Computational Exploration of Bacterial Compounds Targeting Arginine-Specific Mono-Adp-Ribosyl-Transferase 1 (Art1): A Pathway to Novel Therapeutic Anticancer Strategies
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Target and Template Sequences
2.2. Protein Profile Analysis
2.3. Homogy Modeling of Target Protein
2.4. Validation of 3D Target Protein
2.5. Analysis of Physicochemical Characteristics
2.6. Molecular Dynamics Simulation
2.7. Prediction of Active Site
2.8. Preparation of Target Protein
2.9. Preparation of Ligands
2.10. Molecular Docking
2.11. Visualization of Protein–Ligand Interaction
2.12. Biological Activity Prediction
2.13. Pharmacophore Study
2.14. Drug-Likeness and ADMET Study
3. Results and Discussion
3.1. Selection of Target and Template Sequences
3.2. Protein Profile Analysis
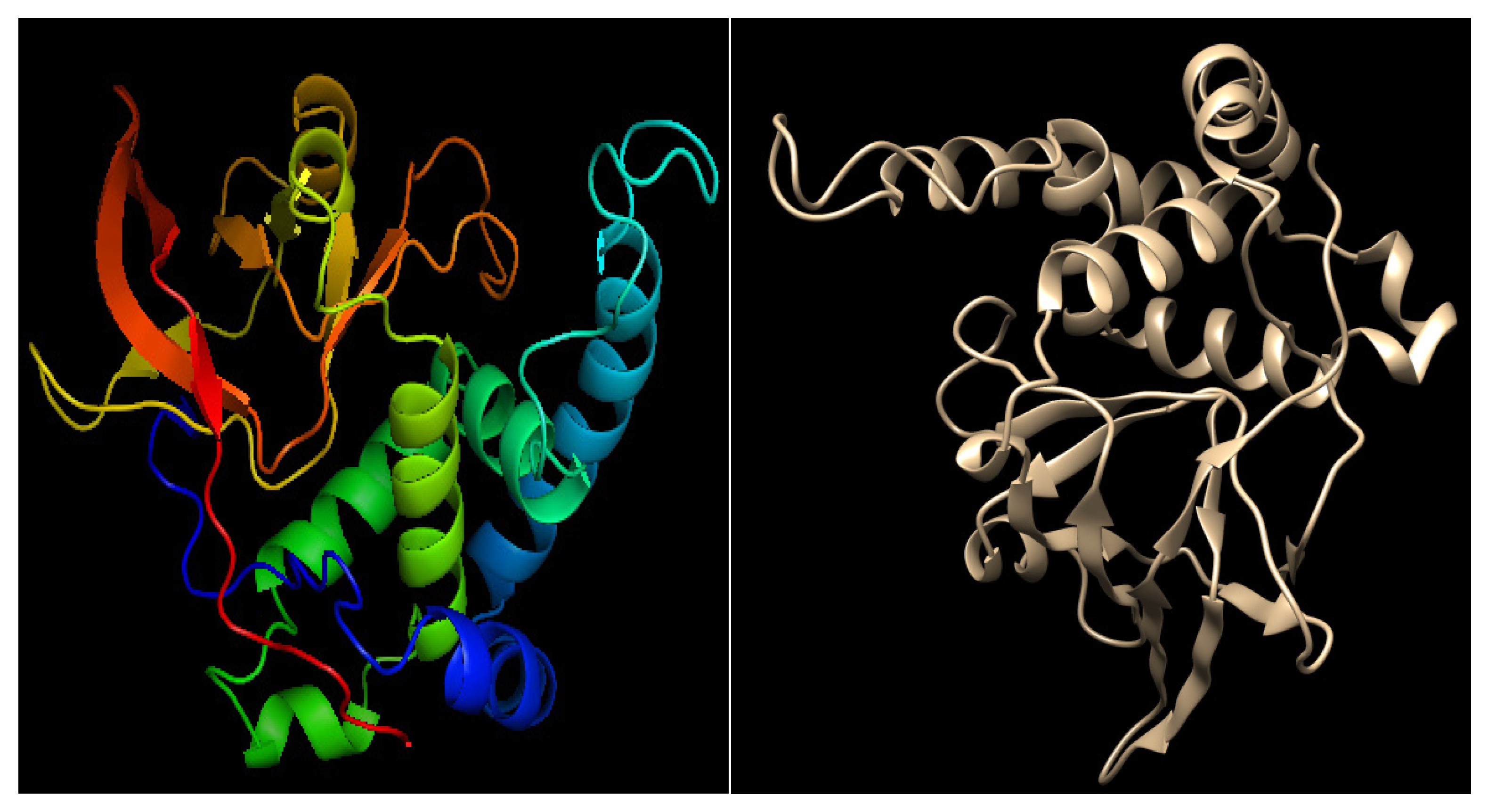
3.3. Homology Modeling and Validation
3.4. Analysis of Physicochemical Characteristics
3.5. Molecular Dynamic Simulation
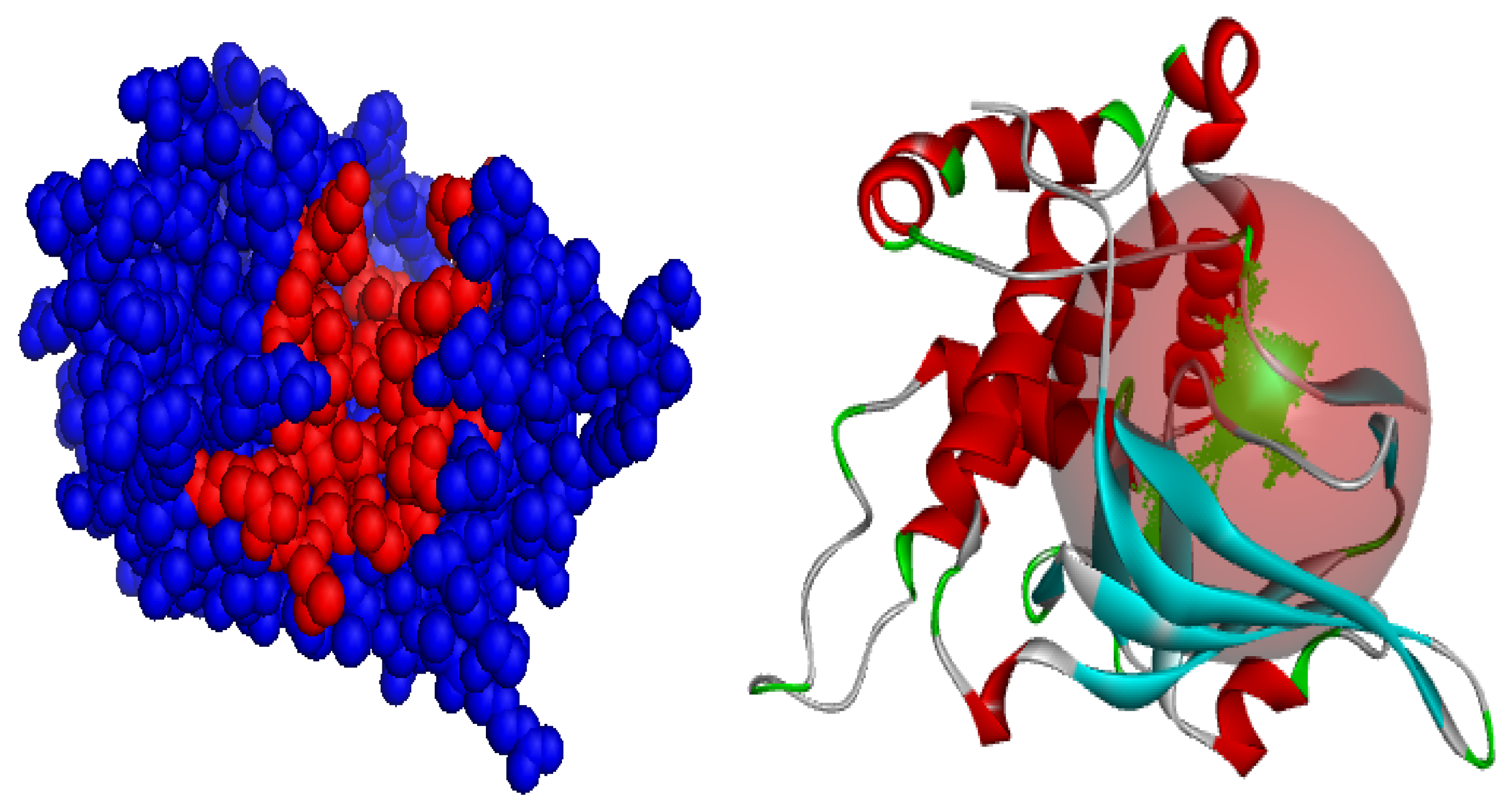
3.6. Prediction of Active Site
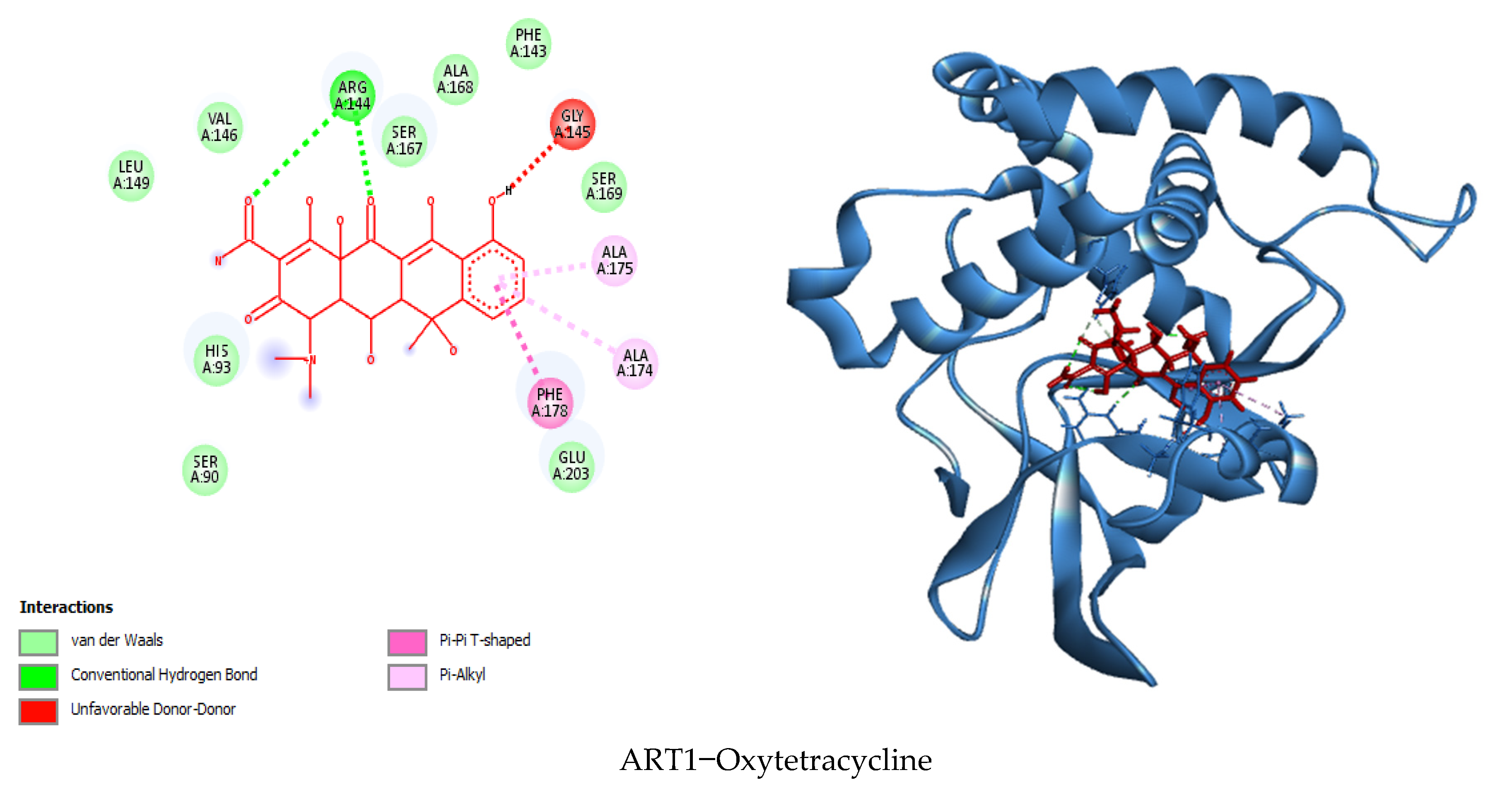
3.7. Visualization of Molecular Docking and Interaction
3.8. Prediction of Biological Activity
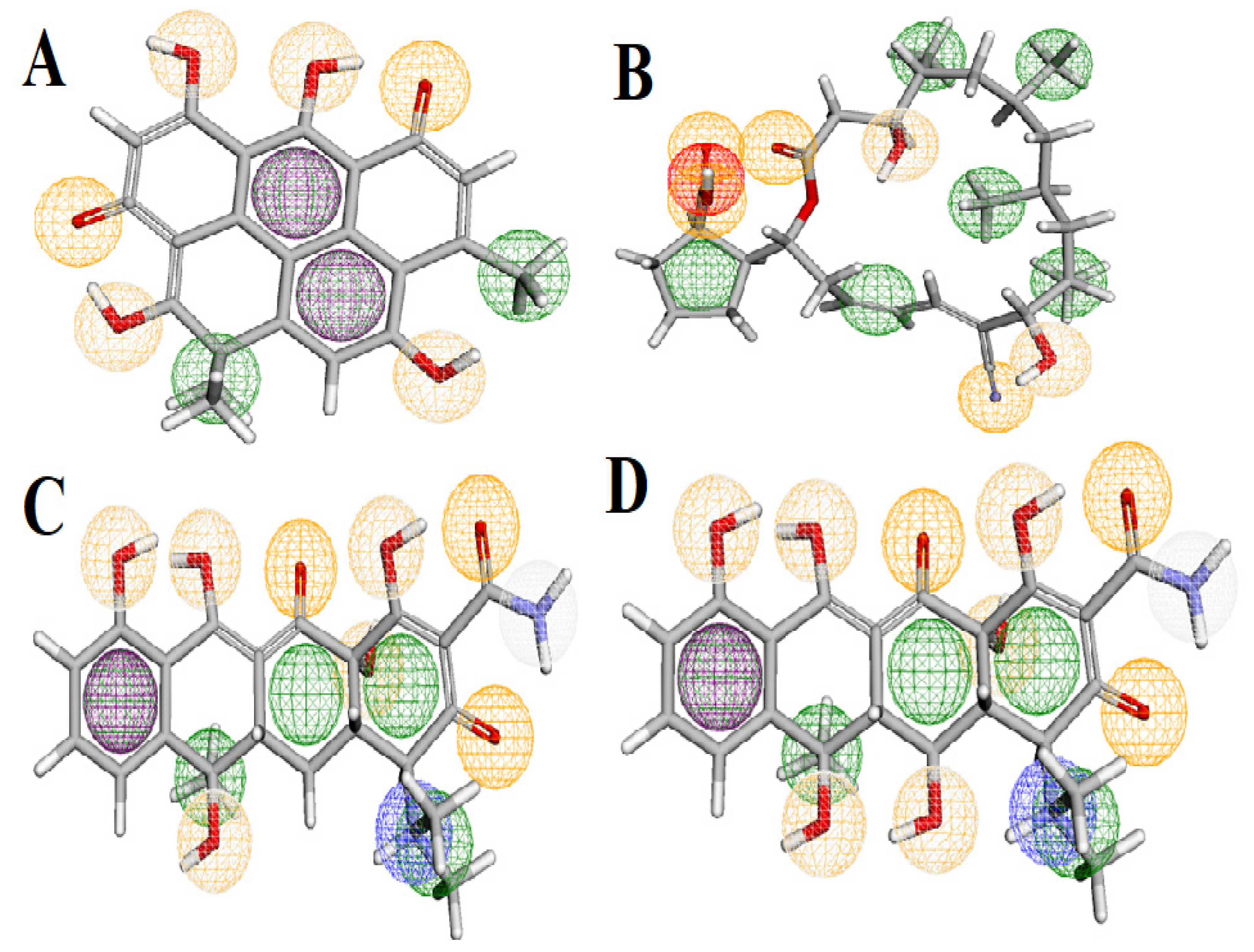
3.9. Pharmacophore Study
3.10. Drug-Likeness and ADMET Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lei, W.; Yang, C.; Wu, Y.; Ru, G.; He, X.; Tong, X.; Wang, S. Nanocarriers Surface Engineered with Cell Membranes for Cancer Targeted Chemotherapy. J. Nanobiotechnol. 2022, 20, 45. [Google Scholar] [CrossRef]
- Lekmine, S.; Benslama, O.; Kadi, K.; Martín García, A.I.; Shamsul Ola, M.; Abdullah Yilmaz, M.; Ali, A. Therapeutic Potential of Hyoscyamus niger-Derived Compounds: Targeting Ovarian Cancer through Antioxidant Activity and EGFR Tyrosine Kinase Inhibition. J. King Saud Univ. Sci. 2024, 36, 103103. [Google Scholar] [CrossRef]
- Li, Z.; Fan, J.; Xiao, Y.; Wang, W.; Zhen, C.; Pan, J.; Wu, W.; Liu, Y.; Chen, Z.; Yan, Q.; et al. Essential Role of Dhx16-Mediated Ribosome Assembly in Maintenance of Hematopoietic Stem Cells. Leukemia 2024, 38, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liao, Y.; Chen, X.; Long, D.; Yu, T.; Shen, F. Regulation of Oncoprotein 18/Stathmin Signaling by ERK Concerns the Resistance to Taxol in Nonsmall Cell Lung Cancer Cells. Cancer Biother. Radiopharm. 2016, 31, 37–43. [Google Scholar] [CrossRef]
- Wang, K.; Ning, S.; Zhang, S.; Jiang, M.; Huang, Y.; Pei, H.; Li, M.; Tan, F. Extracellular Matrix Stiffness Regulates Colorectal Cancer Progression via HSF4. J. Exp. Clin. Cancer Res. 2025, 44, 30. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Tang, Y.; Li, M.; Li, Q.-S.; Li, X.; Yang, L.; Zhao, W.; Jin, C.-C.; Zeng, Z.; Liu, C.; et al. Mono-ADP-Ribosylation of Histone 3 at Arginine-117 Promotes Proliferation through Its Interaction with P300. Oncotarget 2017, 8, 72773–72785. [Google Scholar] [CrossRef]
- Wennerberg, E.; Mukherjee, S.; Spada, S.; Hung, C.; Agrusa, C.J.; Chen, C.; Valeta-Magara, A.; Rudqvist, N.-P.; Van Nest, S.J.; Kamel, M.K.; et al. Expression of the Mono-ADP-Ribosyltransferase ART1 by Tumor Cells Mediates Immune Resistance in Non–Small Cell Lung Cancer. Sci. Transl. Med. 2022, 14, eabe8195. [Google Scholar] [CrossRef]
- Sun, D.Y.; Hu, Y.J.; Li, X.; Peng, J.; Dai, Z.J.; Wang, S. Unlocking the Full Potential of Memory T Cells in Adoptive T Cell Therapy for Hematologic Malignancies. Int. Immunopharmacol. 2025, 144, 113392. [Google Scholar] [CrossRef]
- Lin, T.; Zhang, S.; Tang, Y.; Xiao, M.; Li, M.; Gong, H.; Xie, H.; Wang, Y. ART1 Knockdown Decreases the IL-6-Induced Proliferation of Colorectal Cancer Cells. BMC Cancer 2024, 24, 354. [Google Scholar] [CrossRef]
- Yang, H.; Li, Q.; Chen, X.; Weng, M.; Huang, Y.; Chen, Q.; Liu, X.; Huang, H.; Feng, Y.; Zhou, H.; et al. Targeting SOX13 Inhibits Assembly of Respiratory Chain Supercomplexes to Overcome Ferroptosis Resistance in Gastric Cancer. Nat. Commun. 2024, 15, 4296. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, L.; Tian, J.; Ma, J. Long Non-Coding RNA PART1 Predicts a Poor Prognosis and Promotes the Malignant Progression of Pancreatic Cancer by Sponging miR-122. World J. Surg. Oncol. 2021, 19, 122. [Google Scholar] [CrossRef]
- Boussekine, S.; Lekmine, S.; Gasmi, S.; Benkhedir, A.; Saker, H.; Lidoughi, A. The Protective Effect of Selenium on Diabetic Nephropathy in Wistar Rats. J. Microbiol. Biotechnol. Food Sci. 2022, 12, e5960. [Google Scholar] [CrossRef]
- Mekersi, N.; Addad, D.; Kadi, K.; Casini, S.; Hackenberger, D.K.; Boumaza, A.; Lekmine, S. Effects of Olive Mill Wastewater and Olive Mill Pomace on Soil Physicochemical Properties and Soil Polyphenols. J. Mater. Cycles Waste Manag. 2023, 25, 1404–1416. [Google Scholar] [CrossRef]
- Aftab, U.; Zechel, D.L.; Sajid, I. Antitumor Compounds from Streptomyces sp. KML-2, Isolated from Khewra Salt Mines, Pakistan. Biol. Res. 2015, 48, 58. [Google Scholar] [CrossRef]
- Moussa, H.; Hamid, S.; Mameri, A.; Lekmine, S.; Tahraoui, H.; Kebir, M.; Touzout, N.; Dahmoune, F.; Ola, M.S.; Zhang, J.; et al. From Green Chemistry to Healthy Environments: Silver Nanoparticles as a Dual Antioxidant and Antibacterial Agents for Advancing Biomedicine and Sustainable Wastewater Treatment. Bioengineering 2024, 11, 1205. [Google Scholar] [CrossRef]
- Priyanka, S.; Jayashree, M.; Shivani, R.; Anwesha, S.; Rao, K.V.B.; Arnold, E.I. Characterisation and Identification of Antibacterial Compound from Marine Actinobacteria: In Vitro and In Silico Analysis. J. Infect. Public Health 2019, 12, 83–89. [Google Scholar] [CrossRef]
- Norbakhsh, R.; Moradi Gardeshi, T.; Dalvand, S.; Boroughani, Z. Soil Actinomycetes-Derived Secondary Metabolites-Induced Apoptosis in Human Lung Cancer Cells. Int. J. Med. Lab. 2022, 9, 226–236. [Google Scholar] [CrossRef]
- Ghosh, S. Computational Immunology: Applications; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar] [CrossRef]
- Xia, X. Bioinformatics and Drug Discovery. Curr. Top. Med. Chem. 2017, 17, 1709–1726. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Hou, F.; Ren, C.; Bing, P.; Xiao, X. A Review of Current In Silico Methods for Repositioning Drugs and Chemical Compounds. Front. Oncol. 2021, 11, 711225. [Google Scholar] [CrossRef]
- Gagic, Z.; Ruzic, D.; Djokovic, N.; Djikic, T.; Nikolic, K. In Silico Methods for Design of Kinase Inhibitors as Anticancer Drugs. Front. Chem. 2020, 7, 873. [Google Scholar] [CrossRef]
- Guo, L.; Fu, Z.; Li, H.; Wei, R.; Guo, J.; Wang, H.; Qi, J. Smart Hydrogel: A New Platform for Cancer Therapy. Adv. Colloid Interface Sci. 2025, 323, 103470. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Burley, S.K. Protein Data Bank (PDB): Fifty-Three Years Young and Having a Transformative Impact on Science and Society. Q. Rev. Biophys. 2024, 57, e2. [Google Scholar] [CrossRef]
- Sigrist, C.J.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, version 1.8; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Garzón, J.I.; Chacón, P. iMOD: Multipurpose Normal Mode Analysis in Internal Coordinates. Bioinformatics 2011, 27, 2843–2850. [Google Scholar] [CrossRef] [PubMed]
- Binkowski, T.A.; Naghibzadeh, S.; Liang, J. CASTp: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2003, 31, 3352–3355. [Google Scholar] [CrossRef]
- BIOVIA, D.S. Discovery Studio Modeling Environment; release 2016; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Kang, L.; Gao, X.H.; Liu, H.R.; Men, X.; Wu, H.N.; Cui, P.W.; Oldfield, E.; Yan, J.Y. Structure–Activity Relationship Investigation of Coumarin–Chalcone Hybrids with Diverse Side-Chains as Acetylcholinesterase and Butyrylcholinesterase Inhibitors. Mol. Divers. 2018, 22, 893–906. [Google Scholar] [CrossRef]
- Benslama, O.; Lekmine, S.; Mansouri, N. Phytochemical Constituents of Astragalus monspessulanus and Integrative Analysis for Its Antioxidant, Photoprotective, and Antityrosinase Activities: Experimental and Computational Investigation. Eur. J. Integr. Med. 2023, 60, 102247. [Google Scholar] [CrossRef]
- Djeghim, H.; Bellil, I.; Benslama, O.; Lekmine, S.; Temim, E.; Boufendi, H.; Postigo, I.; Sánchez, P.; Khelifi, D. Effects of Genetic Diversity on the Allergenicity of Peanut (Arachis hypogaea) Proteins: Identification of the Hypoallergenic Accessions Using BALB/c Mice Model and In Silico Analysis of Ara h 3 Allergen Cross-Reactivity. J. Proteom. 2024, 306, 105264. [Google Scholar] [CrossRef]
- Serseg, T.; Benarous, K.; Serseg, M.; Rehman, H.M.; El Bakri, Y.; Goumri-Said, S. Discovery of Inhibitors against SARS-CoV-2 Associated Fungal Coinfections via Virtual Screening, ADMET Evaluation, PASS, Molecular Docking, Dynamics and Pharmacophore Studies. Arab. J. Basic Appl. Sci. 2022, 29, 337–350. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A Platform for Systematic ADMET Evaluation Based on a Comprehensively Collected ADMET Database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Adebiyi, M.; Olugbara, O. Binding Site Identification of COVID-19 Main Protease 3D Structure by Homology Modeling. Indones. J. Electr. Eng. Comput. Sci. 2021, 21, 1713–1719. [Google Scholar] [CrossRef]
- Mansouri, N.; Benslama, O.; Arhab, R. Homology Modeling, Docking and Molecular Dynamics Studies of Some Secondary Metabolites of Actinomycetes as Biocontrol Agents against the 3HNR Enzyme of the Phytopathogenic Fungus Alternaria alternata. J. Biomol. Struct. Dyn. 2023, 41, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Lavadié-González, C.E.; Serrat-Díaz, M.d.J.; Azcanio-Fuentes, L. Homology Modelling and In Silico Structural Characterization of Lanosterol 14α-Demethylase from Cryptococcus neoformans var. grubii. Rev. Cubana Quím. 2021, 33, 198–226. [Google Scholar]
- Elsliger, M.-A.; Wilson, I. 1.8 Structure Validation and Analysis. Compr. Biophys. 2012, 116–135. [Google Scholar] [CrossRef]
- Moussa, H.; Dahmoune, F.; Lekmine, S.; Mameri, A.; Tahraoui, H.; Hamid, S.; Benzitoune, N.; Moula, N.; Zhang, J.; Amrane, A. Optimization of Ultrasound-Assisted Extraction of Bioactive Compounds from Carthamus caeruleus L. Rhizome: Integrating Central Composite Design, Gaussian Process Regression, and Multi-Objective Grey Wolf Optimization Approaches. Process Biochem. 2024, 147, 476–488. [Google Scholar] [CrossRef]
- Sunil, L.; Vasu, P. In silico designing of therapeutic protein enriched with branched-chain amino acids for the dietary treatment of chronic liver disease. J. Mol. Graph. Modell. 2017, 76, 192–204. [Google Scholar] [CrossRef]
- Hasan, A.; Mazumder, H.H.; Khan, A.; Hossain, M.U.; Chowdhury, H.K. Molecular characterization of legionellosis drug target candidate enzyme phosphoglucosamine mutase from Legionella pneumophila (strain Paris): An in silico approach. Genom. Inform. 2014, 12, 268. [Google Scholar] [CrossRef]
- Sun, W.; Jang, M.S.; Zhan, S.; Liu, C.; Sheng, L.; Lee, J.H.; Fu, Y.; Yang, H.Y. Tumor-Targeting and Redox-Responsive Photo-Cross-Linked Nanogel Derived from Multifunctional Hyaluronic Acid–Lipoic Acid Conjugates for Enhanced In Vivo Protein Delivery. Int. J. Biol. Macromol. 2025, 314, 144444. [Google Scholar] [CrossRef]
- Elengoe, A.; Naser, M.A.; Hamdan, S. Modeling and docking studies on novel mutants (K71L and T204V) of the ATPase domain of human heat shock 70 kDa protein 1. Int. J. Mol. Sci. 2014, 15, 6797–6814. [Google Scholar] [CrossRef]
- Beg, A.; Shivangi, F.A.; Meena, L.S. Structural and functional annotation of Rv1514c gene of Mycobacterium tuberculosis H37Rv as glycosyl transferases. J. Adv. Res. Biotechnol. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- De Oliveira, C.C.S.; Pereira, G.R.C.; De Alcantara, J.Y.S.; Antunes, D.; Caffarena, E.R.; De Mesquita, J.F. In silico analysis of the V66M variant of human BDNF in psychiatric disorders: An approach to precision medicine. PLoS ONE 2019, 14, e0215508. [Google Scholar] [CrossRef]
- Kadi, K.; Mrah, R.; Hamli, S.; Lekmine, S.; Dib, D.; Addad, D.; Boukeria, S.; Gueboudji, Z.; Hafsaoui, I. Evaluation of the Anticoagulant Activity of Margins from Olives Extraction in the Khenchela Region. J. Fundam. Appl. Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Gupta, S.; Agnihotri, S.; Ali, S.M.; Yadav, R. Physicochemical analysis and homology modeling of antioxidant proteins of foxtail millet (Setaria italica). Vegetos 2016, 29, 1–6. [Google Scholar] [CrossRef]
- Mhade, S.; Panse, S.; Tendulkar, G.; Awate, R.; Narasimhan, Y.; Kadam, S.; Yennamalli, R.M.; Kaushik, K.S. Amping up the search: A structural and functional repository of antimicrobial peptides for biofilm studies, and a case study of its application to Corynebacterium striatum, an emerging pathogen. Front. Cell. Infect. Microbiol. 2021, 11, 803774. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Upadhayay, A.; Roy, A. In silico analysis and characterization of fresh water fish ATPases and homology modelling. Ann. Proteom. Bioinform. 2017, 1, 018–024. [Google Scholar] [CrossRef]
- Khedraoui, M.; Abchir, O.; Nour, H.; Yamari, I.; Errougui, A.; Samadi, A.; Chtita, S. An in silico study based on QSAR and molecular docking and molecular dynamics simulation for the discovery of novel potent inhibitor against AChE. Pharmaceuticals 2024, 17, 830. [Google Scholar] [CrossRef]
- Tama, F.; Sanejouand, Y.-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001, 14, 1–6. [Google Scholar] [CrossRef]
- Collier, T.A.; Piggot, T.J.; Allison, J.R. Molecular dynamics simulation of proteins. In Protein Nanotechnology: Protocols, Instrumentation, and Applications; Academic Press: Cambridge, MA, USA, 2020; pp. 311–327. [Google Scholar] [CrossRef]
- Hinsen, K. Analysis of domain motions by approximate normal mode calculations. Proteins Struct. Funct. Bioinform. 1998, 33, 417–429. [Google Scholar] [CrossRef]
- Kondrashov, D.A.; Van Wynsberghe, A.W.; Bannen, R.M.; Cui, Q.; Phillips, G.N. Protein structural variation in computational models and crystallographic data. Structure 2007, 15, 169–177, Erratum in Structure 2007, 15, 637. [Google Scholar] [CrossRef]
- Kitao, A.; Go, N. Investigating protein dynamics in collective coordinate space. Curr. Opin. Struct. Biol. 1999, 9, 164–169. [Google Scholar] [CrossRef]
- Strang, G. Computational Science and Engineering; Wellesley-Cambridge Press: Wellesley, MA, USA, 2007. [Google Scholar]
- Dashti, A.; Mashayekhi, G.; Shekhar, M.; Ben Hail, D.; Salah, S.; Schwander, P.; Georges, A.; Singharoy, A.; Frank, J.; Ourmazd, A. Retrieving functional pathways of biomolecules from single-particle snapshots. Nat. Commun. 2020, 11, 4734. [Google Scholar] [CrossRef]
- Atilgan, A.R.; Durell, S.; Jernigan, R.L.; Demirel, M.C.; Keskin, O.; Bahar, I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001, 80, 505–515. [Google Scholar] [CrossRef]
- Skjaerven, L.; Martinez, A.; Reuter, N. Principal component and normal mode analysis of proteins: A quantitative comparison using the GroEL subunit. Proteins Struct. Funct. Bioinform. 2011, 79, 232–243. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the AMBER ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Lindahl, E.; Delarue, M. Refinement of docked protein–ligand and protein–DNA structures using low frequency normal mode amplitude optimization. Nucleic Acids Res. 2005, 33, 4496–4506. [Google Scholar] [CrossRef] [PubMed]
- Toumi, S.; Lekmine, S.; Touzout, N.; Moussa, H.; Elboughdiri, N.; Boudraa, R.; Benslama, O.; Kebir, M.; Danish, S.; Zhang, J.; et al. Harnessing Deep Learning for Real-Time Water Quality Assessment: A Sustainable Solution. Water 2024, 16, 3380. [Google Scholar] [CrossRef]
- Prakasia, P.P. Modeling and structural analysis of acetylcholinesterase enzyme of fishes. Int. J. Pharm. Pharm. Sci. 2020, 12, 36–44. [Google Scholar] [CrossRef]
- Kumarachari, R.K.; Peta, S.; Surur, A.S.; Mekonnen, Y.T. Synthesis, characterization and in silico biological activity of some 2-(N,N-dimethyl guanidinyl)-4,6-diaryl pyrimidines. J. Pharm. Bioallied Sci. 2016, 8, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Lekmine, S.; Benslama, O.; Kadi, K.; Brik, A.; Djeffali, O.; Ounissi, M.; Slimani, M.; Ola, M.S.; Eldahshan, O.A.; Martín-García, A.I.; et al. Preliminary Investigation of Astragalus arpilobus subsp. hauarensis: LC-MS/MS Chemical Profiling, In Vitro Evaluation of Antioxidant, Anti-Inflammatory Properties, Cytotoxicity, and In Silico Analysis against COX-2. Antioxidants 2024, 13, 654. [Google Scholar] [CrossRef]
- Sunseri, J.; Koes, D.R. Pharmit: Interactive exploration of chemical space. Nucleic Acids Res. 2016, 44, 442–448. [Google Scholar] [CrossRef]
- Dammene Debbih, O.; Mazouz, W.; Benslama, O.; Zouchoune, B.; Selatnia, I.; Bouchene, R.; Sid, A.; Bouacida, S.; Mosset, P. Hydrazone analogs as DNA gyrase inhibitors and antioxidant agents: Structure–activity relationship and pharmacophore modeling. J. Chem. Sci. 2024, 136, 32. [Google Scholar] [CrossRef]
- Dotolo, S.; Cervellera, C.; Russo, M.; Russo, G.L.; Facchiano, A. Virtual screening of natural compounds as potential PI3K-AKT1 signaling pathway inhibitors and experimental validation. Molecules 2021, 26, 492. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Z.Y.; Uzairu, A.; Shallangwa, G.; Abechi, S. Molecular docking studies, drug-likeness and in silico ADMET prediction of some novel β-amino alcohol grafted 1,4,5-trisubstituted 1,2,3-triazoles derivatives as elevators of p53 protein levels. Sci. Afr. 2020, 10, e00570. [Google Scholar] [CrossRef]
- Lekmine, S.; Benslama, O.; Kadi, K.; Martín-García, A.I.; Yilmaz, M.A.; Akkal, S.; Boumegoura, A.; Alhomida, A.S.; Ola, M.S.; Ali, A. LC/MS-MS analysis of phenolic compounds in Hyoscyamus albus L. extract: In vitro antidiabetic activity, in silico molecular docking, and In Vivo investigation against STZ-induced diabetic mice. Pharmaceuticals 2023, 16, 1015. [Google Scholar] [CrossRef]
- Akşöz, B.E.; Onurdağ, F.K.K.; Akşöz, E.; Özgen, S. In silico ADME screening and evaluation of antimicrobial and antimycobacterial activities of 3,5-diphenyl pyrazoline derivatives. Süleyman Demirel Univ. Sağlık Bilim. Derg. 2021, 12, 184–191. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Sarkar, B.; Prottoy, M.N.I.; Araf, Y.; Taniya, M.A.; Ullah, M.A. Thrombolytic activity, drug likeness property and ADME/T analysis of isolated phytochemicals from ginger (Zingiber officinale) using in silico approaches. Mod. Res. Inflamm. 2019, 8, 29–43. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K.; et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef]
- Wang, N.-N.; Dong, J.; Deng, Y.-H.; Zhu, M.-F.; Wen, M.; Yao, Z.-J.; Lu, A.-P.; Wang, J.-B.; Cao, D.-S. ADME properties evaluation in drug discovery: Prediction of Caco-2 cell permeability using a combination of NSGA-II and boosting. J. Chem. Inf. Model. 2016, 56, 763–773. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, J.; Hu, C.Q.; Zhang, X.; Ma, B.; Zhang, P. In silico ADME and toxicity prediction of ceftazidime and its impurities. Front. Pharmacol. 2019, 10, 434. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Yuan, H.; Chen, Y.; Hu, Y.; Li, Y.; Zhang, H.; Zhang, S.; Chen, Q.; Zhou, W.; Sun, J.; He, Z.; et al. Disulfide Bond-Driven Nanoassembly of Lipophilic Epirubicin Prodrugs for Breast Cancer Therapy. J. Pharm. Investig. 2025, 55. online ahead of print. [Google Scholar] [CrossRef]
- Piechota, P. Development of In Silico Models for the Prediction of Toxicity Incorporating ADME Information. Ph.D. Thesis, Liverpool John Moores University, Liverpool, UK, 2015. [Google Scholar] [CrossRef]
- Pardridge, W.M. Drug transport across the blood–brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1959–1972. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, T.; Amin, Z. QSAR models for the prediction of plasma protein binding. BioImpacts 2013, 3, 21. [Google Scholar] [CrossRef]
- Li, J.; Yanagisawa, K.; Yoshikawa, Y.; Ohue, M.; Akiyama, Y. Plasma protein binding prediction focusing on residue-level features and circularity of cyclic peptides by deep learning. Bioinformatics 2022, 38, 1110–1117. [Google Scholar] [CrossRef]
- Zadorozhnii, P.V.; Kiselev, V.V.; Kharchenko, A.V. In silico ADME profiling of salubrinal and its analogues. Future Pharmacol. 2022, 2, 160–197. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Computational pharmacokinetics report, ADMET study and conceptual DFT-based estimation of the chemical reactivity properties of marine cyclopeptides. ChemistryOpen 2021, 10, 1142–1149. [Google Scholar] [CrossRef]
- Malik, A.; Manan, A.; Mirza, M.U. Molecular docking and in silico ADMET studies of silibinin and glycyrrhetic acid anti-inflammatory activity. Trop. J. Pharm. Res. 2017, 16, 67–74. [Google Scholar] [CrossRef]
- Duran-Iturbide, N.A.; Díaz-Eufracio, B.I.; Medina-Franco, J.L. In silico ADME/Tox profiling of natural products: A focus on BioFAsqquim. ACS Omega 2020, 5, 16076–16084. [Google Scholar] [CrossRef]
- Halder, S.; Narayana, P.V. A comprehensive review of current and emerging analytical techniques for the identification, quantification, and assessment of genotoxic impurities in drug substances. Eur. Chem. Bull. 2023, 12, 12526–12564. [Google Scholar]
- Li, X.; Du, Z.; Wang, J.; Wu, Z.; Li, W.; Liu, G.; Shen, X.; Tang, Y. In silico estimation of chemical carcinogenicity with binary and ternary classification methods. Mol. Inform. 2015, 34, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Wang, J.; Chen, L.; Zhang, L.; Yu, H.; Hou, T. ADMET evaluation in drug discovery. 12. Development of binary classification models for prediction of hERG potassium channel blockage. Mol. Pharm. 2012, 9, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Dixon, S.L. In silico models for the prediction of dose-dependent human hepatotoxicity. J. Comput. Aided Mol. Des. 2003, 17, 811–823. [Google Scholar] [CrossRef] [PubMed]






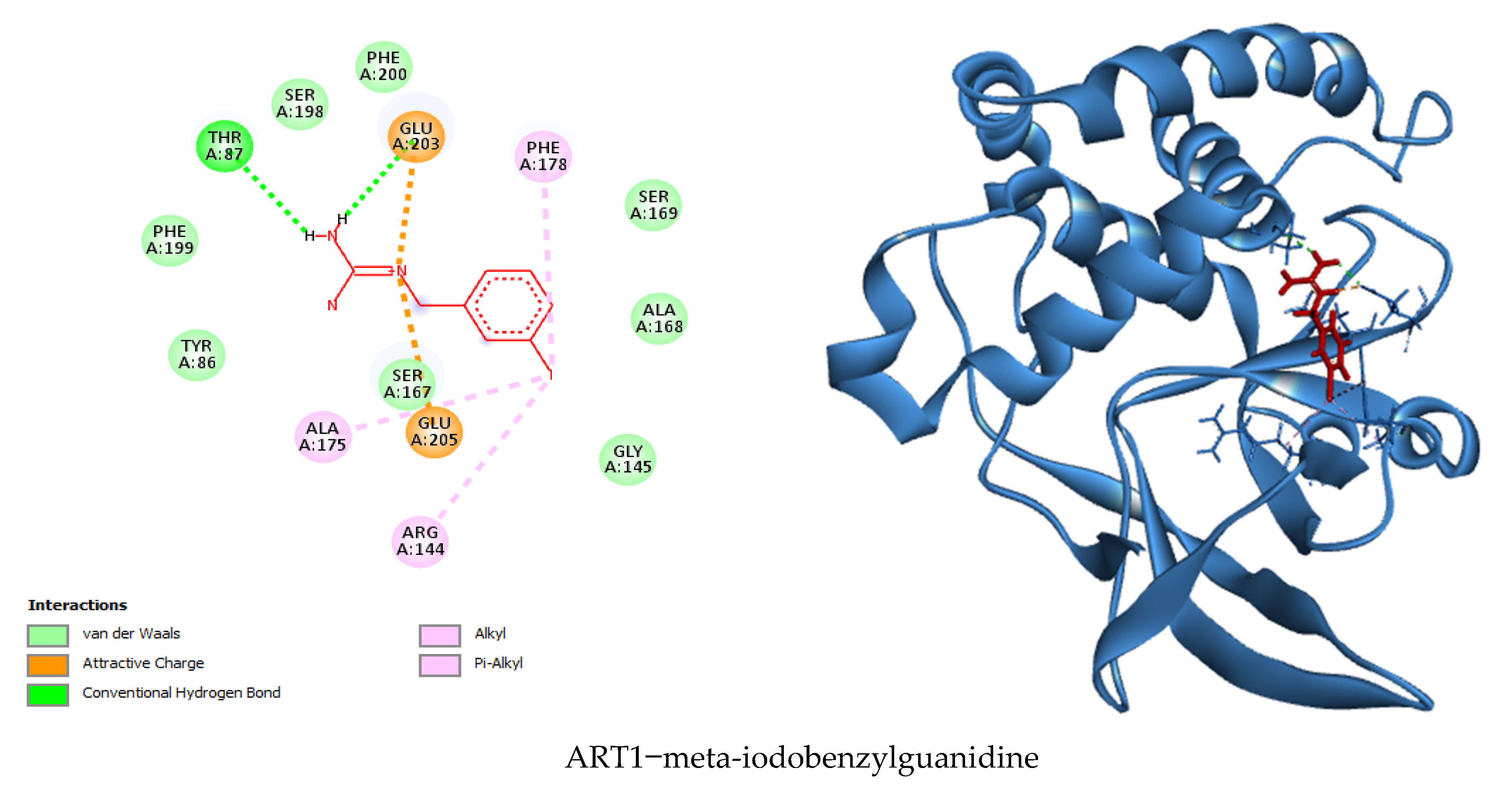
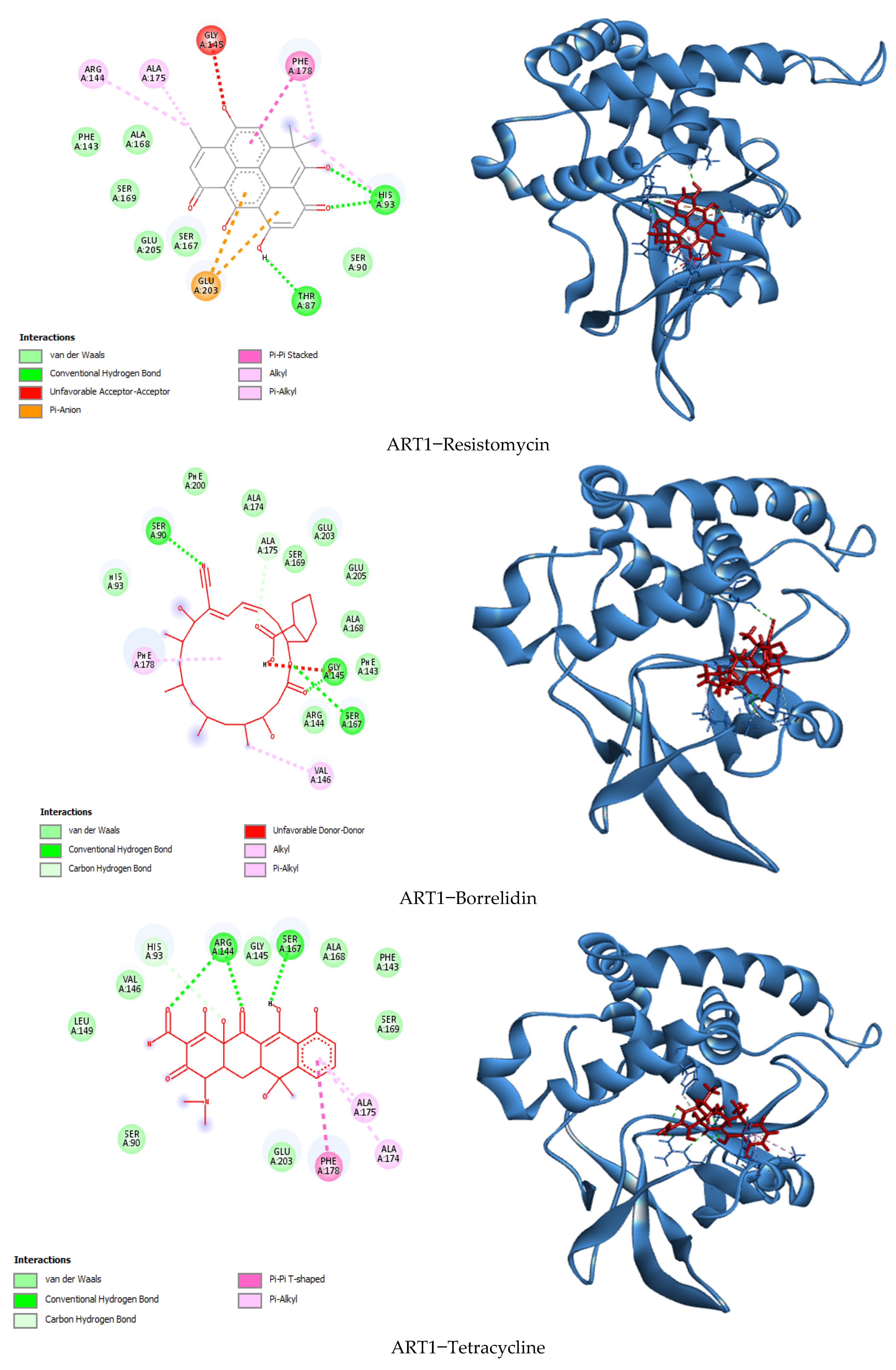
| Binding Energy (Kcal/mol) | Hydrogen Bonds | Hydrophobic Bonds | Electrostatic Bonds | Van der Waals Bonds | Total Number of Bonds | |
|---|---|---|---|---|---|---|
| Meta-iodo-benzyl-guanidine | −6.1 | Thr87, Glu203 | Phe178, Ala175, Arg144 | Glu203, Glu205 | Gly145, Ala168, Ser169, Phe200, Ser198, Phe199, Tyr86, Ser167 | 15 |
| Resistomycin | −9.3 | Thr87, His93, His93 | His93, Phe178, Phe178, Ala175, Arg144 | Glu203, Glu203 | Ser90, Ser167, Glu205, Ser169, Ala168, Phe143. | 16 |
| Borrelidin | −9.0 | Gly145, Ser167, Ser90, Ala175 | Val146, Phe178 | / | Arg144, Phe143, Ala168, Glu205, Glu203, Ser169, Ala174, Phe200, His93 | 15 |
| Tetracycline | −9.0 | Ser167, Arg144, Arg144, His93 | Ala175, Ala175, Phe178 | / | Ser169, Phe143, Ala163, Gly145, Val146, Leu149, Ser90, Glu203 | 15 |
| Oxytetracycline | −8.9 | Arg144, Arg144 | Ala174, Ala175, Phe178 | / | Ser169, Phe143, Ala168, Ser167, Val146, Leu149, His93, Ser90, Glu203 | 14 |
| Biological Activity | Resistomycin | Borrelidin | Tetracycline | Oxytetracycline | ||||
|---|---|---|---|---|---|---|---|---|
| Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | |
| Anticancer activity | 0.896 | 0.005 | 0.799 | 0.012 | 0.529 | 0.063 | 0.465 | 0.082 |
| Molar Weight (g/mol) | LogP | LogS | HBA | HBD | TPSA (Å2) | AMR | nRB | Lpinski | Ghose | Veber | Egan | Muegge | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistomycin | 376.36 | 2.89 | −4.61 MS | 6 | 4 | 115.06 | 104.73 | 0 | Yes | Yes | Yes | Yes | Yes |
| Borrelidin | 489.64 | 3.61 | −2.53 S | 7 | 3 | 127.85 | 136.66 | 2 | Yes | No | Yes | Yes | No |
| Tetracycline | 444.43 | −0.34 | −1.82 S | 9 | 6 | 181.62 | 110.79 | 2 | Yes | No | No | No | No |
| Oxytetracycline | 460.63 | −1.01 | −1.0 S | 10 | 7 | 201.85 | 111.95 | 2 | No | No | No | No | No |
| Resistomycin | Borrelidin | Tetracycline | Oxytetracycline | ||
|---|---|---|---|---|---|
| Absorption | Caco2 | Yes | Yes | Yes | No |
| HIA | Yes | No | Yes | Yes | |
| Distribution | BBB | No | No | No | No |
| PPB | Yes | No | Yes | Yes | |
| Metabolism | CYP1A2 inhibitor | Yes | No | No | No |
| CYP2C19 inhibitor | No | No | No | No | |
| CYP2C9 inhibitor | Yes | No | No | No | |
| CYP2D6 inhibitory | No | No | No | No | |
| CYP3A4 inhibitor | Yes | Yes | No | No | |
| Excretion | Cl | 0.493 (L) | 7.91 (M) | 2.238 (L) | 1.704 (L) |
| Toxicity | Ames test | Yes | No | No | No |
| Carcinogencity | No | No | No | No | |
| hERG Blockers | No | Yes | No | No | |
| H-HT | Yes | Yes | Yes | No | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansouri, N.; Benslama, O.; Lekmine, S.; Tahraoui, H.; Ola, M.S.; Zhang, J.; Amrane, A. Computational Exploration of Bacterial Compounds Targeting Arginine-Specific Mono-Adp-Ribosyl-Transferase 1 (Art1): A Pathway to Novel Therapeutic Anticancer Strategies. Curr. Issues Mol. Biol. 2025, 47, 634. https://doi.org/10.3390/cimb47080634
Mansouri N, Benslama O, Lekmine S, Tahraoui H, Ola MS, Zhang J, Amrane A. Computational Exploration of Bacterial Compounds Targeting Arginine-Specific Mono-Adp-Ribosyl-Transferase 1 (Art1): A Pathway to Novel Therapeutic Anticancer Strategies. Current Issues in Molecular Biology. 2025; 47(8):634. https://doi.org/10.3390/cimb47080634
Chicago/Turabian StyleMansouri, Nedjwa, Ouided Benslama, Sabrina Lekmine, Hichem Tahraoui, Mohammad Shamsul Ola, Jie Zhang, and Abdeltif Amrane. 2025. "Computational Exploration of Bacterial Compounds Targeting Arginine-Specific Mono-Adp-Ribosyl-Transferase 1 (Art1): A Pathway to Novel Therapeutic Anticancer Strategies" Current Issues in Molecular Biology 47, no. 8: 634. https://doi.org/10.3390/cimb47080634
APA StyleMansouri, N., Benslama, O., Lekmine, S., Tahraoui, H., Ola, M. S., Zhang, J., & Amrane, A. (2025). Computational Exploration of Bacterial Compounds Targeting Arginine-Specific Mono-Adp-Ribosyl-Transferase 1 (Art1): A Pathway to Novel Therapeutic Anticancer Strategies. Current Issues in Molecular Biology, 47(8), 634. https://doi.org/10.3390/cimb47080634









