DeLeire, T.; et al. The Burden of Endometriosis: Costs and Quality of Life of Women with Endometriosis and Treated in Referral Centres. Hum. Reprod. Oxf. Engl. 2012, 27, 1292–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisenblat, V.; Prentice, L.; Bossuyt, P.M.; Farquhar, C.; Hull, M.L.; Johnson, N. Combination of the Non-Invasive Tests for the Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016, 2016, CD012281. [Google Scholar] [CrossRef]
- Nisenblat, V.; Bossuyt, P.M.; Shaikh, R.; Farquhar, C.; Jordan, V.; Scheffers, C.S.; Mol, B.W.J.; Johnson, N.; Hull, M.L. Blood Biomarkers for the Non-Invasive Diagnosis of Endometriosis. Cochrane Database Syst. Rev. 2016, 2016, CD012179. [Google Scholar] [CrossRef] [Green Version]
- Surrey, E.; Carter, C.M.; Soliman, A.M.; Khan, S.; DiBenedetti, D.B.; Snabes, M.C. Patient-Completed or Symptom-Based Screening Tools for Endometriosis: A Scoping Review. Arch. Gynecol. Obstet. 2017, 296, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Akter, S.; Xu, D.; Nagel, S.C.; Bromfield, J.J.; Pelch, K.E.; Wilshire, G.B.; Joshi, T. GenomeForest: An Ensemble Machine Learning Classifier for Endometriosis. AMIA Jt. Summits Transl. Sci. Proc. AMIA Jt. Summits Transl. Sci. 2020, 2020, 33–42. [Google Scholar]
- Kleczyk, E.J.; Peri, A.; Yadav, T.; Komera, R.; Peri, M.; Guduru, V.; Amirtharaj, S.; Huang, M. Predicting Endometriosis Onset Using Machine Learning Algorithms. Res. Sq. 2020. in review. [Google Scholar] [CrossRef]
- Vanhie, A.; O, D.; Peterse, D.; Beckers, A.; Cuéllar, A.; Fassbender, A.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T. Plasma MiRNAs as Biomarkers for Endometriosis. Hum. Reprod. Oxf. Engl. 2019, 34, 1650–1660. [Google Scholar] [CrossRef]
- Agrawal, S.; Tapmeier, T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.; Becker, C. The MiRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef] [Green Version]
- Santamaria-Martos, F.; Benítez, I.; Ortega, F.; Zapater, A.; Giron, C.; Pinilla, L.; Pascual, L.; Cortijo, A.; Dalmases, M.; Fernandez-Real, J.M.; et al. Circulating MicroRNA Profile as a Potential Biomarker for Obstructive Sleep Apnea Diagnosis. Sci. Rep. 2019, 9, 13456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. Role of Non-Coding RNAs in the Pathogenesis of Endometriosis. Front. Oncol. 2020, 10, 1370. [Google Scholar] [CrossRef] [PubMed]
- Panir, K.; Schjenken, J.E.; Robertson, S.A.; Hull, M.L. Non-Coding RNAs in Endometriosis: A Narrative Review. Hum. Reprod. Update 2018, 24, 497–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M. RNAi, MicroRNAs, and Human Disease. Cancer Chemother. Pharmacol. 2006, 58 (Suppl. 1), S63–S68. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Nematian, S.E.; Mamillapalli, R.; Kadakia, T.S.; Majidi Zolbin, M.; Moustafa, S.; Taylor, H.S. Systemic Inflammation Induced by MicroRNAs: Endometriosis-Derived Alterations in Circulating MicroRNA 125b-5p and Let-7b-5p Regulate Macrophage Cytokine Production. J. Clin. Endocrinol. Metab. 2018, 103, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Anastasiu, C.V.; Moga, M.A.; Elena Neculau, A.; Bălan, A.; Scârneciu, I.; Dragomir, R.M.; Dull, A.-M.; Chicea, L.-M. Biomarkers for the Noninvasive Diagnosis of Endometriosis: State of the Art and Future Perspectives. Int. J. Mol. Sci. 2020, 21, 1750. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-W.; Eun, Y.-G.; Lee, Y.-C. Diagnostic Value of Salivary MiRNA in Head and Neck Squamous Cell Cancer: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2021, 22, 7026. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Majem, B.; Álvarez-Castro, A.; Díaz-Peña, R.; Abalo, A.; Suárez-Cabrera, L.; Gil-Moreno, A.; Santamaría, A.; López-López, R.; Muinelo-Romay, L.; et al. A Novel Saliva-Based MiRNA Signature for Colorectal Cancer Diagnosis. J. Clin. Med. 2019, 8, 2029. [Google Scholar] [CrossRef] [Green Version]
- Setti, G.; Pezzi, M.E.; Viani, M.V.; Pertinhez, T.A.; Cassi, D.; Magnoni, C.; Bellini, P.; Musolino, A.; Vescovi, P.; Meleti, M. Salivary MicroRNA for Diagnosis of Cancer and Systemic Diseases: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R.S.; Jakymiw, A.; Yao, B.; Pauley, B.A.; Carcamo, W.C.; Katz, J.; Cheng, J.Q.; Chan, E.K.L. High Resolution of MicroRNA Signatures in Human Whole Saliva. Arch. Oral Biol. 2011, 56, 1506–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.W.; et al. STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies. BMJ 2015, 351, h5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreiseitl, S.; Ohno-Machado, L. Logistic Regression and Artificial Neural Network Classification Models: A Methodology Review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Crown, W.H. Potential Application of Machine Learning in Health Outcomes Research and Some Statistical Cautions. Value Health J. Int. Soc. Pharm. Outcomes Res. 2015, 18, 137–140. [Google Scholar] [CrossRef] [Green Version]
- Ghassemi, M.; Naumann, T.; Schulam, P.; Beam, A.L.; Chen, I.Y.; Ranganath, R. A Review of Challenges and Opportunities in Machine Learning for Health. AMIA 2020, 2020, 191–200. [Google Scholar]
- Sanal, M.G.; Paul, K.; Kumar, S.; Ganguly, N.K. Artificial Intelligence and Deep Learning: The Future of Medicine and Medical Practice. J. Assoc. Physicians India 2019, 67, 71–73. [Google Scholar]
- Goyal, A.; Kuchana, M.; Ayyagari, K.P.R. Machine Learning Predicts Live-Birth Occurrence before in-Vitro Fertilization Treatment. Sci. Rep. 2020, 10, 20925. [Google Scholar] [CrossRef]
- Subramanian, M.; Wojtusciszyn, A.; Favre, L.; Boughorbel, S.; Shan, J.; Letaief, K.B.; Pitteloud, N.; Chouchane, L. Precision Medicine in the Era of Artificial Intelligence: Implications in Chronic Disease Management. J. Transl. Med. 2020, 18, 472. [Google Scholar] [CrossRef]
- Lopez-Rincon, A.; Mendoza-Maldonado, L.; Martinez-Archundia, M.; Schönhuth, A.; Kraneveld, A.D.; Garssen, J.; Tonda, A. Machine Learning-Based Ensemble Recursive Feature Selection of Circulating MiRNAs for Cancer Tumor Classification. Cancers 2020, 12, 1785. [Google Scholar] [CrossRef]
- Lopez-Rincon, A.; Martinez-Archundia, M.; Martinez-Ruiz, G.U.; Schoenhuth, A.; Tonda, A. Automatic Discovery of 100-MiRNA Signature for Cancer Classification Using Ensemble Feature Selection. BMC Bioinform. 2019, 20, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metselaar, P.I.; Mendoza-Maldonado, L.; Li Yim, A.Y.F.; Abarkan, I.; Henneman, P.; Te Velde, A.A.; Schönhuth, A.; Bosch, J.A.; Kraneveld, A.D.; Lopez-Rincon, A. Recursive Ensemble Feature Selection Provides a Robust MRNA Expression Signature for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Sci. Rep. 2021, 11, 4541. [Google Scholar] [CrossRef] [PubMed]
- Metzemaekers, J.; Haazebroek, P.; Smeets, M.J.G.H.; English, J.; Blikkendaal, M.D.; Twijnstra, A.R.H.; Adamson, G.D.; Keckstein, J.; Jansen, F.W. EQUSUM: Endometriosis QUality and Grading Instrument for SUrgical Performance: Proof of Concept Study for Automatic Digital Registration and Classification Scoring for r-ASRM, EFI and Enzian. Hum. Reprod. Open 2020, 2020, hoaa053. [Google Scholar] [CrossRef] [PubMed]
- Potla, P.; Ali, S.A.; Kapoor, M. A Bioinformatics Approach to MicroRNA-Sequencing Analysis. Osteoarthr. Cartil. Open 2021, 3, 100131. [Google Scholar] [CrossRef]
- LaRocca, D.; Barns, S.; Hicks, S.D.; Brindle, A.; Williams, J.; Uhlig, R.; Johnson, P.; Neville, C.; Middleton, F.A. Comparison of Serum and Saliva MiRNAs for Identification and Characterization of MTBI in Adult Mixed Martial Arts Fighters. PLoS ONE 2019, 14, e0207785. [Google Scholar] [CrossRef] [Green Version]
- Gyvyte, U.; Juzenas, S.; Salteniene, V.; Kupcinskas, J.; Poskiene, L.; Kucinskas, L.; Jarmalaite, S.; Stuopelyte, K.; Steponaitiene, R.; Hemmrich-Stanisak, G.; et al. MiRNA Profiling of Gastrointestinal Stromal Tumors by Next-Generation Sequencing. Oncotarget 2017, 8, 37225–37238. [Google Scholar] [CrossRef]
- Gyvyte, U.; Kupcinskas, J.; Juzenas, S.; Inciuraite, R.; Poskiene, L.; Salteniene, V.; Link, A.; Fassan, M.; Franke, A.; Kupcinskas, L.; et al. Identification of Long Intergenic Non-Coding RNAs (LincRNAs) Deregulated in Gastrointestinal Stromal Tumors (GISTs). PLoS ONE 2018, 13, e0209342. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. MiRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhang, L. Construction and Comprehensive Analysis of a CeRNA Network to Reveal Potential Prognostic Biomarkers for Lung Adenocarcinoma. BMC Cancer 2021, 21, 849. [Google Scholar] [CrossRef]
- Li, Q.; Liu, G.; Bao, Y.; Wu, Y.; You, Q. Evaluation and Application of Tools for the Identification of Known MicroRNAs in Plants. Appl. Plant Sci. 2021, 9, e11414. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bargaje, R.; Hariharan, M.; Scaria, V.; Pillai, B. Consensus MiRNA Expression Profiles Derived from Interplatform Normalization of Microarray Data. RNA 2010, 16, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, J.-M.; Jézéquel, P.; Gillois, P.; Silva, L.; Ben Azzouz, F.; Lambert-Lacroix, S.; Juin, P.; Campone, M.; Gaultier, A.; Moreau-Gaudry, A.; et al. Random Forest of Perfect Trees: Concept, Performance, Applications, and Perspectives. Bioinform. Oxf. Engl. 2021. [Google Scholar] [CrossRef] [PubMed]
- Lecointre, L.; Bund, V.; Sangnier, E.; Ouldamer, L.; Bendifallah, S.; Koskas, M.; Bolze, P.-A.; Collinet, P.; Canlorbe, G.; Touboul, C.; et al. Status of Surgical Management of Borderline Ovarian Tumors in France: Are Recommendations Being Followed? Multicentric French Study by the FRANCOGYN Group. Ann. Surg. Oncol. 2021, 28, 7616–7623. [Google Scholar] [CrossRef] [PubMed]
- Geoffron, S.; Lier, A.; de Kermadec, E.; Sermondade, N.; Varinot, J.; Thomassin-Naggara, I.; Bendifallah, S.; Daraï, E.; Chabbert-Buffet, N.; Kolanska, K. Fertility Preservation in Women with Malignant and Borderline Ovarian Tumors: Experience of the French ESGO-Certified Center and Pregnancy-Associated Cancer Network (CALG). Gynecol. Oncol. 2021, 161, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Harrell, F.E.J.; Lee, K.L.; Mark, D.B. Multivariable Prognostic Models: Issues in Developing Models, Evaluating Assumptions and Adequacy, and Measuring and Reducing Errors. Stat. Med. 1996, 15, 361–387. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Eijkemans, M.J.; Harrell, F.E.J.; Habbema, J.D. Prognostic Modelling with Logistic Regression Analysis: A Comparison of Selection and Estimation Methods in Small Data Sets. Stat. Med. 2000, 19, 1059–1079. [Google Scholar] [CrossRef]
- Kern, F.; Krammes, L.; Danz, K.; Diener, C.; Kehl, T.; Küchler, O.; Fehlmann, T.; Kahraman, M.; Rheinheimer, S.; Aparicio-Puerta, E.; et al. Validation of Human MicroRNA Target Pathways Enables Evaluation of Target Prediction Tools. Nucleic Acids Res. 2021, 49, 127–144. [Google Scholar] [CrossRef]
- Wan, N.; Weinberg, D.; Liu, T.-Y.; Niehaus, K.; Ariazi, E.A.; Delubac, D.; Kannan, A.; White, B.; Bailey, M.; Bertin, M.; et al. Machine Learning Enables Detection of Early-Stage Colorectal Cancer by Whole-Genome Sequencing of Plasma Cell-Free DNA. BMC Cancer 2019, 19, 832. [Google Scholar] [CrossRef] [Green Version]
- Monnaka, V.U.; Hernandes, C.; Heller, D.; Podgaec, S. Overview of MiRNAs for the Non-Invasive Diagnosis of Endometriosis: Evidence, Challenges and Strategies. A Systematic Review. Einstein Sao Paulo Braz. 2021, 19, eRW5704. [Google Scholar] [CrossRef] [PubMed]
- Courts, C.; Madea, B. Specific Micro-RNA Signatures for the Detection of Saliva and Blood in Forensic Body-fluid Identification. J. Forensic Sci. 2011, 56, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- El-Mogy, M.; Lam, B.; Haj-Ahmad, T.A.; McGowan, S.; Yu, D.; Nosal, L.; Rghei, N.; Roberts, P.; Haj-Ahmad, Y. Diversity and Signature of Small RNA in Different Bodily Fluids Using next Generation Sequencing. BMC Genom. 2018, 19, 408. [Google Scholar] [CrossRef] [PubMed]
- Kraus, V.B.; Burnett, B.; Coindreau, J.; Cottrell, S.; Eyre, D.; Gendreau, M.; Gardiner, J.; Garnero, P.; Hardin, J.; Henrotin, Y.; et al. Application of Biomarkers in the Development of Drugs Intended for the Treatment of Osteoarthritis. Osteoarthr. Cartil. 2011, 19, 515–542. [Google Scholar] [CrossRef] [Green Version]
- Moustafa, S.; Burn, M.; Mamillapalli, R.; Nematian, S.; Flores, V.; Taylor, H.S. Accurate Diagnosis of Endometriosis Using Serum MicroRNAs. Am. J. Obstet. Gynecol. 2020, 223, 557.e1–557.e11. [Google Scholar] [CrossRef]
- Maged, A.M.; Deeb, W.S.; El Amir, A.; Zaki, S.S.; El Sawah, H.; Al Mohamady, M.; Metwally, A.A.; Katta, M.A. Diagnostic Accuracy of Serum MiR-122 and MiR-199a in Women with Endometriosis. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 2018, 141, 14–19. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Yao, H.; Luo, H.; Lin, B.; Zhang, X.; Liang, X.; Sun, R.; Zhao, S.; Li, Y.; et al. MiRNA Expression Profile of Saliva in Subjects of Yang Deficiency Constitution and Yin Deficiency Constitution. Cell. Physiol. Biochem. 2018, 49, 2088–2098. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).