1. Introduction
The immune system is an intricate network of cells, tissues, and signaling molecules that collectively protect the body from infections, eliminate harmful agents, and maintain internal stability or homeostasis [
1]. While traditional research models have contributed significantly to our understanding of immune function, they present critical limitations that immune organoids are uniquely positioned to address [
2].
Traditional two-dimensional (2D) cell cultures, though offering simplicity and reproducibility, fundamentally lack the cellular interactions and three-dimensional architecture essential for replicating in vivo immune environments [
3,
4]. Animal models, despite their systemic complexity, often exhibit species-specific differences that limit direct translation to human immune responses [
5]. These limitations create a critical gap in our ability to accurately model human immune pathogenesis and predict therapeutic responses.
Immune organoids bridge this translational gap by providing intermediate complexity that combines the experimental control of in vitro systems with the physiological relevance approaching in vivo conditions. These 3D structures are multicellular self-organized tissues that mimic the architecture, functions, and complexities of their corresponding in vivo organs [
6,
7]. Conceptually, immune organoids occupy a unique position in the research model continuum, offering higher physiological relevance than 2D cultures while maintaining greater experimental control than animal models [
8,
9,
10].
In recent years, advances in bioengineering and stem cell biology have enabled the development of immune organoids that partially recreate the microenvironments of lymphoid tissues, such as lymph nodes, spleen, and thymus. These models facilitate the study of specific immune processes, including antigen presentation, T-cell activation, B-cell maturation, and cytokine production [
11,
12]. However, current immune organoid models typically replicate only selected features of immune tissues rather than their full cellular and structural complexity [
13,
14,
15].
The transformative potential of immune organoids lies in their applications across cancer immunotherapy and autoimmune disease research. In oncology, they enable testing of checkpoint inhibitors [
16] and CAR-T cells under human-relevant conditions [
17]. For autoimmune diseases, they provide platforms to explore immune dysregulation mechanisms [
18] and screen immunomodulatory therapies [
19]. Additionally, they show promise in infectious disease research and vaccine development [
20].
While immune organoids successfully address the physiological irrelevance of 2D cultures and the species-specificity limitations of animal models, critical challenges remain in replicating systemic immune interactions, achieving standardization of fabrication protocols, and scaling production for clinical applications. These limitations currently restrict their widespread adoption as predictive models for immunotherapy development. This narrative review provides a comprehensive analysis of immune organoid development and applications, with particular emphasis on cancer immunotherapy and autoimmune disease research. We examine current fabrication techniques, therapeutic applications, and the challenges limiting clinical translation, while proposing evidence-based solutions and future directions for this rapidly evolving field.
2. Methods
This narrative review was conducted to synthesize and critically appraise the existing literature on the development and applications of immune organoids in cancer immunotherapy and autoimmune disease research. The methodology followed a structured yet flexible approach consistent with accepted practices for narrative reviews, aiming to offer comprehensive insights into recent advancements, current challenges, and future directions in the field.
2.1. Search Strategy and Sources
An extensive literature search was conducted across multiple electronic databases, including PubMed, Scopus, Web of Science, Google Scholar, and ScienceDirect, covering publications from 2000 to April 2025. Search terms included combinations of keywords such as “immune organoids,” “cancer immunotherapy,” “autoimmune diseases,” “immune-oncology,” “organoid models,” “3D culture,” “CAR-T cells,” “checkpoint inhibitors,” and “immune system modeling.” Boolean operators (AND, OR) were used to expand or limit search queries. Priority was given to peer-reviewed original research articles, systematic reviews, and high-impact narrative reviews. Supplementary searches included the manual screening of reference lists from relevant articles and gray literature, including preprints and regulatory documents.
2.2. Inclusion and Exclusion Criteria
Studies were included if they:
Focused on the design, fabrication, or application of immune organoids;
Explored immune organoids in relation to cancer immunotherapy, autoimmune disease modeling, or drug testing;
Were published in English and available in full text.
Exclusion criteria comprised:
Studies limited solely to non-immune organoid systems (e.g., brain or liver organoids without immune components);
Articles not related to human immune modeling (e.g., plant or microbial organoids);
Editorials, opinion pieces, and news articles without substantive scientific data.
2.3. Data Extraction and Synthesis
Relevant information was extracted from selected studies, including study objectives, methodologies, organoid type (stem cell-derived, tissue-derived, or bioprinted), cell sources, immune cell components, applications in immunotherapy or disease modeling, reported outcomes, limitations, and scalability challenges. Data synthesis involved thematic analysis, whereby studies were grouped under conceptual themes, such as organoid development, immunotherapy applications, fabrication techniques, and technological limitations. Tables were generated to summarize key features of immune organoids, comparative advantages over traditional models, advances in co-culture systems, and integration with artificial intelligence (AI) and biomaterials.
2.4. Quality Appraisal
Although formal risk of bias tools is not standard in narrative reviews, efforts were made to prioritize high-quality studies from reputable journals. Where possible, findings were cross-referenced with existing systematic reviews or meta-analyses to assess the consistency. Recent developments and innovations were critically evaluated for maturity, reproducibility, and translational potential.
3. Development of Immune Organoids
The development of immune organoids has revolutionized the study of human immune function by providing a sophisticated platform to replicate selected aspects of the complex interactions within the immune system. Unlike traditional models, immune organoids enable researchers to recreate 3D structures with cellular diversity, structural organization, and functional features reflective of in vivo systems, though a full replication of organ-level complexity remains a future goal [
21].
Table 1 compares immune organoids with traditional 2D cultures and animal models, emphasizing their superior physiological relevance and predictive value for human responses.
3.1. Definition and Characteristics
Immune organoids are 3D bioengineered constructs designed to mimic critical features of immune tissues [
6,
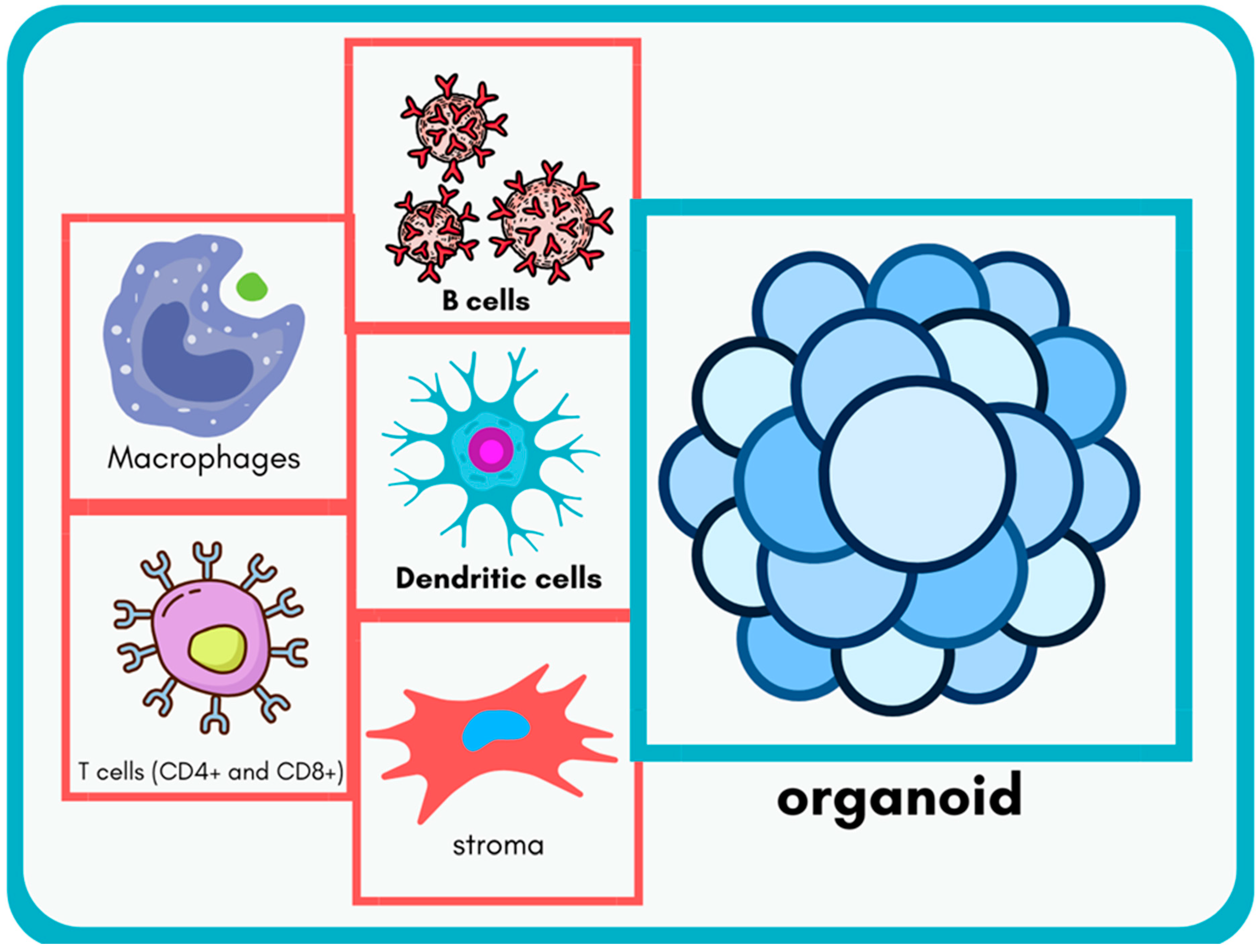
7]. They are derived from stem cells or primary tissues and emulate key structures, such as lymph nodes, spleen, and thymus. One defining feature is their cellular diversity, which includes immune cell types, like T cells, B cells, macrophages, dendritic cells, and stromal cells. This heterogeneity is essential for reproducing immune processes, such as antigen presentation, T-cell activation, and antibody production. For instance, B cells within these organoids can undergo class-switch recombination and somatic hypermutation, crucial steps for producing high-affinity antibodies [
26].
Another hallmark is structural organization, which facilitates cellular interactions in a spatially defined manner, akin to natural immune tissues. For example, lymphoid organoids can recreate follicular structures where B cells interact with T follicular helper cells, providing insights into germinal center dynamics [
27]. Functionally, immune organoids simulate key immune processes, including cytokine secretion and antigen-specific T-cell proliferation. Nonetheless, it must be emphasized that current organoid models generally allow the exploration of a subset of immune functions rather than fully replicating immune tissue complexity. The defining features of immune organoids, including their cellular diversity and structural organization, are depicted in
Figure 1, which illustrates the main immune cell types and stromal elements that collectively recapitulate the complexity of natural immune tissues. On a molecular level, immune organoids enable the study of intracellular signaling cascades central to immune activation. For instance, antigen presentation by dendritic cells and macrophages triggers T-cell receptor (TCR) signaling in CD4
+ and CD8
+ T cells via the ZAP-70–LAT–SLP76 axis, leading to NF-κB, AP-1, and NFAT activation [
28,
29]. B-cell maturation within organoids is governed by signals through the B-cell receptor (BCR) complex and co-stimulation via CD40-CD40L interactions, activating downstream pathways, such as PI3K/AKT and MAPK, to drive class-switch recombination and somatic hypermutation [
30]. Furthermore, cytokine secretion (e.g., IL-2, IL-21, IFN-γ) and receptor engagement (e.g., IL-2Rα) trigger the JAK/STAT signaling axis, shaping T-cell expansion and fate decisions [
31].
3.2. Fabrication Techniques
The fabrication of immune organoids involves sophisticated bioengineering techniques, each offering distinct advantages depending on research objectives. To guide method selection, we present each approach using a unified framework of advantages, limitations, and key applications.
3.2.1. Stem Cell-Derived Models
The advantages of stem cell-derived models include their exceptional scalability, as iPSCs and ESCs provide unlimited cell sources for large-scale production, and their potential for standardization through consistent genetic backgrounds that reduce donor variability. These models offer superior controllability through defined differentiation protocols that enable precise lineage specification, and they excel in disease modeling by incorporating patient-specific mutations for personalized studies.
However, several limitations constrain their utility. The extended maturation time, requiring weeks to months for functional immune cell development, poses logistical challenges for time-sensitive applications. Additionally, these models may exhibit incomplete differentiation, lacking full phenotypic maturation compared to primary cells, and they require substantial technical complexity demanding specialized expertise in stem cell culture and differentiation protocols.
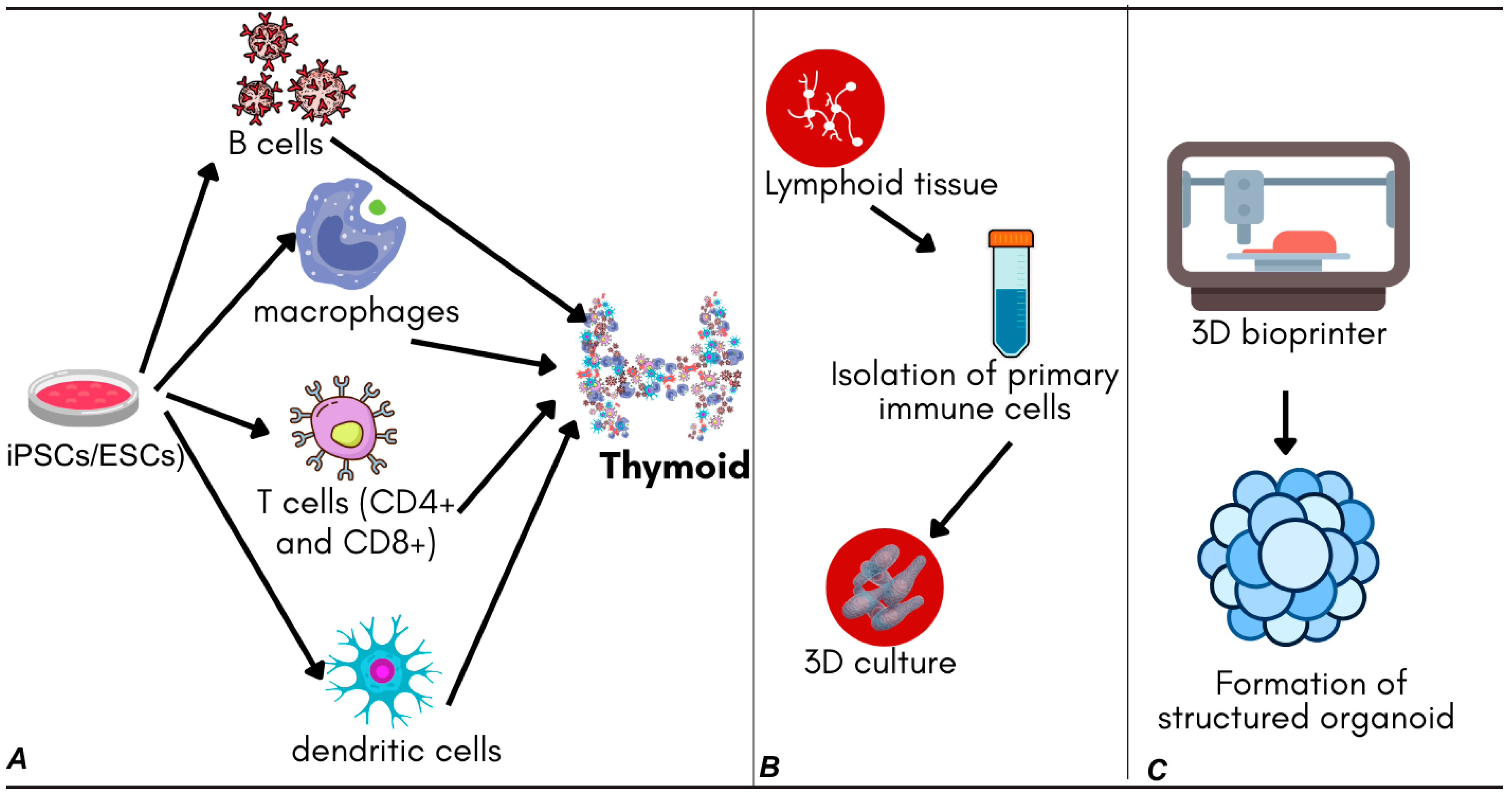
Stem cell-derived models rely on the pluripotent capacity of induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs), which are directed to differentiate into immune cell lineages under tightly controlled conditions. Recent advances in differentiation protocols have enabled the generation of thymic organoids that support T-cell maturation, using defined combinations of growth factors and cytokines to mimic thymic stromal environments [
32,
33]. These models provide valuable insights into immune cell development and are especially useful for studying congenital immunodeficiencies and T-cell-based immunotherapies.
Molecular control over lineage specification is achieved using stage-specific exposure to morphogens and transcriptional regulators. For example, differentiation into thymic epithelial lineage involves the activation of FOXN1, a master regulator of thymic organogenesis, modulated by BMP4 and FGF7 signaling [
34]. Similarly, Notch signaling via DLL4 and Notch1 engagement is critical for T-cell lineage commitment from hematopoietic progenitors, activating downstream genes, such as GATA3, BCL11B, and TCF7 [
35].
3.2.2. Tissue-Derived Models
Tissue-derived models offer significant advantages through their preservation of native complexity, maintaining endogenous cellular heterogeneity and tissue architecture that closely reflects physiological conditions. These models provide superior physiological relevance by preserving natural cell–cell interactions and signaling networks, enable rapid establishment with shorter timeframes to achieve functional organoids compared to stem cell approaches, and demonstrate excellent disease specificity by capturing patient-specific pathological features.
Nevertheless, these models face important limitations, including substantial donor variability that creates significant heterogeneity between different tissue sources, limited scalability due to dependence on primary tissue availability, standardization challenges that make it difficult to achieve consistent protocols across donors, and ethical considerations requiring fresh tissue procurement from patients.
Tissue-derived models use primary lymphoid tissues, such as the tonsils, lymph nodes, or spleen, to isolate and culture immune cells within a three-dimensional framework. This approach retains much of the native cellular heterogeneity and tissue architecture, allowing for more physiologically accurate modeling of immune responses. Tonsil-derived organoids, for example, have demonstrated the ability to recapitulate key immunological features, such as germinal center formation and antigen-driven antibody production, making them suitable for studying humoral immunity [
4,
36].
3.2.3. Three-Dimensional Bioprinting Approaches
Three-dimensional bioprinting approaches provide unique advantages through their spatial precision, enabling the controlled positioning of different cell types and ECM components with unprecedented accuracy. These methods offer exceptional architectural control, allowing the recreation of complex tissue zonation and compartmentalization, provide extensive customization opportunities for designing organoids with specific structural features, and enhance reproducibility through standardized printing protocols that reduce batch-to-batch variation.
However, significant limitations constrain their widespread adoption. Technical barriers require specialized equipment and bioink formulation expertise that may not be readily available in all laboratories. The printing process itself may compromise cell viability and function, while the current resolution limits prevent the achievement of single-cell precision. Additionally, cost considerations include higher initial investment and operational expenses compared to conventional organoid culture methods.
Three-dimensional bioprinting represents a cutting-edge technique that allows the precise spatial arrangement of immune cells and extracellular matrix (ECM) components through the layer-by-layer deposition of bio-inks. This method has enabled the creation of organoid structures that mimic lymphoid tissue compartments, facilitating the study of immune cell migration, antigen presentation, and vaccine efficacy. Bioprinted models have already been used to evaluate vaccine candidates by reproducing functional zones akin to lymphoid follicles [
26,
37,
38].
3.2.4. Method Selection Decision Framework
To assist researchers in selecting the optimal fabrication approach, we propose a comprehensive decision framework based on specific research requirements and constraints. Stem cell-derived models should be chosen when large-scale, standardized production is required, when studying genetic diseases or rare immune disorders, when long-term scalability is a priority, or when standardization across multiple laboratories is essential for reproducible results.
Tissue-derived models represent the optimal choice when maximum physiological relevance is critical for the research question, when studying patient-specific responses or disease heterogeneity, when rapid organoid establishment is needed for time-sensitive applications, or when native immune architecture preservation is essential for accurate modeling.
Three-dimensional bioprinting approaches should be selected when precise spatial control over cell positioning is required for the experimental design, when studying immune cell migration or tissue zonation phenomena, when custom organoid architectures are needed for specific applications, or when standardized, reproducible protocols are a priority for multi-site studies.
Figure 2 provides a visual overview of these fabrication strategies and their respective applications.
Despite these advancements, several challenges persist across all approaches, including achieving full functional maturation of immune cells, ensuring reproducibility between batches, and developing protocols that allow for large-scale production suitable for translational research and drug screening applications.
4. Applications for Immunotherapy Development
The development of immune organoids has opened transformative avenues in immunotherapy research, providing a human-relevant platform to study aspects of immune responses, develop novel therapies, and optimize existing treatments. Although immune organoids replicate important elements of the immune microenvironment, it is important to recognize that their current complexity allows the exploration of selected immune functions rather than full replication of in vivo immune tissues. In the context of cancer and autoimmune disease research, immune organoids, often in combination with tumor-derived organoids or co-cultures, have emerged as promising tools to enhance precision medicine efforts.
Table 2 categorizes different types of immune organoids, such as lymphoid and thymic organoids, highlighting their source materials and specific applications.
4.1. Cancer Immunotherapy
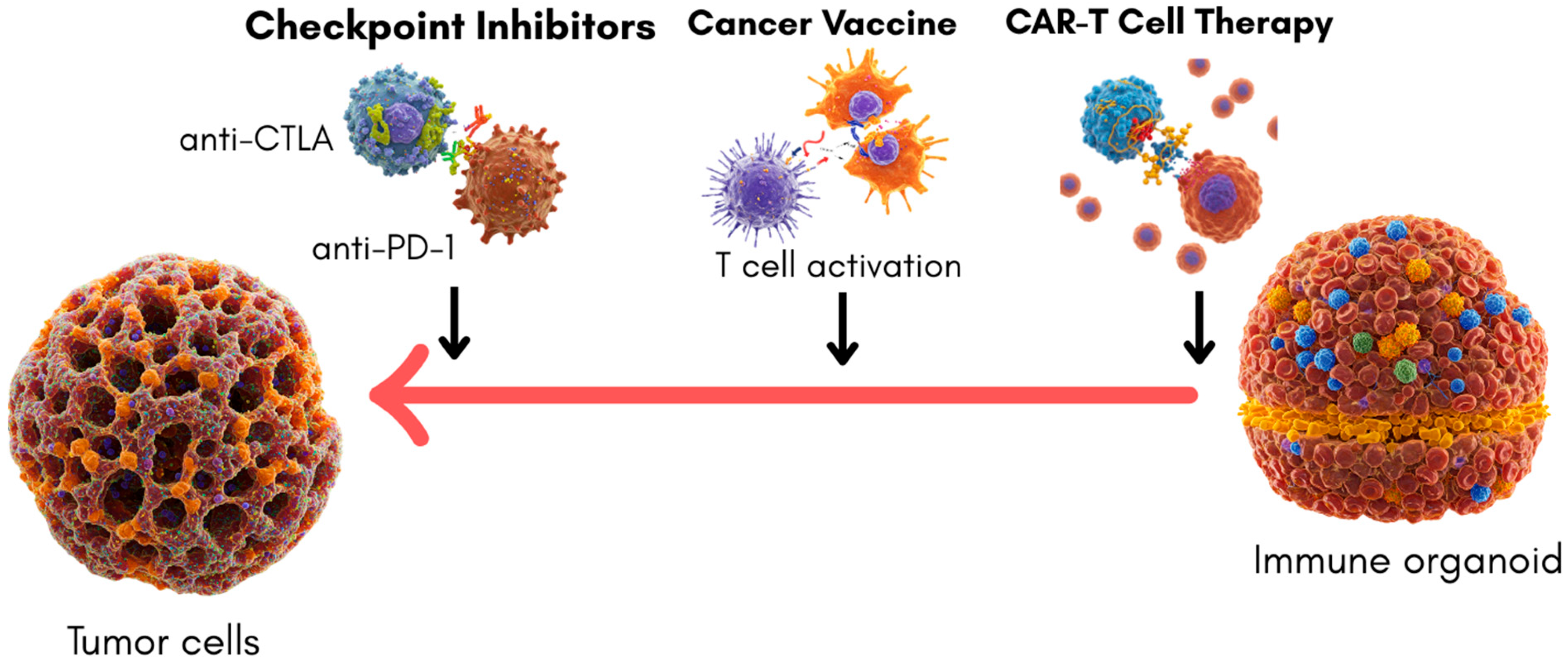
Immune organoids offer new opportunities for studying tumor–immune interactions, albeit primarily through co-culture systems with tumor organoids rather than as stand-alone immune structures. Their ability to recreate specific immune microenvironments provides valuable insights into therapeutic efficacy and mechanism of action, although they currently model only a subset of the complex tumor–immune dynamics. Notably, they allow the interrogation of immune checkpoint blockade, CAR-T-cell activation, and, to a lesser extent, cancer vaccine responses. However, the translational utility of immune organoids depends on how well these models predict actual clinical outcomes.
Checkpoint Inhibitors, such as those targeting PD-1/PD-L1 and CTLA-4 pathways, have revolutionized cancer therapy. Co-culturing tumor-derived organoids with immune cells or immune organoids has enabled researchers to evaluate checkpoint inhibitor activity in a setting that approximates human physiology [
44]. However, pure immune organoid models for studying checkpoint blockade without the presence of tumor cells remain largely undeveloped. Molecular interrogation within co-cultured immune organoids allows for the quantification of PD-1/PD-L1 pathway suppression of TCR-mediated signaling [
45]. Engagement of PD-1 with PD-L1 inhibits PI3K and Ras-MAPK pathways through SHP2 recruitment, attenuating T-cell activation and cytokine secretion [
46]. The restoration of IL-2, IFN-γ, and granzyme B expression following anti-PD-1 treatment serves as a readout for functional reactivation [
47]. These represent robust proof-of-concept findings. However, only a few studies have correlated ex vivo immune organoid responses with clinical treatment outcomes. Dijkstra et al. [
48] demonstrated that patient-derived tumor organoid co-cultures with autologous T cells could predict immune checkpoint blockade responsiveness, including resistance patterns that mirrored clinical non-response. While promising, these findings remain preliminary.
CAR-T-cell therapy similarly benefits from co-culture approaches. On a molecular level, CAR-T cells in immune organoids have shown intracellular calcium flux and the upregulation of effector genes (e.g., GZMB, IFNG, TNF) upon antigen recognition, reflecting real-time activation through synthetic CD3ζ and 4-1BB/CD28 domains [
49]. Organoid systems permit the analysis of early transcriptional events using single-cell RNA-seq, capturing signatures of T-cell exhaustion or memory differentiation post-therapy [
50]. Immune cells, including CAR-T cells, have been tested for cytotoxic activity against tumor organoids, providing a functional readout of efficacy, persistence, and off-target effects [
51]. Despite these mechanistic insights, the current studies remain preclinical and have not yet demonstrated a predictive correlation with CAR-T efficacy or toxicity in patients.
Cancer vaccines represent another promising area where immune organoid systems could eventually be impactful. Current models typically involve the use of tumor organoids or lymphoid-like structures in co-culture settings to simulate antigen presentation and T-cell activation [
52,
53,
54,
55]. For example, inspired by lymph node architecture, 3D lymphoid aggregates have been used to study dendritic cell maturation and vaccine-driven T-cell activation [
56,
57]. Yet, these remain early-stage platforms, with no validated correlation between in vitro vaccine responses and actual patient immunogenicity or tumor control. Thus, while conceptually promising, vaccine-related organoid systems remain in early exploratory phases. A summary of how immune organoids are used to model and evaluate major cancer immunotherapy strategies and also “clinical Readiness” scale to guide translational interpretation is illustrated in
Figure 3 and
Table 3, respectively.
Overall, while these 3D models represent a substantial improvement over traditional 2D assays, their current use in translational immunotherapy research is largely exploratory. Further progress will require harmonized protocols, larger patient cohorts, and longitudinal validation of organoid-derived biomarkers in prospective clinical trials.
4.2. Autoimmune Disease Research
Autoimmune diseases present unique research challenges due to their complex pathophysiology and significant patient-to-patient variability [
58]. Immune organoids offer promising but still developing platforms for studying specific aspects of autoimmune dysregulation, with applications varying considerably across different disease contexts.
4.2.1. Disease-Specific Organoid Applications
Rheumatoid Arthritis (RA) Models: RA organoid models primarily focus on synovial tissue–immune cell interactions, emphasizing the role of Th17/Treg imbalances in joint destruction. These models typically incorporate synovial fibroblasts, macrophages, and T-cell subsets to study inflammatory cascades involving TNF-α, IL-1β, and IL-17 [
59]. Success rates for establishing functional RA organoids are in the range of 60–80% depending on tissue source and culture conditions. Key cellular components include elevated Th17:Treg ratios (typically 3:1 compared to 1:1 in healthy controls) and activated M1 macrophages expressing high levels of CD68 and TNF-α [
43].
Systemic Lupus Erythematosus (SLE) Models: SLE organoids emphasize autoantibody production and B-cell dysregulation, often incorporating patient-derived B cells, plasma cells, and dendritic cells. These models focus on germinal center dysfunction and aberrant class-switch recombination leading to anti-nuclear antibody production. Establishment success rates are lower (40–60%) due to the complexity of recreating systemic autoimmune processes in vitro. Key features include elevated BAFF/APRIL signaling and defective regulatory B-cell (Breg) populations [
60].
Multiple Sclerosis (MS) Models: MS organoids typically combine neural tissue organoids with immune cell infiltrates, modeling neuroinflammation and demyelination processes. These models emphasize oligodendrocyte–microglia interactions and Th1/Th17 cell infiltration across blood–brain barrier mimics [
61,
62].
4.2.2. Patient Variability and Personalized Approaches
Immune organoids show promise in capturing heterogeneous clinical phenotypes observed in autoimmune diseases. Studies utilizing patient-derived organoids have demonstrated significant variability in drug responses that correlate with clinical treatment outcomes. For example, patient-derived organoids from seven individuals with epithelial ovarian cancer were shown to closely replicate clinical responses to platinum-based chemotherapy, supporting their predictive validity [
63]. Similarly, organoids generated from patients with colorectal cancer peritoneal metastases demonstrated the capacity to forecast patient outcomes following cytoreductive surgery combined with hyperthermic intraperitoneal chemotherapy [
64]. However, variability in response depends on individual clonal properties [
65]. This heterogeneity underscores the potential value of organoids for personalized medicine approaches.
Table 4 summarizes disease-specific organoid characteristics, success rates, and key findings.
Modeling Disease Pathogenesis has primarily involved generating organoids from affected tissues, such as the gut, pancreas, or synovium, rather than from immune tissues themselves [
56]. Some co-culture studies incorporating immune cells with these organoid systems have been used to model aspects of autoimmunity, such as T-cell infiltration and cytokine dysregulation [
18,
43]. Mechanistic studies using immune organoids have begun to characterize autoimmune T-cell activation via hyperresponsiveness to self-antigens presented by MHC-II complexes. This involves the dysregulation of central tolerance pathways, including impaired autoimmune regulator (AIRE) expression in thymic epithelial cells and overactivation of STAT1/STAT3 in response to cytokines, such as IL-6 and IFN-γ [
66,
67,
68]. Aberrant Th17/Treg balance, mediated through the differential activation of RORγt and FOXP3, is another hallmark that can be recapitulated in gut-associated immune organoids responding to IL-1β and IL-23 [
69]. Nonetheless, the literature on pure immune organoids recreating autoimmune pathogenesis remains scarce currently.
Drug screening in autoimmune diseases using organoid models is an emerging field, though it currently has limited examples. While there is substantial research involving organoids to study disease pathogenesis and treatment responses, such as testing TNF-α inhibitors in rheumatoid arthritis models [
59], applications specifically for the high-throughput drug screening of autoimmune therapies using immune organoids have not yet been widely explored. However, patient-derived organoids show significant potential for advancing personalized therapies, particularly in cancer. These models are already being employed in preclinical drug screening and for predicting patient-specific treatment responses [
70,
71].
Most drug screening efforts have primarily focused on tissue-derived organoids, particularly those derived from tumors [
72,
73], with only a few studies examining organoids derived from pluripotent stem cells [
74,
75]. The novelty of using patient-specific immune organoids lies in their potential to screen for sensitive drugs and offer tailored treatment options for cancer. These personalized immune organoids provide a theoretical framework for predicting therapeutic responses and identifying biomarkers. Furthermore, integrating omics technologies could enable more individualized interventions [
76], though the clinical application of these approaches remains a goal for the future rather than an immediate reality.
Table 4.
Disease-specific autoimmune organoid models.
Table 4.
Disease-specific autoimmune organoid models.
| Disease | Organoid Model Types | Key Cellular Components | Methods Used | Clinical Application | Success Rate | Drug Screening Outcomes | Therapeutic Response | Patient Variability Captured |
|---|
| Rheumatoid Arthritis [77,78,79] | Synovial–immune organoid | Synovial fibroblast-like synoviocytes (FLS), M1-like macrophages, endothelial cells | 3D co-culture of synovial fibroblasts, endothelial cells, macrophages | Pathogenesis modeling, drug screening | High (reproducible co-cultures with FLS and immune cells) | Inflammatory readouts, including upregulated TNF-α and IL-6; relevant for drug evaluation though response rates in organoids not quantified | Recapitulates TNF-α/IL-1β-driven inflammation | Significant variability in FLS behavior across patients; RA-FLS from different individuals show heterogeneity in cytokine production and matrix remodeling |
| Systemic Lupus Erythematosus [57,80,81] | Germinal center-like B-cell organoid | B cells, plasma cells, dendritic cells, defective Bregs, BAFF/APRIL | Patient B cells/plasma cells + DCs in scaffold | Autoantibody study, screening B-cell-targeted therapy | In vivo variability high in BAFF expression and autoantibody profiles | Belimumab (anti-BAFF) reduces naïve/transitional B-cell fractions (~90% depletion in T3 cells), correlating with modest clinical responses (~58% achieving SRI-4 vs. ~46% placebo in trials) | BAFF-driven B-cell dysregulation; supports belimumab modeling with significant outcome | High variability in autoantibody production (10-fold difference between patients) |
| Multiple Sclerosis [61,62] | Neural immune assembloid | Th1/Th17 T cells, CD20+ B cells, activated microglia, oligodendrocytes | iPSC-derived cerebral organoid ± immune module | Pathogenesis modeling, targets for therapeutic outcome | Moderate (CNS assembloids with iPSC-derived organoids increasingly used) | Anti-CD20 therapies (e.g., rituximab, ocrelizumab) reduce B-cell activation; modeled via CD20+ T-cell depletion | Decrease oligodendrocyte differentiation, decrease proliferation markers, neuroinflammation modeling | Progressive vs. relapsing-remitting phenotypes show distinct organoid characteristics |
| Type 1 Diabetes [82,83] | Islet–immune organoid | Islet β cells, autoreactive CD8+/CD4+ T cells, Tregs | Engineered pluripotent stem cell transplant | Autoimmunity modeling, β-cell therapy screening | Moderate (islet-like organoids are established, immune integration ongoing) | Teplizumab delays T1D onset; organoid-based screening in development; β-cell destruction modeled in co-cultures | Insulin independence within 75 days post-transplantation and maintained glycemic stability for over a year | Variable degrees of insulitis among donors; age-dependent changes in immune infiltration and β-cell autoimmunity |
5. Challenges and Limitations
While immune organoids hold immense promise as transformative platforms for studying immune responses and developing immunotherapies, their widespread adoption is hindered by several biological and technical challenges. These limitations stem from the complexity of replicating immune system functions, limited maturation of organoid models, heterogeneity among organoids, scalability challenges, and, to a lesser extent, resource demands.
5.1. Complexity of Immune Responses
One of the primary challenges in developing immune organoids is replicating the systemic complexity of immune interactions. The immune system operates as a dynamic, multi-layered network involving crosstalk between various cell types and organs, such as the lymph nodes, spleen, thymus, and bone marrow. Current immune organoid models often focus on localized processes, but lack the systemic integration necessary to fully recapitulate immune cell trafficking, antigen presentation across tissues, and systemic cytokine dissemination [
8,
26]. Moreover, interactions between the immune system and non-immune tissues, such as epithelial barriers in the gut or skin, are challenging to model without integrated multi-organoid or organ-on-a-chip approaches [
84]. Additionally, many immune organoid systems are still limited by incomplete tissue complexity, missing specific immune subpopulations (e.g., stromal cells, follicular dendritic cells), or architectural features critical for faithful immune responses [
85]. As a result, while immune organoids provide valuable insights, they currently model only a subset of in vivo immune functionality. Furthermore, the molecular maturation of immune cells within organoids remains incomplete. For instance, insufficient upregulation of activation markers (e.g., CD69, CD25), cytokine transcription (e.g., IL2, IFNG), and memory-associated surface proteins (e.g., CD45RO, CCR7) suggests a lack of full phenotypic and transcriptional differentiation [
86]. Attempts to address this involve optimizing cytokine cocktails (e.g., IL-7, IL-15, IL-21) and ECM stiffness, which affect nuclear translocation of STATs and other transcriptional regulators [
87,
88,
89].
5.2. Lack of Standardization
A significant barrier to the broader application of immune organoids is the lack of standardized protocols for their generation and functional validation. Differences in the source of cells (e.g., iPSCs, ESCs, or primary tissues), culture conditions, extracellular matrices, and differentiation protocols lead to variability in organoid structure, cellular composition, and functionality.
5.2.1. Emerging Standardization Solutions
Several initiatives are addressing standardization challenges through coordinated efforts across the research community. Consensus extracellular matrix (ECM) formulations represent a major advance, with recent efforts focusing on developing standardized, defined ECM hydrogels to replace variable Matrigel preparations. Some studies have proposed guidelines for synthetic hydrogel compositions containing defined concentrations of laminin, collagen IV, and fibronectin for immune organoid culture [
90,
91]. These standardized matrices show 15–20% improvement in batch-to-batch reproducibility compared to undefined natural matrices.
Standardized cytokine cocktails are emerging through consensus protocols for cytokine supplementation during immune organoid differentiation. For thymic organoids, standardized combinations include BMP4 (10 ng/mL), FGF7 (25 ng/mL), and FGF10 (50 ng/mL) for epithelial development, followed by IL-7 (5 ng/mL) and SCF (100 ng/mL) for T-cell maturation [
32,
33]. These protocols show >80% success rates across multiple laboratories when followed precisely.
5.2.2. Mitigating Donor-Specific Effects
Laboratories are implementing several strategies to reduce donor variability through systematic approaches to cell sourcing and culture standardization. iPSC standardization involves using well-characterized iPSC lines from defined genetic backgrounds to reduce donor-specific variation. Pooling multiple iPSC lines (typically 3–5 donors) in organoid cultures provides more representative population responses while maintaining experimental control [
76].
For tissue-derived organoids, primary cell protocols incorporate standardized approaches, including defined tissue dissociation methods using standardized enzyme concentrations, cell sorting protocols to ensure consistent starting populations, and cryopreservation standards to maintain cell viability across experiments. Computational correction methods represent an emerging solution, with advanced analytical approaches using machine learning to identify and correct for donor-specific batch effects in organoid data, thereby improving cross-laboratory reproducibility [
92,
93].
Without universally accepted benchmarks, such as standardized markers for immune cell activation, antigen presentation, or cytokine production, reproducibility across laboratories remains challenging [
26]. Establishing robust criteria for organoid quality control is critical for their acceptance in both academic and translational settings. A further challenge lies in the inconsistent expression of molecular markers, such as FOXP3 (regulatory T cells), T-BET (Th1 cells), and BCL6 (germinal center B cells), which can compromise functional interpretation [
94,
95]. Molecular phenotyping using flow cytometry or single-cell transcriptomics is essential for validation, but is not yet universally adopted in organoid workflows [
96,
97].
5.3. Scalability
Scaling up immune organoid production remains a significant barrier to their broader application in high-throughput drug screening and personalized therapeutic strategies. While manufacturing costs are an important consideration, the challenges of scalability are multifaceted, affecting the consistency, efficiency, and clinical utility of immune organoid platforms.
One critical challenge is the success rate of establishment. Creating functional immune organoids from patient-derived cells or pluripotent stem cells often yields inconsistent results. Success rates can vary widely depending on donor variability, cell quality, and the technical precision required during organoid fabrication. In some protocols, only partial efficiency is achieved, which limits the feasibility of producing immune organoids at scale for clinical or industrial use [
98]. This variability underscores the need for more robust and standardized protocols that can accommodate biological heterogeneity.
Another concern is the time required for generation. The maturation of immune organoids into fully functional structures capable of simulating in vivo immune responses typically spans several weeks to months. This extended timeline poses logistical challenges for applications requiring rapid turnaround, such as patient-specific immunotherapy testing or emergency pathogen response [
99]. Shortening the time to maturity without compromising organoid functionality is a key area for future development.
Heterogeneity within and between organoid batches is also a substantial limitation. Even organoids derived from the same donor or cell line can vary in size, morphology, cellular composition, and immune functionality. These inconsistencies complicate data interpretation and reduce the reliability of organoids in high-throughput or comparative studies [
100]. Addressing heterogeneity through quality control measures and manufacturing standardization is crucial for increasing the translational potential of immune organoids.
A further technical limitation lies in the lack of vascularization. Most immune organoids currently lack blood vessel-like networks, which restrict nutrient and oxygen diffusion to their inner cores. This results in size constraints and the development of hypoxic regions, impeding long-term viability and functional maturation [
101]. While recent efforts to introduce vascularization into organoid systems show promise, such as co-culturing immune organoids with endothelial cells, integrating vascular organoids, using co-differentiation approaches, employing microfluidic organ-on-a-chip systems, or utilizing 3D bioprinting to create perfusable channels, these methods have not yet been widely adopted or optimized for immune organoid applications [
102].
However, the scalability of immune organoid systems is constrained by biological variability, time-intensive protocols, intra-batch inconsistency, and structural limitations, such as the absence of vasculature. Addressing these issues will require an interdisciplinary innovation and collaborative refinement of both the biological and engineering aspects of organoid technology.
5.4. Limited Complexity and Tissue Architecture
Immune organoids currently lack the full tissue complexity and organization of native immune organs. Critical structural features, such as the zonation seen in lymph nodes or the medulla–cortex organization of the thymus, are often only partially replicated [
103]. Efforts are ongoing to mimic the zonation process using the oxygen biosensor device [
104]. Moreover, key supporting cell types, such as fibroblastic reticular cells, endothelial cells, or specialized antigen-presenting cells, may be absent or insufficiently integrated into the organoids. These cells are crucial for adequate functioning of the immune response to organoids. This limits the fidelity of immune education, activation, and tolerance mechanisms modeled in vitro. Current efforts are aimed at improving architectural fidelity include controlling initial cell seeding density, using bioprinting approaches, and integrating stromal compartments [
105], but these remain areas of active research and development.
5.5. Resource Demands
While the cost of materials (e.g., matrices, cytokines) and infrastructure (e.g., 3D bioreactors, advanced imaging) is non-trivial, relatively few studies have provided detailed analyses of cost as a primary barrier in immune organoid adoption. Thus, while cost may pose challenges, especially for resource-limited settings, biological and scalability challenges currently represent more significant obstacles [
99].
5.6. Addressing the Challenges
Efforts to overcome the limitations of immune organoid development are progressing rapidly. One major barrier, the lack of vascularization, is being actively addressed through microfluidic organ-on-chip systems and perfused vascular networks, which aim to mimic physiological fluid dynamics and improve nutrient and oxygen diffusion [
8,
102]. Recent advances in microfluidic integration have enabled the continuous perfusion of immune–tumor co-cultures, supporting longer-term viability and more stable cytokine gradients [
106]. These systems have enhanced the representation of vascularized tissue niches, yet they still fall short in the complete replication of the lymphatic drainage, which is essential for immune cell recirculation and trafficking. Studies that have attempted to do so are yet to achieve complete anatomical resemblance [
107,
108,
109]. The absence of a functional lymphatic component may impair studies of dendritic cell migration, antigen transport, and T-cell egress, thus limiting the translational fidelity of immune activation and checkpoint response modeling.
In parallel, standardization efforts are being deployed to enhance reproducibility across laboratories. Initiatives focused on harmonizing protocols for cell sourcing, media composition, matrix selection, and functional assays are gaining momentum [
89]. Such measures are critical to reducing variability and ensuring reproducible readouts, particularly in applications like drug screening and immunotherapy response prediction.
From a scalability standpoint, emerging technologies, like high-throughput 3D bioprinting, microfabrication, and robotic culture systems, are being developed to improve the uniformity and throughput of immune organoid generation [
24]. Although production costs remain a barrier, collaborative efforts between academia, biotech, and regulatory agencies are fostering platforms that are more cost-efficient and industrially scalable. These advances are key to accelerating the integration of immune organoids into mainstream preclinical pipelines and, ultimately, clinical decision-making.
6. Future Directions
Immune organoids possess immense potential to revolutionize immunotherapy and immunological research. Fully realizing this potential will require addressing the current limitations through a systematic advancement across multiple fronts, organized into achievable timelines and addressing specific barriers to clinical adoption.
6.1. Timeline-Based Development Strategy
6.1.1. Short-Term Goals (1–3 Years)
Priority development of standardized ECM hydrogels focuses on creating consensus extracellular matrix formulations to replace variable Matrigel preparations. A standardized concentration of ECM to fit all preparation may not be feasible as the concentration of different organoids varies and rightly depends on the organ of interest and the physical properties modeled [
110].
Quality control systems for immune organoid reproducibility increasingly emphasize standardized protocols to ensure consistency across batches. Key criteria include maintaining consistent organoid size, viability, and functional capacity. For example, microwell-based fabrication methods have been employed to reduce variability in organoid morphology and ensure uniform starting cell densities, with intra-batch variation in organoid diameter often maintained below 10% to reduce necrosis and enhance functional reliability [
111]. Viability thresholds of ≥70–80% post-thaw or after extended culture periods are commonly used to define batch acceptance, particularly in protocols aligned with biobanking and clinical-grade production standards [
112]. These practices reflect a growing consensus toward standardizing immune organoid platforms for translational and clinical application.
The establishment of consensus cytokine protocols involves developing standardized differentiation media compositions for major organoid types, with defined concentrations, timing, and supplier specifications. Initial thymic organoid protocols have demonstrated promising levels of reproducibility under standardized laboratory conditions, with studies showing a consistent generation of thymic epithelial cell phenotypes and support for T-cell maturation in vitro. For instance, Liu et al. [
113] reported a robust and uniform formation of miniaturized thymic organoids (mTOs) using a microwell-array system, suitable for medium-throughput applications. While these results affirm protocol stability within controlled settings, large-scale, multi-center validation are highly recommended.
6.1.2. Medium-Term Goals (3–5 Years)
Development of perfusable organoid systems incorporating endothelial networks represents a critical advancement for vascularization integration. Current approaches, including co-culture with human umbilical vein endothelial cells (HUVECs) and bioprinting with vascular channels, show promising outcomes [
102].
Integration of immune organoids with tumor, gut, and skin models will produce multi-organoid platforms that simulate systemic immune responses. Early proof-of-concept studies demonstrate successful immune cell trafficking between connected organoid compartments [
84,
114]. Machine learning algorithms for the real-time optimization of culture conditions represent AI-optimized production capabilities, enabling the prediction of organoid quality metrics and the identification of optimal differentiation protocols. Current systems show 25–30% improvement in success rates when compared to standard protocols [
92,
93,
115,
116].
6.1.3. Long-Term Goals (5+ Years)
Complete organoid platforms with integrated immune niches, perfusion networks, and systemic circulation mimics will enable fully vascularized multi-organoid systems. These systems would facilitate the comprehensive study of immune cell trafficking, systemic cytokine responses, and organ-specific immune interactions [
102,
114].
Patient-derived organoid systems for routine immunotherapy prediction and personalized treatment selection represent the goal of clinical integration. A recent review asserts that the development of immune cell-integrated organoid models will enable a faithful simulation of the tumor microenvironment and integration into clinical decision-making workflows [
117].
Fully automated organoid manufacturing platforms capable of producing thousands of standardized organoids per week will support industrial drug screening applications through automated production systems [
118].
6.2. Multi-Organoid Systems
A promising avenue for advancing immune organoid research is the integration with other organ systems to create multi-organoid platforms. Such systems can more accurately model the systemic nature of immune responses, including interactions between immune cells and non-immune tissues, such as tumors, gut models, and skin. For example, coupling immune organoids with tumor-on-a-chip models enables an investigation of immune surveillance, immune evasion mechanisms, and immunotherapy responses in cancer [
114]. Similarly, integrating immune organoids with gut models could elucidate the influence of the microbiome on immune function [
119], offering insights into autoimmune and inflammatory diseases. Also, incorporating sphingosine-1-phosphate (S1P) signaling gradients into organoid systems may enhance the fidelity of immune cell trafficking and compartmentalization, enabling more physiologic modeling of lymphoid zonation [
120]. These complex, interconnected models would also facilitate the study of systemic immune-related adverse effects, such as cytokine release syndrome, thereby improving drug safety evaluation.
6.3. Advanced Biomaterials
The development of advanced biomaterials tailored to support immune tissue architecture is critical for the next generation of immune organoids. The Wnt/β-catenin signaling pathway plays a critical role in maintaining progenitor cell populations and supporting the self-renewal capacity of immune organoids [
121]. Fine-tuning this pathway may improve organoid yield and viability, especially during scale-up processes. Traditional matrices, such as Matrigel, while supportive, lack the biochemical and mechanical specificity required for optimal immune organoid development [
90]. Future biomaterials should mimic the ECM of lymphoid tissues, supporting appropriate immune cell migration, aggregation, and activation. Innovations such as synthetic hydrogels with tunable stiffness, biofunctionalized scaffolds, and dynamic ECMs responsive to cytokine signals are particularly promising [
90,
122,
123,
124,
125]. These biomaterials would enable the formation of more physiologically relevant structures, such as germinal centers, thus improving the predictive power and clinical relevance of immune organoid systems.
6.4. AI Integration
AI offers transformative potential for optimizing immune organoid research. AI-driven analyses can efficiently process large-scale datasets generated from immune organoid experiments, including transcriptomics, proteomics, and imaging data [
115,
116]. Machine learning algorithms can identify novel biomarkers, predict patient-specific responses to therapies such as checkpoint inhibitors or CAR-T-cell treatments, and guide the optimization of organoid fabrication by identifying key variables that impact reproducibility and functionality [
92,
93]. Furthermore, AI could assist in the real-time monitoring of organoid development and immune responses, enabling the dynamic adjustment of culture conditions to better mimic physiological states [
117]. Integrating AI into the immune organoid research will thus accelerate both discovery and clinical translation.
6.5. Barriers to Clinical Adoption
6.5.1. Regulatory Challenges
The translation of immune organoids into clinical practice faces significant regulatory hurdles that must be systematically addressed through coordinated efforts with regulatory agencies. Currently, no standardized regulatory framework exists for organoid-based diagnostic or therapeutic applications. The FDA’s 2019 guidance on “Tissue-Engineered Medical Products” provides some direction, but specific guidelines for immune organoids remain undefined. Key regulatory requirements likely include the validation of organoid-derived predictions against clinical outcomes in prospective studies, standardized manufacturing protocols meeting Good Manufacturing Practice (GMP) standards, defined quality control metrics and release criteria for clinical-grade organoids, and long-term stability and reproducibility data across multiple sites.
For organoids to be used as predictive biomarkers, they must meet companion diagnostic requirements, including analytical validity, clinical validity, and clinical utility. This process typically requires 3–5 years and substantial investment (running into millions of USD) for regulatory approval [
126].
Coordination between the FDA, EMA, and other international regulatory bodies will be essential for global adoption through international harmonization efforts. The International Council for Harmonisation (ICH) guidelines may need updating to address organoid-specific considerations.
6.5.2. Economic Considerations
While precise cost estimates vary between centers, it is understood that establishing patient-derived organoid lines for drug sensitivity assays entails substantial initial expenses due to specialized infrastructure and reagents. Importantly, if organoid testing helps prevent high-cost treatment failures, particularly in immunotherapy, where expenditures can exceed six figures per patient, the approach may become economically justifiable over a defined patient cohort. Efforts toward automation and scale-up are projected to reduce per-organoid costs and improve consistency, although quantifiable capital and operational metrics are not yet publicly documented. Ultimately, integration into standard care pathways will hinge on the outcomes of prospective clinical validation studies and health technology assessments demonstrating both clinical utility and cost-effectiveness.
6.5.3. Ongoing Clinical Validation Studies
Several clinical studies are actively evaluating the translational potential of organoid technology, particularly in oncology, though the clinical validation of immune organoids remains limited.
One notable effort is the SENSOR Trial (Single-Patient Experimental Therapy Using Organoids in Refractory Cancer), a prospective pilot study in the Netherlands investigating the feasibility of using patient-derived tumor organoids to inform drug selection in metastatic colorectal cancer patients [
127]. Among 54 eligible patients, organoids were successfully generated in 31 cases, 25 cultures underwent drug screening, and 19 exhibited in vitro responses to at least one compound. However, six patients treated based on these results did not achieve objective clinical responses, highlighting the current limitations of organoid-guided therapy in complex metastatic diseases.
In Europe, coordinated efforts, such as the Human Cancer Models Initiative (HCMI), and similar consortium-based programs aim to standardize tumor organoid generation and implement a large-scale, multicenter validation of organoid-based drug sensitivity testing [
128]. These initiatives focus primarily on solid tumors, with growing interest in expanding toward immunotherapy applications, although trials specifically targeting immune organoid models remain at an early stage.
In the United States, organoid research is supported by various NIH programs, including the NIH 4D Nucleome, Cancer Moonshot, and Common Fund initiatives. Multiple NIH-funded consortia support the development of organoid models for disease modeling, therapeutic screening, and regulatory readiness, including over two dozen projects listed in public NIH portfolios [
129].
Together, these programs underscore the translational promise of organoid platforms while also emphasizing the need for larger, well-controlled clinical studies to establish predictive accuracy and reproducibility across disease contexts, including immune-mediated conditions.
6.6. Clinical Translation
Achieving clinical translation of immune organoids requires overcoming the current limitations in their scalability, standardization, and reproducibility. While preclinical studies have shown their potential, particularly in personalized immunotherapy and disease modeling, they need to be translated into practical clinical tools and validated through clinical trials. For example, patient-derived immune organoids could be used to test responses to immunotherapies ex vivo, predicting which patients are most likely to benefit from specific treatments [
130]. Large-scale validation studies, along with the establishment of robust biobanking practices [
131,
132], will be necessary to integrate organoid technologies into clinical workflows. Moreover, regulatory agencies are increasingly interested in 3D models for drug development [
126], but standardized manufacturing practices and reproducibility benchmarks must be achieved to gain widespread regulatory acceptance.
6.7. Long-Term Vision
The long-term vision for immune organoids is their incorporation into personalized medicine frameworks, where patient-specific organoid models inform real-time therapeutic decisions. Multi-organoid systems simulating interactions between different tissues, advanced biomaterials mimicking native tissue ECMs, AI-driven optimization, and rigorous clinical validation will collectively enable the widespread clinical use of immune organoids. Such developments would revolutionize precision immunotherapy, enhance our understanding of immune aging, improve vaccine development pipelines, and offer new models for studying emerging infectious diseases. For example, the modeling of host–pathogen interactions can be improved by engineering immune organoids to express toll-like receptors (TLRs), facilitating innate immune activation in response to microbial ligands or synthetic adjuvants [
133]. As interdisciplinary collaboration across bioengineering, immunology, and computational biology continues to evolve, immune organoids are poised to become indispensable tools in next-generation healthcare. Future work may also explore epigenetic regulation, including chromatin accessibility and histone modifications, to more accurately reproduce lineage commitment and immune memory formation in organoid-based models.
Table 5 summarizes key signaling pathways relevant to immune organoid development and potential applications, now expanded to include therapeutic targeting potential.
7. Limitations of the Review
This narrative review provides a broad and integrative overview of immune organoids and their applications in cancer immunotherapy and autoimmune disease research. However, several limitations should be acknowledged. First, the review is not systematic and therefore lacks the methodological rigor of a formal systematic review or meta-analysis. The selection of studies was based on thematic relevance and expert judgment, which may introduce selection bias and limit reproducibility. Although efforts were made to include high-impact and peer-reviewed literature, the absence of a formal quality assessment tool means that variability in the methodological quality of included studies could influence the conclusions drawn.
Second, the rapidly evolving nature of organoid research poses a challenge to capturing the most recent advancements. Some studies, especially those in preprint or early publication stages, may not have been included despite offering valuable insights. Furthermore, given the diversity of immune organoid models, applications, and fabrication methods, this review could not exhaustively cover all technological variations or disease-specific implementations.
Third, while this review draws connections between immune organoid systems and their translational potential, it does not include a quantitative evaluation of outcomes, such as effect sizes or comparative efficacy, due to the narrative approach. This limits its utility for guiding clinical decision-making or for comparing different organoid models on a standardized scale.
Finally, this review focuses predominantly on human models and does not account for developments in animal-derived organoid systems, which may also contribute valuable insights, particularly in early-stage immunological research. These limitations underscore the need for complementary systematic reviews and experimental studies to build on the themes discussed and validate the translational promise of immune organoids.
8. Conclusions
Immune organoids represent a ground-breaking advancement in immunology, offering high-fidelity, physiologically relevant models to study human immune responses. By bridging the critical gaps left by traditional two-dimensional culture and animal models, immune organoids enable the detailed investigation of immune mechanisms, disease pathogenesis, and therapeutic interventions. Their applications in cancer immunotherapy, autoimmune disease research, and infectious disease modeling highlight their transformative potential, allowing researchers to develop, optimize, and personalize treatments with unprecedented precision. While challenges related to complexity, scalability, heterogeneity, standardization, and biological maturation remain, ongoing advancements in bioengineering, biomaterials, AI, and clinical integration are steadily addressing these limitations through systematic, timeline-based approaches.
The integration of immune organoids into multi-organ systems, coupled with emerging solutions for standardization and scalability challenges, positions these platforms for clinical translation within the next decade. Regulatory frameworks are evolving to accommodate organoid-based diagnostics, while economic analyses support their cost-effectiveness for personalized medicine applications. As the field progresses, immune organoids are poised not only to revolutionize immunotherapy and autoimmune disease management, but also to reshape broader areas of biomedical research and clinical care, including regenerative medicine and vaccine development. Ultimately, these innovative systems hold the promise of improving patient outcomes and ushering in a new era of personalized, effective medical care, with concrete pathways toward clinical implementation now clearly defined.
Author Contributions
Conceptualization, D.B.O., E.O.O. and S.B.; methodology, E.E.; software, S.V.O.; validation, E.O.O. and S.V.O.; formal analysis, S.B.; investigation, D.B.O.; resources, S.B.; data curation, D.B.O. and E.O.O.; writing—original draft preparation, D.B.O. and E.O.O.; writing—review and editing, D.B.O., E.O.O., E.E., S.V.O. and S.B.; visualization, D.B.O.; supervision, S.V.O. and S.B.; project administration, E.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
This work was supported by the Department of Research and Innovation, Medway NHS Foundation Trust, Gillingham, Kent, UK.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nicholson, L.B. The immune system. Essays Biochem. 2016, 60, 275–301. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.S.; Warrington, R.; Watson, W.; Kim, H.L. An introduction to immunology and immunopathology. Allergy. Asthma. Clin. Immunol. 2018, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Chwalek, K.; Bray, L.J.; Werner, C. Tissue-engineered 3D tumor angiogenesis models: Potential technologies for anti-cancer drug discovery. Adv. Drug Deliv. Rev. 2014, 79–80, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hammel, J.H.; Vasudevan, S.A.; Kleinstreuer, N.C. Modeling immunity in vitro: Slices, chips and engineered tissues. Annu. Rev. Biomed. Eng. 2021, 23, 461–491. [Google Scholar] [CrossRef]
- Wagar, L.E.; DiFazio, R.M.; Davis, M.M. Advanced model systems and tools for basic and translational human immunology. Genome Med. 2018, 10, 73. [Google Scholar] [CrossRef]
- Corrò, C.; Novellasdemunt, L.; Li, V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef]
- de Souza, N. Organoids. Nat. Methods 2018, 15, 23. [Google Scholar] [CrossRef]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Papp, D.; Korcsmaros, T.; Hautefort, I. Revolutionizing immune research with organoid-based co-culture and chip systems. Clin. Exp. Immunol. 2024, 218, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Ghose, A.; Naicker, K.; Sanchez, E.; Chargari, C.; Rassy, E.; Boussios, S. Advances in adoptive T-cell therapy for metastatic melanoma. Curr. Res. Transl. Med. 2023, 71, 103404. [Google Scholar] [CrossRef]
- Mastrangeli, M.; van den Eijnden-van Raaij, J. Organs-on-chip: The way forward. Stem Cell Rep. 2021, 16, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Piergiovanni, M.; Leite, S.B.; Corvi, R.; Whelan, M. Standardisation needs for organ on chip devices. Lab Chip 2021, 21, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Paloschi, V.; Sabater-Lleal, M.; Middelkamp, H.; Vivas, A.; Johansson, S.; van der Meer, A.; Tenje, M.; Maegdefessel, L. Organ-on-a-chip technology: A novel approach to investigate cardiovascular diseases. Cardiovasc. Res. 2021, 117, 2742–2754. [Google Scholar] [CrossRef]
- Scognamiglio, G.; De Chiara, A.; Parafioriti, A.; Armiraglio, E.; Fazioli, F.; Gallo, M.; Aversa, L.; Camerlingo, R.; Cacciatore, F.; Colella, G.; et al. Patient-derived organoids as a potential model to predict response to PD-1/PD-L1 checkpoint inhibitors. Br. J. Cancer 2019, 121, 979–982. [Google Scholar] [CrossRef]
- Ahmadi, M.; Putnam, N.; Dotson, M.; Hayoun, D.; Padilla, J.; Fatima, N.; Bhanap, P.; Nonterah, G.; de Mollerat du Jeu, X.; Ji, Y. Accelerating CAR T cell manufacturing with an automated next-day process. Curr. Res. Transl. Med. 2024, 73, 103489. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, Y.; Xu, P.; Feng, X.; Liu, Y.; Lu, Y. The functions and applications of organoids in rheumatic immune diseases. J. Holist. Integr. Pharm. 2024, 5, 141–147. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Assante Miranda, L.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef]
- de Oliveira, L.F.; Filho, D.M.; Marques, B.L.; Maciel, G.F.; Parreira, R.C.; do Carmo Neto, J.R.; Da Silva, P.E.F.; Guerra, R.O.; da Silva, M.V.; Santiago, H.D.C.; et al. Organoids as a novel tool in modelling infectious diseases. Semin. Cell Dev. Biol. 2023, 144, 87–96. [Google Scholar] [CrossRef]
- Ye, W.; Luo, C.; Li, C.; Huang, J.; Liu, F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 2020, 477, 31–40. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Kim, S.; Shah, S.B.; Graney, P.L.; Singh, A. Multiscale engineering of immune cells and lymphoid organs. Nat. Rev. Mater. 2019, 4, 355–378. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Yang, G.-H.; Lee, J.; Kim, G. 3D bioprinting and its in vivo applications. J. Biomed. Mater. Res. B Appl. Biomater. 2018, 106, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; van der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging organoid-immune co-culture models for cancer research: From oncoimmunology to personalized immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef] [PubMed]
- Suhito, I.R.; Chaudhary, S.; Tay, A. Engineering human immune organoids for translational immunology. Bioact. Mater. 2025, 44, 164–183. [Google Scholar] [CrossRef]
- Purwada, A.; Jaiswal, M.K.; Ahn, H.; Nojima, T.; Kitamura, D.; Gaharwar, A.K.; Cerchietti, L.; Singh, A. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 2015, 63, 24–34. [Google Scholar] [CrossRef]
- Hwang, J.R.; Byeon, Y.; Kim, D.; Park, S.-G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020, 52, 750–761. [Google Scholar] [CrossRef]
- Lo, W.L.; Shah, N.H.; Ahsan, N.; Horkova, V.; Stepanek, O.; Salomon, A.R.; Kuriyan, J.; Weiss, J. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 2018, 19, 733–741. [Google Scholar] [CrossRef]
- Luo, W.; Weisel, F.; Shlomchik, M.J. B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity 2018, 48, 313–326.e5. [Google Scholar] [CrossRef]
- Lv, Y.; Qi, J.; Babon, J.J.; Cao, L.; Fan, G.; Lang, J.; Zhang, J.; Mi, P.; Kobe, B.; Wang, F. The JAK-STAT pathway: From structural biology to cytokine engineering. Signal Transduct. Target. Ther. 2024, 9, 221. [Google Scholar] [CrossRef]
- Azar, J.; Donabedian, L.; Karajgikar, J.; Dahle, T.; Li, P.; Samadani, R. The use of stem cell-derived organoids in disease modeling: An update. Int. J. Mol. Sci. 2021, 22, 7667. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.A.; Armitage, L.H.; Morton, J.J.; Alzofon, N.; Handler, D.; Kelly, G.; Homann, D.; Jimeno, A.; Russ, H.A. Generation of functional thymic organoids from human pluripotent stem cells. Stem Cell Rep. 2023, 18, 829–840. [Google Scholar] [CrossRef]
- Tsai, P.T.; Lee, R.A.; Wu, H. BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood 2003, 102, 3947–3953. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, E.V.; Scripture-Adams, D.D. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin. Immunol. 2008, 20, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Wagar, L.E.; Salahudeen, A.; Constantz, C.M.; Wendel, B.S.; Lyons, M.M.; Mallajosyula, V.; Jatt, L.P.; Adamska, J.Z.; Blum, L.K.; Gupta, N.; et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021, 27, 125–135. [Google Scholar] [CrossRef]
- Brassard, J.A.; Nikolaev, M.; Hübscher, T.; Hofer, M.; Lutolf, M.P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021, 20, 22–29. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, W.; Zhao, Q.; Feng, R. Developments and opportunities for 3D bioprinted organoids. Int. J. Bioprint. 2021, 7, 364. [Google Scholar] [CrossRef]
- Seet, C.S.; He, C.; Bethune, M.T.; Li, S.; Chick, B.; Gschweng, E.H.; Zhu, Y.; Kim, K.; Kohn, D.B.; Baltimore, D.; et al. Generation of mature T cells from human hematopoietic stem/progenitor cells in artificial thymic organoids. Nat. Methods 2017, 14, 521–530. [Google Scholar] [CrossRef]
- Wang, J.; Tao, X.; Zhu, J.; Dai, Z.; Du, Y.; Xie, Y.; Chu, X.; Fu, G.; Lei, Z. Tumor organoid-immune co-culture models: Exploring a new perspective of tumor immunity. Cell Death Discov. 2025, 11, 195. [Google Scholar] [CrossRef]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood. Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef]
- Kromann, E.H.; Cearra, A.P.; Neves, J.F. Organoids as a tool to study homeostatic and pathological immune–epithelial interactions in the gut. Clin. Exp. Immunol. 2024, 218, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Moten, D.; Teneva, I.; Apostolova, D.; Batsalova, T.; Dzhambazov, B. Molecular mimicry of the rheumatoid arthritis-related immunodominant T-cell epitope within type II collagen (CII260-270) by the bacterial L-asparaginase. Int. J. Mol. Sci. 2022, 23, 9149. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid models of tumor immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Sugiura, D.; Shimizu, K.; Maruhashi, T.; Watada, M.; Okazaki, I.M.; Okazaki, T. PD-1 Primarily Targets TCR Signal in the Inhibition of Functional T Cell Activation. Front. Immunol. 2019, 10, 630. [Google Scholar] [CrossRef]
- Lin, X.; Kang, K.; Chen, P.; Zeng, Z.; Li, G.; Xiong, W.; Yi, M.; Xiang, B. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer 2024, 23, 108. [Google Scholar] [CrossRef]
- Kaptein, P.; Slingerland, N.; Metoikidou, C.; Prinz, F.; Brokamp, S.; Machuca-Ostos, M.; de Roo, G.; Schumacher, T.N.M.; Yeung, Y.A.; Moynihan, K.D.; et al. CD8-targeted IL2 unleashes tumor-specific immunity in human cancer tissue by reviving the dysfunctional T cell pool. Cancer Discov. 2024, 14, 1226–1251. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Smirnov, S.; Mateikovich, P.; Samochernykh, K.; Shlyakhto, E. Recent advances on CAR-T signaling pave the way for prolonged persistence and new modalities in clinic. Front. Immunol. 2024, 15, 1335424. [Google Scholar] [CrossRef]
- Ginhoux, F.; Yalin, A.; Dutertre, C.A.; Amit, I. Single-cell immunology: Past, present, and future. Immunity 2022, 55, 393–404. [Google Scholar] [CrossRef]
- Ning, R.X.; Wang, H.-Y.; Wang, L. Application status and optimization suggestions of tumor organoids and CAR-T cell co-culture models. Cancer Cell Int. 2024, 24, 98. [Google Scholar] [CrossRef]
- Linares, C.A.; Varghese, A.; Ghose, A.; Shinde, S.D.; Adeleke, S.; Sanchez, E.; Sheriff, M.; Chargari, C.; Rassy, E.; Boussios, S. Hallmarks of the Tumour Microenvironment of Gliomas and Its Interaction with Emerging Immunotherapy Modalities. Int. J. Mol. Sci. 2023, 24, 13215. [Google Scholar] [CrossRef]
- Demmers, L.C.; Kretzschmar, K.; Van Hoeck, A.; Bar-Epraïm, Y.E.; van den Toorn, H.W.P.; Koomen, M.; van Son, G.; van Gorp, J.; Pronk, A.; Smakman, N.; et al. Single-cell derived tumor organoids display diversity in HLA class I peptide presentation. Nat. Commun. 2020, 11, 5338. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, S.; Lin, X.; Chen, G.; Kang, J.; Ma, Z.; Wang, Y.; Li, Z.; Xiao, X.; He, A.; et al. Application of organoids in carcinogenesis modeling and tumor vaccination. Front. Oncol. 2022, 12, 855996. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.Y.; Masi, R.; Gasmi, B.; Hanada, K.I.; Parkhurst, M.; Gartner, J.; Sindiri, S.; Prickett, T.; Robbins, P.; Zacharakis, N.; et al. Using patient-derived tumor organoids from common epithelial cancers to analyze personalized T-cell responses to neoantigens. Cancer Immunol. Immunother. 2023, 72, 3149–3162. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.; Johnson, S.C.; Quek, Y.J.; Li, X.; Tay, A. Integrative lymph node-mimicking models created with biomaterials and computational tools to study the immune system. Mater. Today Bio 2022, 14, 100269. [Google Scholar] [CrossRef]
- Wagar, L.E. Human immune organoids: A tool to study vaccine responses. Nat. Rev. Immunol. 2023, 23, 699. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S. Pathogenesis of autoimmune disease. Nat. Rev. Nephrol. 2023, 19, 509–524. [Google Scholar] [CrossRef]
- Badillo-Mata, J.A.; Camacho-Villegas, T.A.; Lugo-Fabres, P.H. 3D cell culture as tools to characterize rheumatoid arthritis signaling and development of new treatments. Cells 2022, 11, 3410. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, R.; Liang, Q.; Ni, S.; Yang, M.; Qiu, L.; Ji, J.; Gu, Z.; Dong, C. Organ-based characterization of B cells in patients with systemic lupus erythematosus. Front. Immunol. 2025, 16, 1509033. [Google Scholar] [CrossRef]
- Daviaud, N.; Chen, E.; Edwards, T.; Sadiq, S.A. Cerebral organoids in primary progressive multiple sclerosis reveal stem cell and oligodendrocyte differentiation defect. Biol. Open 2023, 12, bio059845. [Google Scholar] [CrossRef] [PubMed]
- de Sèze, J.; Maillart, E.; Gueguen, A.; Laplaud, D.A.; Michel, L.; Thouvenot, E.; Zephir, H.; Zimmer, L.; Biotti, D.; Liblau, R. Anti-CD20 therapies in multiple sclerosis: From pathology to the clinic. Front. Immunol. 2023, 14, 1004795. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Sun, F.; Wang, J.; Wang, Y.; Zhu, H.; Chen, M.; Liu, L.; Liu, L.; Lin, H.; Wu, X. Developing patient-derived organoids to predict PARP inhibitor response and explore resistance overcoming strategies in ovarian cancer. Pharmacol. Res. 2022, 179, 106232. [Google Scholar] [CrossRef] [PubMed]
- Ubink, I.; Bolhaqueiro, A.C.F.; Elias, S.G.; Raats, D.A.E.; Constantinides, A.; Peters, N.A.; Wassenaar, E.C.E.; de Hingh, I.H.J.T.; Rovers, K.P.; van Grevenstein, W.M.U.; et al. Organoids from colorectal peritoneal metastases as a platform for improving hyperthermic intraperitoneal chemotherapy. Br. J. Surg. 2019, 106, 1404–1414. [Google Scholar] [CrossRef]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.K.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable Clonal Repopulation Dynamics Influence Chemotherapy Response in Colorectal Cancer. Science 2012, 339, 543–548. [Google Scholar] [CrossRef]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid models of human gastrointestinal development and disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef]
- Peterson, P.; Org, T.; Rebane, A. Transcriptional regulation by AIRE: Molecular mechanisms of central tolerance. Nat. Rev. Immunol. 2008, 8, 948–957. [Google Scholar] [CrossRef]
- Shirafkan, F.; Hensel, L.; Rattay, K. Immune tolerance and the prevention of autoimmune diseases essentially depend on thymic tissue homeostasis. Front. Immunol. 2024, 15, 1339714. [Google Scholar] [CrossRef]
- Lee, G.R. The Balance of Th17 versus Treg Cells in Autoimmunity. Int. J. Mol. Sci. 2018, 19, 730. [Google Scholar] [CrossRef]
- Zhu, J.; Ji, L.; Chen, Y.; Li, H.; Huang, M.; Dai, Z.; Wang, J.; Xiang, D.; Fu, G.; Lei, Z.; et al. Organoids and organs-on-chips: Insights into predicting the efficacy of systemic treatment in colorectal cancer. Cell Death Discov. 2023, 9, 72. [Google Scholar] [CrossRef]
- Mazzocchi, A.R.; Rajan, S.A.P.; Votanopoulos, K.I.; Hall, A.R.; Skardal, A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci. Rep. 2018, 8, 2886. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Maru, Y.; Tanaka, N.; Itami, M.; Hippo, Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol. Oncol. 2019, 154, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Berkers, G.; van Mourik, P.; Vonk, A.M.; Kruisselbrink, E.; Dekkers, J.F.; de Winter-de Groot, K.M.; Arets, H.G.M.; Marck-van der Wilt, R.E.P.; Dijkema, J.S.; Vanderschuren, M.M.; et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 2019, 26, 1701–1708.e3. [Google Scholar] [CrossRef] [PubMed]
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388. [Google Scholar] [CrossRef]
- Heinzelmann, E.; Piraino, F.; Costa, M.; Roch, A.; Norkin, M.; Garnier, V.; Homicsko, K.; Brandenberg, N. iPSC-derived and patient-derived organoids: Applications and challenges in scalability and reproducibility as pre-clinical models. Curr. Res. Toxicol. 2024, 7, 100197. [Google Scholar] [CrossRef]
- Jin, X.; Huang, L.; Wang, X.; Tan, Y.; Huang, M.; Fu, H.; Wen, C.; Zhou, M. Synovial organoids: From fundamental construction to groundbreaking applications in arthritic disorders. J. Orthop. Transl. 2025, 54, 26–36. [Google Scholar] [CrossRef]
- Philippon, E.M.L.; van Rooijen, L.J.E.; Khodadust, F.; van Hamburg, J.P.; van der Laken, C.J.; Tas, S.W. A novel 3D spheroid model of rheumatoid arthritis synovial tissue incorporating fibroblasts, endothelial cells, and macrophages. Front. Immunol. 2023, 14, 1188835. [Google Scholar] [CrossRef]
- Wang, X.; He, J.; Zhang, Q.; He, J.; Wang, Q. Constructing a 3D co-culture in vitro synovial tissue model for rheumatoid arthritis research. Mater. Today Bio 2025, 31, 101492. [Google Scholar] [CrossRef]
- Stohl, W.; Hiepe, F.; Latinis, K.M.; Thomas, M.; Scheinberg, M.A.; Clarke, A.; Aranow, C.; Wellborne, F.R.; Abud-Mendoza, C.; Hough, D.R.; et al. Belimumab Reduces Autoantibodies, Normalizes Low Complement, and Reduces Select B-Cell Populations in Patients with Systemic Lupus Erythematosus. Arthritis Rheum. 2012, 64, 2328–2337. [Google Scholar] [CrossRef]
- Huang, W.; Quach, T.D.; Dascalu, C.; Liu, Z.; Leung, T.; Byrne-Steele, M.; Pan, W.; Yang, Q.; Han, J.; Lesser, M.; et al. Belimumab promotes negative selection of activated autoreactive B cells in systemic lupus erythematosus patients. JCI Insight 2018, 3, e122525. [Google Scholar] [CrossRef]
- Yu, X.; Liu, C.; Kuang, Z.; Song, S.; Tian, L.; Wang, Y. Islet organoids: A new hope for islet transplantation in diabetes. Front. Immunol. 2025, 15, 1540209. [Google Scholar] [CrossRef]
- Kokori, E.; Olatunji, G.; Ogieuhi, I.J.; Aboje, J.E.; Olatunji, D.; Aremu, S.A.; Igwe, S.C.; Moradeyo, A.; Ajayi, Y.I.; Aderinto, N. Teplizumab’s immunomodulatory effects on pancreatic β-cell function in type 1 diabetes mellitus. Clin. Diabetes Endocrinol. 2024, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Picollet-D’hahan, N.; Zuchowska, A.; Lemeunier, I.; Le Gac, S. Multiorgan-on-a-chip: A systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021, 39, 788–810. [Google Scholar] [CrossRef] [PubMed]
- Wörsdörfer, P.; Takashi, I.; Asahina, I.; Sumita, Y.; Ergün, S. Do not keep it simple: Recent advances in the generation of complex organoids. J. Neural. Transm. 2020, 127, 1569–1577. [Google Scholar] [CrossRef]
- Alvarez, F.; Liu, Z.; Bay, A.; Piccirillo, C.A. Deciphering the developmental trajectory of tissue-resident Foxp3+ regulatory T cells. Front. Immunol. 2024, 15, 1331846. [Google Scholar] [CrossRef]
- Kim, J.K.; Han, S.B.; Park, S.I.; Kim, I.S.; Kim, D.H. Nuclear transport of STAT6 determines the matrix rigidity dependent M2 activation of macrophages. Biomaterials 2022, 290, 121859. [Google Scholar] [CrossRef]
- Rochman, Y.; Spolski, R.; Leonard, W.J. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol. 2009, 9, 480–490. [Google Scholar] [CrossRef]
- Yu, F.; Hunziker, W.; Choudhury, D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines 2019, 10, 165. [Google Scholar] [CrossRef]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.H.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.G.; et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, L.; Zhang, Y. Advances in Organoid Culture Research. Glob. Med. Genet. 2022, 9, 268–276. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, X.; Zeng, S.; Ren, X.; Yan, Y.; Gong, Z. Applying artificial intelligence for cancer immunotherapy. Acta. Pharm. Sin. B 2021, 11, 3393–3405. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Fang, D.; Luo, Y. Informing immunotherapy with multi-omics driven machine learning. NPJ. Digit. Med. 2024, 7, 67. [Google Scholar] [CrossRef]
- Amarnath, S.; Laurence, A.; Zhu, N.; Cunha, R.; Eckhaus, M.A.; Taylor, S.; Foley, J.E.; Ghosh, M.; Felizardo, T.C.; Fowler, D.H. Tbet is a critical modulator of FoxP3 expression in autoimmune graft-versus-host disease. Haematologica 2017, 102, 1446–1456. [Google Scholar] [CrossRef][Green Version]
- Sawant, D.V.; Wu, H.; Yao, W.; Sehra, S.; Kaplan, M.H.; Dent, A.L. The transcriptional repressor Bcl6 controls the stability of regulatory T cells by intrinsic and extrinsic pathways. Immunology 2015, 145, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Li, B.D.; Timm, E.A.; Riedy, M.C.; Harlow, S.P.; Stewart, C.C. Chapter 7 Molecular Phenotyping by Flow Cytometry. Methods Cell Biol. 1994, 42, 95–130. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Fisch, A.S.; Pestana, A.; Sachse, V.; Doll, C.; Hofmann, E.; Heiland, M.; Obermueller, T.; Heidemann, J.; Dommerich, S.; Schoppe, D.; et al. Feasibility analysis of using patient-derived tumour organoids for treatment decision guidance in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2024, 213, 115100. [Google Scholar] [CrossRef]
- Bose, S.; Clevers, H.; Shen, X. Promises and challenges of organoid-guided precision medicine. Med 2021, 2, 1011–1026. [Google Scholar] [CrossRef]
- Jensen, K.B.; Little, M.H. Organoids are not organs: Sources of variation and misinformation in organoid biology. Stem. Cell Rep. 2023, 18, 1255–1270. [Google Scholar] [CrossRef]
- Zhao, Z.; Hardwick, L.; Kornfeld, J.; Grün, D. Organoids. Nat. Rev. Methods Primers 2022, 2, 94. [Google Scholar] [CrossRef]
- Nwokoye, P.N.; Abilez, O.J. Bioengineering methods for vascularizing organoids. Cell Rep. Methods 2024, 4, 100779. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Nishikawa, M.; Kutsuzawa, N.; Tokito, F.; Kobayashi, T.; Kurniawan, D.A.; Shioda, H.; Cao, W.; Shinha, K.; Nakamura, H.; et al. Advancements in Microphysiological systems: Exploring organoids and organ-on-a-chip technologies in drug development -focus on pharmacokinetics related organs. Drug Metab. Pharmacokinet. 2025, 60, 101046. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Safitri, A.R.; Danoy, M.; Maekawa, T.; Kinoshita, H.; Shinohara, M.; Sakai, Y.; Fujii, T.; Leclerc, E. Investigation of the hepatic respiration and liver zonation on rat hepatocytes using an integrated oxygen biosensor in a microscale device. Biotechnol. Prog. 2019, 35, e2854. [Google Scholar] [CrossRef]
- Mirshafiei, M.; Rashedi, H.; Yazdian, F.; Rahdar, A.; Baino, F. Advancements in tissue and organ 3D bioprinting: Current techniques, applications, and future perspectives. Mater. Des. 2024, 240, 112853. [Google Scholar] [CrossRef]
- Gaebler, D.; Hachey, S.J.; Hughes, C.C.W. Improving tumor microenvironment assessment in chip systems through next-generation technology integration. Front. Bioeng. Biotechnol. 2024, 12, 1462293. [Google Scholar] [CrossRef]
- Kim, S.; Chung, M.; Jeon, N.L. Three-dimensional biomimetic model to reconstitute sprouting lymphangiogenesis in vitro. Biomaterials 2016, 78, 115–128. [Google Scholar] [CrossRef]
- Chang, C.-W.; Seibel, A.J.; Song, J.W. Application of microscale culture technologies for studying lymphatic vessel biology. Microcirculation 2019, 26, e12547. [Google Scholar] [CrossRef]
- Lee, S.; Kang, H.; Park, D.; Yu, J.; Koh, S.K.; Cho, D.; Kim, D.; Kang, K.; Jeon, N.L. Modeling 3D Human Tumor Lymphatic Vessel Network Using High-Throughput Platform. Adv. Biol. 2021, 5, 2000195. [Google Scholar] [CrossRef]
- Cai, D.; Weng, W. Development potential of extracellular matrix hydrogels as hemostatic materials. Front. Bioeng. Biotechnol. 2023, 11, 1187474. [Google Scholar] [CrossRef]
- Guo, L.; Li, C.; Gong, W. Toward reproducible tumor organoid culture: Focusing on primary liver cancer. Front. Immunol. 2024, 15, 1290504. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.-R.; Mun, S.J.; Kim, T.H.; Kim, H.; Kang, D.; Kim, S.; Shin, J.H.; Choi, D.; Ahn, S.-J.; Son, M.J. Guidelines for Manufacturing and Application of Organoids: Liver. Int. J. Stem Cells 2024, 17, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fan, Y. A miniaturized approach for robust thymus organoid production. Cell Rep. 2025, 44, 115575. [Google Scholar] [CrossRef] [PubMed]
- Maulana, T.I.; Kromidas, E.; Wallstabe, L.; Cipriano, M.; Alb, M.; Zaupa, C.; Hudecek, M.; Fogal, B.; Loskill, P. Immunocompetent cancer-on-chip models to assess immuno-oncology therapy. Adv. Drug Deliv. Rev. 2021, 173, 281–305. [Google Scholar] [CrossRef]
- Bai, L.; Wang, Y.; Li, G.; Zhang, W.; Zhang, H.; Su, J. AI-enabled organoids: Construction, analysis, and application. Bioact. Mater. 2024, 31, 525–548. [Google Scholar] [CrossRef]
- Maramraju, S.; Kowalczewski, A.; Kaza, A.; Liu, X.; Singaraju, J.P.; Albert, M.V.; Ma, Z.; Yang, H. AI-organoid integrated systems for biomedical studies and applications. Bioeng. Transl. Med. 2024, 9, e10641. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, F.; Zuo, X.; Li, M. Breakthroughs and challenges of organoid models for assessing cancer immunotherapy: A cutting-edge tool for advancing personalised treatments. Cell Death Discov. 2025, 11, 222. [Google Scholar] [CrossRef]
- Renner, H.; Grabos, M.; Becker, K.; Kagermeier, T.; Wu, J.; Otto, M.; Peischard, S.; Zeuschner, D.; Tsytsyura, Y.; Disse, P.; et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. eLife 2020, 9, e52904. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef]
- Aoki, M.; Aoki, H.; Ramanathan, R.; Hait, N.C.; Takabe, K. Sphingosine-1-Phosphate Signaling in Immune Cells and Inflammation: Roles and Therapeutic Potential. Mediat. Inflamm. 2016, 2016, 8606878. [Google Scholar] [CrossRef]
- Xue, C.; Chu, Q.; Shi, Q.; Zeng, Y.; Lu, J.; Li, L. Wnt signaling pathways in biology and disease: Mechanisms and therapeutic advances. Signal Transduct. Target. Ther. 2025, 10, 106. [Google Scholar] [CrossRef]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef]
- Alghuwainem, A.; Alshareeda, A.T.; Alsowayan, B. Scaffold-free 3-D cell sheet technique bridges the gap between 2-D cell culture and animal models. Int. J. Mol. Sci. 2019, 20, 4926. [Google Scholar] [CrossRef]
- Moura, B.S.; Monteiro, M.V.; Ferreira, L.P.; Lavrador, P.; Gaspar, V.M.; Mano, J.F. Advancing tissue decellularized hydrogels for engineering human organoids. Adv. Funct. Mater. 2022, 32, 2202825. [Google Scholar] [CrossRef]
- Gan, Z.; Qin, X.; Liu, H.; Liu, J.; Qin, J. Recent advances in defined hydrogels in organoid research. Bioact. Mater. 2023, 28, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Carter, N. A Three-Pillar Approach to Driving Organoid Adoption in Drug Discovery [Internet]. Biocompare.com. 2024. Available online: https://biocompare.com/Editorial-Articles/610434-A-Three-Pillar-Approach-to-Driving-Organoid-Adoption-in-Drug-Discovery/ (accessed on 11 January 2025).
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Human Cancer Models Initiative (HCMI). Available online: https://www.cancer.gov/ccg/research/functional-genomics/hcmi (accessed on 28 March 2023).
- NIH—Intramural Research Program. Innovation Awards Spark New Intramural Collaborations. Available online: https://irp.nih.gov/blog/post/2020/09/innovation-awards-spark-new-intramural-collaborations (accessed on 1 August 2025).
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef] [PubMed]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; van Ineveld, R.L.; Derakhshan, S.; de Haan, S.; et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020, 11, 1310. [Google Scholar] [CrossRef]
- Sartorius, R.; Trovato, M.; Manco, R.; D’Apice, L.; De Berardinis, P. Exploiting viral sensing mediated by Toll-like receptors to design innovative vaccines. NPJ Vaccines 2021, 6, 127. [Google Scholar] [CrossRef]
- Staal, F.J.T.; Luis, T.C.; Tiemessen, M.M. WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 2008, 8, 581–593. [Google Scholar] [CrossRef]
- Vanderbeck, A.; Maillard, I. Notch signaling at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 2020, 109, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Dibus, M.; Joshi, O.; Ivaska, J. Novel tools to study cell-ECM interactions, cell adhesion dynamics and migration. Curr. Opin. Cell Biol. 2024, 88, 102355. [Google Scholar] [CrossRef] [PubMed]
- Mackay, F.; Schneider, P. Cracking the BAFF code. Nat. Rev. Immunol. 2009, 9, 491–502. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Kanno, Y.; Vahedi, G.; Hirahara, K.; Singleton, K.; O’Shea, J.J. Transcriptional and Epigenetic Control of T Helper Cell Specification: Molecular Mechanisms Underlying Commitment and Plasticity. Annu. Rev. Immunol. 2012, 30, 707–731. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).