Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Analysis of INV Genes in P. deltoides and S. suchowensis
2.2. Chromosomal Localization and Synteny Analysis
2.3. Structural Characterization of INV Genes
2.4. Phylogeny
2.5. Collection and Processing of Plant Materials
2.6. Primers, RNA Extraction, and qRT-PCR
2.7. Analysis of INV Cis-Acting Elements
3. Results and Discussion
3.1. Identification of INV Gene Families in P. deltoides and S. suchowensis
3.2. Chromosomal Distribution and Identification of Gene Duplication
3.3. Phylogenetic Relationships, Conserved Motifs, and Gene Structures of INV Genes in Populus and Salix
3.4. Phylogenetic Analysis of INV Gene Families in 11 Plant Species
3.5. Identification of INVs Associated with the Development of Poplar and Willow Floss
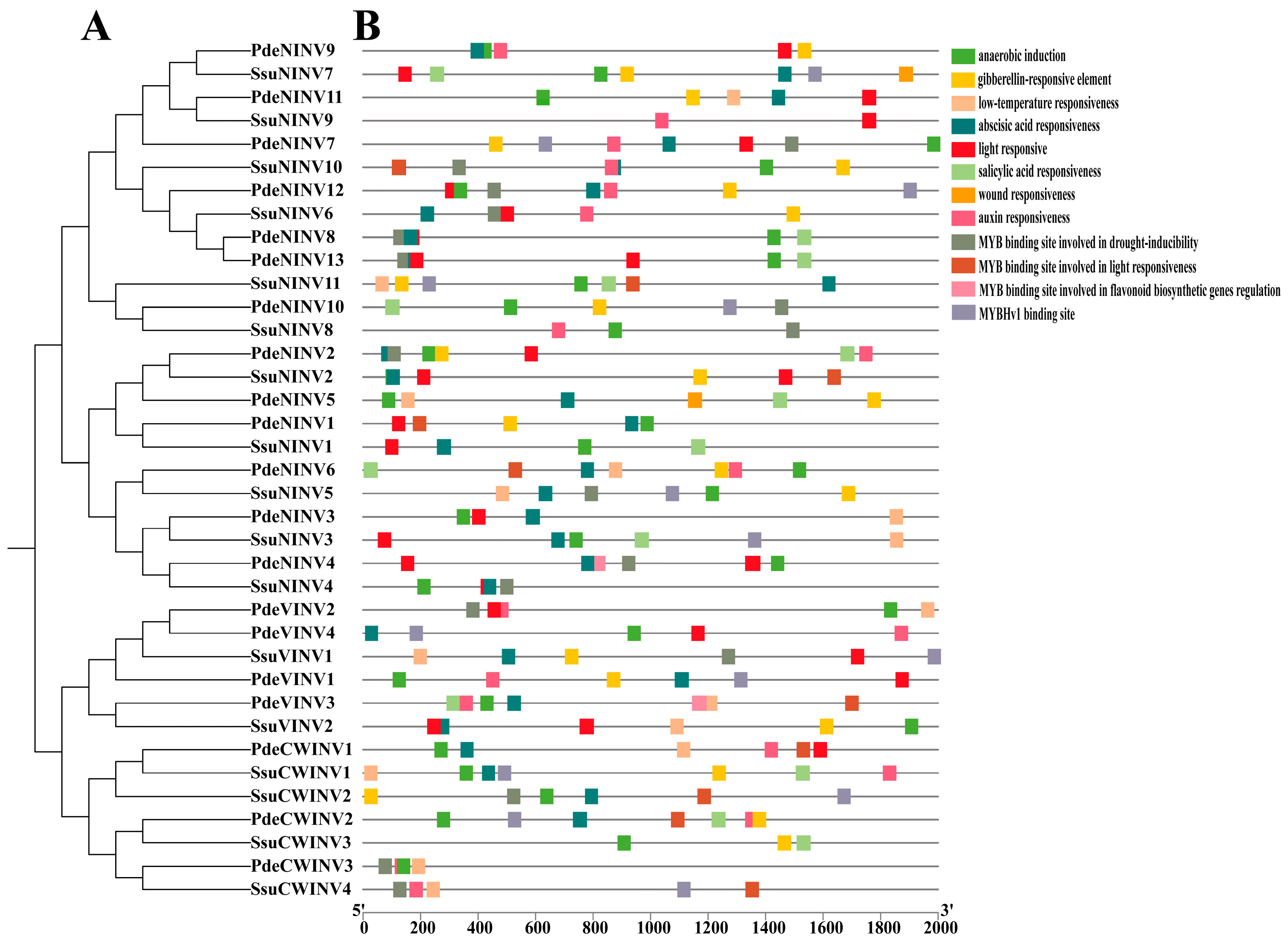
3.6. Promoter Element Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three-letter acronym |
References
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Roitsch, T.; Gonzalez, M.C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Geng, M.T.; Wu, X.H.; Liu, J.; Li, R.M.; Hu, X.W.; Guo, J.C. Genome-wide identification, 3D modeling, expression and enzymatic activity analysis of cell wall invertase gene family from cassava (Manihot esculenta Crantz). Int. J. Mol. Sci. 2014, 15, 7313–7331. [Google Scholar] [CrossRef]
- Ji, X.M.; Van den Ende, W.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Wan, H.; Wu, L.; Yang, Y.; Zhou, G.; Ruan, Y.L. Evolution of sucrose metabolism: The dichotomy of invertases and beyond. Trends Plant Sci. 2018, 23, 163–177. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Feng, J.; Qin, Q.; Huang, J. Overexpression of a loquat (Eriobotrya japonica Lindl.) vacuolar invertase affects sucrose levels and growth. Plant Cell Tiss. Org. 2015, 123, 99–108. [Google Scholar] [CrossRef]
- Abbas, A.; Shah, A.N.; Shah, A.A.; Nadeem, M.A.; Alsaleh, A.; Javed, T.; Alotaibi, S.S.; Abdelsalam, N.R. Genome-wide analysis of invertase gene family, and expression profiling under abiotic stress conditions in potato. Biology 2022, 11, 539. [Google Scholar] [CrossRef]
- Du, C.L.; Cai, C.L.; Lu, Y.; Li, Y.M.; Xie, Z.S. Identification and expression analysis of invertase family genes during grape (Vitis vinifera L.) berry development under CPPU and GA treatment. Mol. Genet Genom. 2023, 298, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Georgelis, N.; Braun, E.L.; Shaw, J.R.; Hannah, L.C. The two AGPase subunits evolve at different rates in angiosperms, yet they are equally sensitive to activity-altering amino acid changes when expressed in bacteria. Plant Cell 2007, 19, 1458–1472. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.-Q.; Lüscher, M.; Sturm, A. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 1999, 11, 177–189. [Google Scholar] [CrossRef]
- Jain, M.; Chourey, P.S.; Boote, K.J.; Allen, L.H.J. Short-term high temperature growth conditions during vegetative-to-reproductive phase transition irreversibly compromise cell wall invertase-mediated sucrose catalysis and microspore meiosis in grain sorghum (Sorghum bicolor). J. Plant Physiol. 2010, 167, 578–582. [Google Scholar] [CrossRef]
- Qi, X.; Wu, Z.; Li, J.; Mo, X.; Wu, S.; Chu, J.; Wu, P. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol. Biol. 2007, 64, 575–587. [Google Scholar] [CrossRef]
- Vargas, W.A.; Salerno, G.L. The Cinderella story of sucrose hydrolysis: Alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Sci. 2010, 178, 1–8. [Google Scholar] [CrossRef]
- Wang, L.; Cook, A.; Patrick, J.W.; Chen, X.Y.; Ruan, Y.L. Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 2014, 78, 686–696. [Google Scholar] [CrossRef]
- Shen, L.B.; Yao, Y.; He, H.; Qin, Y.L.; Liu, Z.J.; Liu, W.X.; Qi, Z.Q.; Yang, L.J.; Cao, Z.M.; Yang, Y. Genome-wide identification, expression, and functional analysis of the alkaline/neutral invertase gene family in pepper. Int. J. Mol. Sci. 2018, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cao, L.; Chen, D.; Chang, R.; Cao, J.; Zhang, Q.; Qin, Y.; Liu, G.; Xu, Z. PtrVINV2 is dispensable for cellulose synthesis but essential for salt tolerance in Populus trichocarpa Torr. and Gray. Plant Biotechnol. J. 2025, 24, 1892–1908. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, T.; Rao, P.; Yang, N.; Li, G.; Jia, L.; An, X.; Chen, Z. Morphology, sucrose metabolism and gene network reveal the molecular mechanism of seed fiber development in poplar. Int. J. Biol. Macromol. 2023, 246, 125633. [Google Scholar] [CrossRef]
- Hamzeh, M.; Dayanandan, S. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast TRNT-TRNF region and nuclear rDNA. Am. J. Bot. 2004, 91, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xia, X.F.; Xu, S.J.; Wu, J.; Peng, L.; Zhao, L. Development, structure and evolutionary significance of seed appendages in Salix matsudana (Salicaceae). PLoS ONE 2018, 13, e0203061. [Google Scholar] [CrossRef]
- Zhou, F.; Wu, H.; Chen, Y.; Wang, M.; Tuskan, G.A.; Yin, T. Function and molecular mechanism of a poplar placenta limited MIXTA gene in regulating differentiation of plant epidermal cells. Int. J. Biol. Macromol. 2023, 242 Pt 2, 124743. [Google Scholar] [CrossRef]
- Wu, H.; Wang, L.; Zhao, S.; Gao, M.; Cao, J.; Hao, Y.; Yu, L.; Zhao, T.; Wang, S.; Han, J.; et al. GhLPF1 associated network is involved with cotton lint percentage regulation revealed by the integrative analysis of spatial transcriptome. Adv. Sci. 2025, 11, e2414175. [Google Scholar] [CrossRef]
- Koornneef, M.; Dellaert, L.W.; van der Veen, J.H. EMS and radiation induced mutation frequencies at individual loci in 21 Arabidopsis thaliana (L.) Heynh. Mutat. Res. 1982, 93, 109–123. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, D.M.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocya-nin.biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef]
- Larkin, J.C.; Walker, J.D.; Bolognesi-Winfield, A.C.; Gray, J.G.; Walker, A.R. Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 1999, 151, 1591–1604. [Google Scholar] [CrossRef]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef] [PubMed]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parish, R.W. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell. 2009, 21, 72–89. [Google Scholar] [CrossRef]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010, 22, 2322–2335. [Google Scholar] [CrossRef]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kartika, D.; Ruan, Y.L. Looking into ’hair tonics’ for cotton fiber initiation. New Phytol. 2021, 229, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Chen, T.; Chong, K.; Xue, Y.; Liu, S.; Wang, T. Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol. Cell. Proteom. 2007, 6, 207–230. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Kaur, N.; Stromvik, M.; Vodkin, L. Transcript profiling reveals expression differences in wild-type and glabrous soybean lines. BMC Plant Biol. 2011, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gao, K.; Su, X.; Rao, P.; An, X. Genome-Wide Identification of the Invertase Gene Family in Populus. PLoS ONE 2015, 10, e0138540. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Wen, X.; Qu, X.; Zhang, Y.; Zhang, X.; Deng, P.; Chen, C.; Ji, W.; Zhang, H. Characteristics and expression analysis of invertase gene family in common wheat (Triticum aestivum L.). Genes 2022, 14, 41. [Google Scholar] [CrossRef]
- Jin, A.; Ozawa, T.; Tajiri, K.; Obata, T.; Kondo, S.; Kinoshita, K.; Kadowaki, S.; Takahashi, K. A rapid and efficient singlecell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat. Med. 2009, 15, 1088–1092. [Google Scholar] [CrossRef]
- Shen, L.B.; Qin, Y.L.; Qi, Z.Q.; Niu, Y.; Liu, Z.J.; Liu, W.X.; He, H.; Cao, Z.M.; Yang, Y. Genome-wide analysis, expression profile, and characterization of the acid invertase gene family in pepper. Int. J. Mol. Sci. 2019, 20, 15. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of bigbiological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, K.; Li, G.; Li, Y.; Gao, Z. Identification and expression analyses of invertase genes in moso bamboo reveal their potential drought stress functions. Front. Genet. 2021, 12, 696300. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C.; et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008, 6, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Han, X.; Zhang, J.; Sun, P.; Lu, M.; Hu, J. Selection of reliable reference genes for gene expression analysis under abiotic stresses in the desert biomass willow, Salix psammophila. Front. Plant Sci. 2016, 7, 1505. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, L.B.; Wang, L.; Zhang, Z.; Ma, M.; Wang, R.Z.; Qian, M.; Zhang, S.L. Technology, genome-wide identification and comparative analysis of the superoxide dismutase gene family in pear and their functions during fruit ripening. Postharvest Biol. Technol. 2018, 143, 68–77. [Google Scholar] [CrossRef]
- Dai, X.; Hu, Q.; Cai, Q.; Feng, K.; Ye, N.; Tuskan, G.A.; Milne, R.; Chen, Y.; Wan, Z.; Wang, Z.; et al. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 2014, 24, 1274–1277. [Google Scholar] [CrossRef]
- Hou, J.; Ye, N.; Dong, Z.; Lu, M.; Li, L.; Yin, T. Major chromosomal rearrangements distinguish willow and poplar after the ancestral “Salicoid” genome duplication. Genome Biol. Evol. 2016, 8, 1868–1875. [Google Scholar] [CrossRef]
- Bocock, P.N.; Morse, A.M.; Dervinis, C.; Davis, J.M. Evolution and diversity of invertase genes in Populus trichocarpa. Planta 2008, 227, 565–576. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhen, L.; Tan, X.; Li, L.; Wang, X. The involvement of hexokinase in the coordinated regulation of glucose and gibberellin on cell wall invertase and sucrose synthesis in grape -berry. Mol. Biol. Rep. 2014, 41, 7899–7910. [Google Scholar] [CrossRef]
- Ahiakpa, J.K.; Magdy, M.; Karikari, B.; Munir, S.; Mumtaz, M.A.; Tamim, S.A.; Mahmood, S.; Liu, G. Genome-wide identification and expression profiling of tomato invertase genes indicate their response to stress and phytohormones. J. Plant Growth Regul. 2022, 41, 1481–1498. Available online: https://link.springer.com/article/10.1007/s00344-021-10384-5 (accessed on 15 November 2024). [CrossRef]
- Zhang, Y.; Zhou, F.; Wang, H.; Chen, Y.; Yin, T.; Wu, H. Genome-wide comparative analysis of the fasciclin-like arabinogalactan proteins (FLAs) in Salicacea and identification of secondary tissue development-related genes. Int. J. Mol. Sci. 2023, 24, 1481. [Google Scholar] [CrossRef] [PubMed]





| Gene Symbol | Gene ID | Chr | Start | End | Amino Acid | PI | Mw |
|---|---|---|---|---|---|---|---|
| PdeCWINV1 | EVM0014612 | chr16 | 5,978,219 | 5,994,019 | 578 | 65,117.66 | 6.58 |
| PdeCWINV2 | EVM0008374 | chr6 | 22,923,121 | 22,926,712 | 576 | 65,562.07 | 5.15 |
| PdeCWINV3 | EVM0008949 | chr6 | 22,919,482 | 22,922,237 | 570 | 64,803.28 | 7.66 |
| PdeVINV1 | EVM0021361 | chr3 | 15,565,345 | 15,566,949 | 534 | 59,632.07 | 5.3 |
| PdeVINV2 | EVM0032360 | chr3 | 14,483,232 | 14,487,904 | 661 | 73,385.74 | 5.92 |
| PdeVINV3 | EVM0005782 | chr15 | 13,459,417 | 13,463,828 | 626 | 70,134.85 | 5.3 |
| PdeVINV4 | EVM0034321 | Contig00422 | 29,267 | 33,178 | 460 | 51,001.51 | 5.36 |
| SsuCWINV1 | KAG5253080 | chr1 | 7,929,888 | 7,932,471 | 513 | 57,757.56 | 8.66 |
| SsuCWINV2 | KAG5243975 | chr6 | 15,728,185 | 15,732,344 | 578 | 65,683.72 | 8.97 |
| SsuCWINV3 | KAG5244133 | chr6 | 16,948,561 | 16,952,254 | 614 | 69,846.19 | 5.7 |
| SsuCWINV4 | KAG5244132 | chr6 | 16,945,043 | 16,947,752 | 547 | 62,500.56 | 6.68 |
| SsuVINV1 | KAG5249197 | chr3 | 8,030,054 | 8,035,528 | 550 | 61,029.28 | 5.92 |
| SsuVINV2 | KAG5230526 | chr15 | 11,591,356 | 11,595,290 | 642 | 72,151.16 | 5.21 |
| PdeNINV1 | EVM0016105 | chr8 | 6,703,225 | 6,707,387 | 692 | 77,974.79 | 6.1 |
| PdeNINV2 | EVM0028453 | chr13 | 472,802 | 477,178 | 617 | 69,982.91 | 5.83 |
| PdeNINV3 | EVM0036036 | chr8 | 1,215,796 | 1,219,873 | 663 | 74,292.93 | 6.45 |
| PdeNINV4 | EVM0037445 | chr10 | 21,183,172 | 21,187,826 | 666 | 74,536.12 | 6.02 |
| PdeNINV5 | EVM0003860 | chr5 | 773,232 | 777,399 | 657 | 74,839.51 | 5.78 |
| PdeNINV6 | EVM0006001 | chr4 | 19,121,245 | 19,126,930 | 617 | 69,484.1 | 7.82 |
| PdeNINV7 | EVM0018844 | chr5 | 24,044,123 | 24,046,098 | 543 | 62,277.55 | 6.01 |
| PdeNINV8 | EVM0008004 | chr19 | 14,355,560 | 14,362,118 | 557 | 63,338.65 | 5.92 |
| PdeNINV9 | EVM0037237 | chr4 | 17,702,452 | 17,705,280 | 573 | 65,529.25 | 6.11 |
| PdeNINV10 | EVM0031867 | chr2 | 6,833,729 | 6,837,875 | 722 | 81,121.68 | 5.1 |
| PdeNINV11 | EVM0022732 | chr9 | 10,672,157 | 10,674,968 | 574 | 65,655.41 | 5.92 |
| PdeNINV12 | EVM0014961 | chr13 | 12,780,570 | 12,788,090 | 557 | 63,568.04 | 6.35 |
| PdeNINV13 | EVM0012606 | Contig01214 | 1913 | 3300 | 245 | 27,475.09 | 5.99 |
| SsuNINV1 | KAG5240243 | chr8 | 5,743,869 | 5,747,374 | 683 | 77,291.94 | 6.14 |
| SsuNINV2 | KAG5232299 | chr13 | 345,772 | 349,410 | 640 | 72,308.86 | 5.97 |
| SsuNINV3 | KAG5239587 | chr8 | 1,071,815 | 1,075,805 | 665 | 74,395.89 | 6.02 |
| SsuNINV4 | KAG5237853 | chr10 | 15,999,564 | 16,002,831 | 667 | 74,351.93 | 5.87 |
| SsuNINV5 | KAG5248034 | chr4 | 14,455,452 | 14,460,986 | 467 | 52,028.75 | 5.37 |
| SsuNINV6 | KAG5224657 | chr19 | 13,831,154 | 13,837,354 | 557 | 63,446.95 | 6.23 |
| SsuNINV7 | KAG5247877 | chr4 | 13,017,770 | 13,021,450 | 574 | 65,729.31 | 6.12 |
| SsuNINV8 | KAG5251679 | chr2 | 13,585,135 | 13,590,231 | 713 | 80,109.69 | 5.18 |
| SsuNINV9 | KAG5239072 | chr9 | 9,074,707 | 9,077,316 | 574 | 65,572.23 | 6.14 |
| SsuNINV10 | KAG5233215 | chr13 | 11,775,507 | 11,781,303 | 557 | 63,485.93 | 6.39 |
| SsuNINV11 | KAG5251679 | Contig02329 | 58,959 | 59,417 | 137 | 15,469.01 | 7.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Tang, Q.; Mao, J.; Jia, C.; Qin, Z.; Chen, Y.; Liang, Q.; Dai, X.; Chen, Y.; Yin, T.; et al. Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development. Curr. Issues Mol. Biol. 2025, 47, 423. https://doi.org/10.3390/cimb47060423
Wang H, Tang Q, Mao J, Jia C, Qin Z, Chen Y, Liang Q, Dai X, Chen Y, Yin T, et al. Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development. Current Issues in Molecular Biology. 2025; 47(6):423. https://doi.org/10.3390/cimb47060423
Chicago/Turabian StyleWang, Hui, Qianhua Tang, Jinyan Mao, Chang Jia, Zilu Qin, Yiqun Chen, Qingqing Liang, Xiaogang Dai, Yingnan Chen, Tongming Yin, and et al. 2025. "Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development" Current Issues in Molecular Biology 47, no. 6: 423. https://doi.org/10.3390/cimb47060423
APA StyleWang, H., Tang, Q., Mao, J., Jia, C., Qin, Z., Chen, Y., Liang, Q., Dai, X., Chen, Y., Yin, T., & Wu, H. (2025). Genome-Wide Comparative Analysis of Invertases in the Salicaceae with the Identification of Genes Involved in Catkin Fiber Initiation and Development. Current Issues in Molecular Biology, 47(6), 423. https://doi.org/10.3390/cimb47060423





