T Cell Repertoire Analysis as a Molecular Signature of the Spectrum of T-LGL Lymphoproliferative Disorders: Tracing the Literature
Abstract
1. Introduction
2. Methods
3. Results
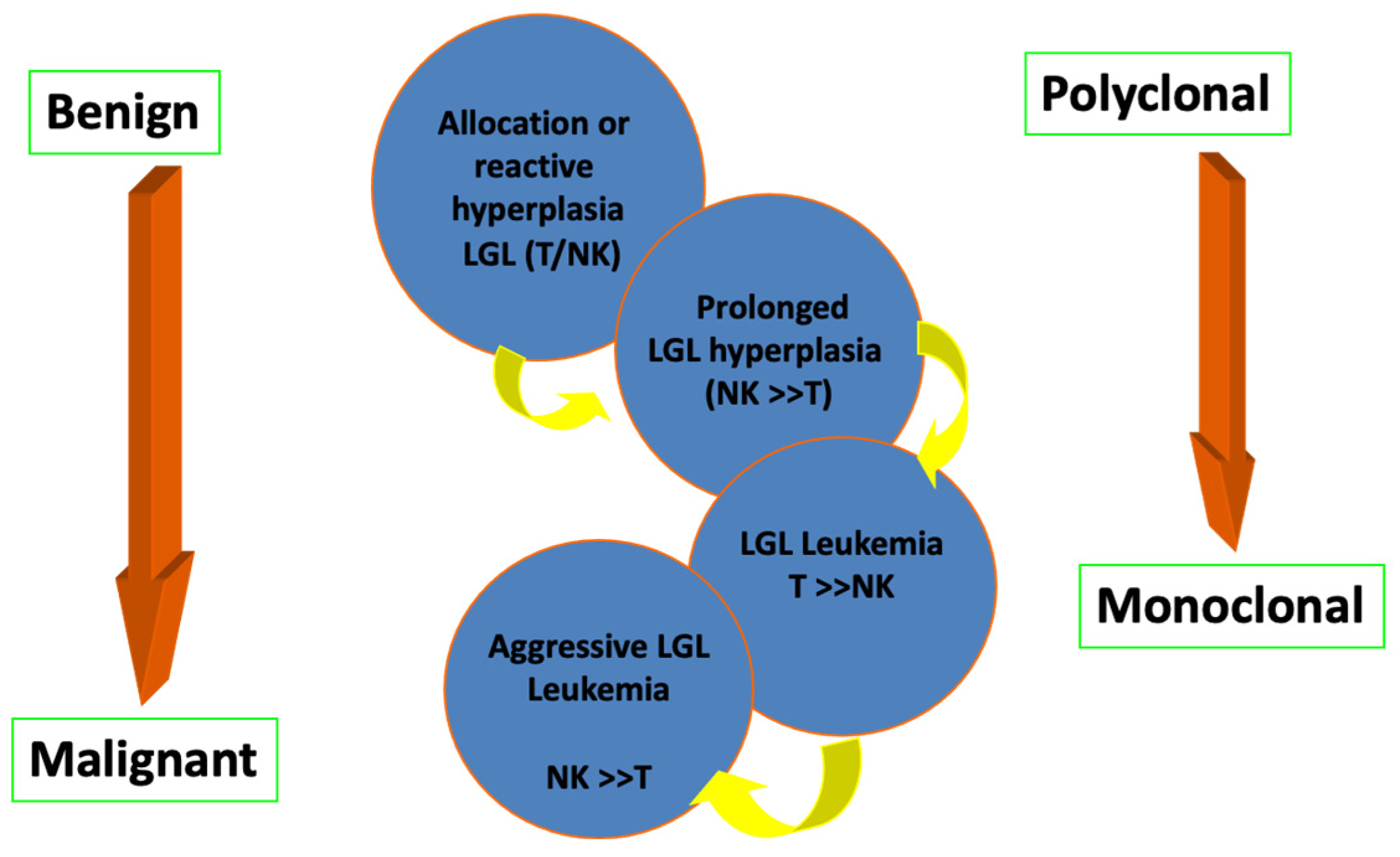
3.1. Spectrum of T-LGL Status and Associations with Other Disorders
3.1.1. Lymphoproliferative Cytotoxic T Lymphocytes and Hematological Diseases
3.1.2. T-LGL Lymphoproliferative Disorders and Myeloid Failure Syndromes: What Is the Exact Association?
Chronic Idiopathic Neutropenia (CIN)
3.1.3. T-LGL Lymphoproliferative Disorders and Allogeneic Hematopoietic Cell Transplantation
3.2. New Insights of the T-LGL Lymphoproliferative Disorders in the Next-Generation Sequencing (NGS) Era
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- O’Keefe, C.L.; Plasilova, M.; Wlodarski, M.; Risitano, A.M.; Rodriguez, A.R.; Howe, E.; Young, N.S.; His, E.; Maciejewski, J.P. Molecular analysis of TCR clonotypes in LGL: A clonal model for polyclonal responses. J. Immunol. 2004, 172, 1960–1969. [Google Scholar] [CrossRef] [PubMed]
- Plasilova, M.; Risitano, A.; Maciejewski, J.P. Application of the Molecular Analysis of the T-Cell Receptor Repertoire in the Study of Immune-Mediated Hematologic Diseases. Hematology 2003, 8, 173–181. [Google Scholar] [CrossRef]
- Calabretto, G.; Teramo, A.; Barilà, G.; Vicenzetto, C.; Gasparini, V.R.; Semenzato, G.; Zambello, R. Neutropenia and Large Granular Lymphocyte Leukemia: From Pathogenesis to Therapeutic Options. Cells 2021, 10, 2800. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mohty, M.; Faucher, C.; Vey, N.; Chabannon, C.; Sainty, D.; Arnoulet, C.; Gaugler, B.; Gastaut, J.A.; Maraninchi, D.; Olive, D.; et al. Features of large granular lymphocytes (LGL) expansion following allogeneic stem cell transplantation: A long-term analysis. Leukemia 2002, 16, 2129–2133. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, T.; Stamatopoulos, K.; Stavroyianni, N.; Paterakis, G.; Phisphis, M.; Stefanoudaki-Sofianatou, K. Evidence for T-large granular lymphocyte-mediated neutropenia in Rituximab-treated lymphoma patients: Report of two cases. Leuk. Res. 2002, 26, 597–600. [Google Scholar] [CrossRef]
- Qiu, Z.-Y.; Tian, G.-Y.; Zhang, Z.; Zhang, Y.-Q.; Xu, W.; Li, J.-Y. Large granular lymphocytosis after transplantation. Oncotarget 2017, 8, 81697–81708. [Google Scholar] [CrossRef]
- Kataria, A.; Cohen, E.; Saad, E.; Atallah, E.; Bresnahan, B. Large Granular Lymphocytic Leukemia Presenting Late After Solid Organ Transplantation: A Case Series of Four Patients and Review of the Literature. Transplant. Proc. 2014, 46, 3278–3281. [Google Scholar] [CrossRef]
- Horiuchi, T.; Hirokawa, M.; Kawabata, Y.; Kitabayashi, A.; Matsutani, T.; Yoshioka, T.; Tsuruta, Y.; Suzuki, R.; Miura, A.B. Identification of the T cell clones expanding within both CD8+CD28+ and CD8+CD28− T cell subsets in recipients of allogeneic hematopoietic cell grafts and its implication in post-transplant skewing of T cell receptor repertoire. Bone Marrow Transplant. 2001, 27, 731–739. [Google Scholar] [CrossRef]
- Liu, X.; Chesnokova, V.; Forman, S.J.; Diamond, D.J. Molecular analysis of T-cell receptor repertoire in bone marrow transplant recipients: Evidence for oligoclonal T-cell expansion in graft-versus-host disease lesions. Blood 1996, 87, 3032–3044. [Google Scholar] [CrossRef]
- Lai, L.; Wang, L.; Chen, H.; Zhang, J.; Yan, Q.; Ou, M.; Lin, H.; Hou, X.; Chen, S.; Dai, Y.; et al. T cell repertoire following kidney transplantation revealed by high-throughput sequencing. Transplant. Immunol. 2016, 39, 34–45. [Google Scholar] [CrossRef]
- Gorochov, G.; Debré, P.; Leblond, V.; Sadat-Sowti, B.; Sigaux, F.; Autran, B. Oligoclonal expansion of CD8+ CD57+ T cells with restricted T-cell receptor beta chain variability after bone marrow transplantation. Blood 1994, 83, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, U.; Parylo, S.; Kedia, S.; Hussein, S.; Atallah, J.P. Large Granular Lymphocytic Leukemia: A Report of Response to Rituximab. Case Rep. Hematol. 2017, 2017, 7506542. [Google Scholar] [CrossRef] [PubMed]
- Lamy, T.; Loughran, T.P. Clinical features of large granular lymphocyte leukemia. Semin. Hematol. 2003, 40, 185–195. [Google Scholar] [CrossRef]
- Ghrenassia, E.; Roulin, L.; Aline-Fardin, A.; Marzac, C.; Féger, F.; Gay, J.; Pacanowski, J.; Hertig, A.; Coppo, P. The Spectrum of Chronic CD8+ T-Cell Expansions: Clinical Features in 14 Patients. PLoS ONE 2014, 9, e91505. [Google Scholar] [CrossRef] [PubMed]
- Delobelp, P.; Godela, A.; Thebaults, S.; Alricl, L.; Duffautm, M. Transient clonal expansion of T-large granular lymphocytes during primary cytomegalovirus infection. J. Infect. 2006, 53, 65–67. [Google Scholar] [CrossRef]
- Neff, J.L.; Howard, M.T.; Morice, W.G. Distinguishing T-cell Large Granular Lymphocytic Leukemia from Reactive Conditions. Surg. Pathol. Clin. 2013, 6, 631–639. [Google Scholar] [CrossRef]
- Rose, M.G.; Berliner, N. T-cell large granular lymphocyte leukemia and related disorders. Oncologist 2004, 9, 247–258. [Google Scholar] [CrossRef]
- Wong, K.F.; Chan, J.C.; Liu, H.S.; Man, C.; Kwong, Y.L. Chromosomal abnormalities in T-cell large granular lymphocyte leukaemia: Report of two cases and review of the literature. Br. J. Haematol. 2002, 116, 598. [Google Scholar] [CrossRef]
- Posnett, D.N.; Sinha, R.; Kabak, S.; Russo, C. Clonal populations of T cells in normal elderly humans: The T cell equivalent to “benign monoclonal gammapathy”. J. Exp. Med. 1994, 179, 609–618. [Google Scholar] [CrossRef]
- Au, W.Y.; Lam, C.C.; Lie, A.K.; Pang, A.; Kwong, Y.L. T-cell large granular lymphocyte leukemia of donor origin after allogeneic bone marrow transplantation. Am. J. Clin. Pathol. 2003, 120, 626–630. [Google Scholar] [CrossRef]
- Kothapalli, R.; Bailey, R.D.; Kusmartseva, I.; Mane, S.; Epling-Burnette, P.K.; Loughran, T.P. Constitutive expression of cytotoxic proteases and down-regulation of protease inhibitors in LGL leukemia. Int. J. Oncol. 2023, 22, 33–39. [Google Scholar] [CrossRef]
- Loughran, T.P., Jr.; Hadlock, K.G.; Perzova, R.; Gentile, T.C.; Yang, Q.; Foung, S.K.; Poiesz, B.J. Epitope mapping of HTLV envelope seroreactivity in LGL leukaemia. Br. J. Haematol. 1998, 101, 318–324. [Google Scholar] [CrossRef]
- Sokol, L.; Loughran, T.P. Large Granular Lymphocyte Leukemia. Oncologist 2006, 11, 263–273. [Google Scholar] [CrossRef]
- O’Malley, D.P. T-Cell Large Granular Leukemia and Related Proliferations. Am. J. Clin. Pathol. 2007, 127, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Rahul, E.; Ningombam, A.; Acharya, S.; Tanwar, P.; Ranjan, A.; Chopra, A. Large granular lymphocytic leukemia: A brief review. Am. J. Blood Res. 2022, 12, 17–32. [Google Scholar] [PubMed] [PubMed Central]
- Lamy, T.; Loughran, T.P. How I treat LGL leukemia. Blood 2011, 117, 2764–2774. [Google Scholar] [CrossRef]
- Langerak, A.W.; Assmann, J.L.J.C. Large granular lymphocyte cells and immune dysregulation diseases—The chicken or the egg? Haematologica 2018, 103, 193–194. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lamy, T.; Moignet, A.; Loughran, T.P. LGL leukemia: From pathogenesis to treatment. Blood 2017, 129, 1082–1094. [Google Scholar] [CrossRef]
- Jerez, A.; Clemente, M.J.; Makishima, H.; Koskela, H.; Leblanc, F.; Peng, N.K.; Olson, T.; Przychodzen, B.; Afable, M.; Gomez-Segui, I.; et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 2012, 120, 3048–3057. [Google Scholar] [CrossRef]
- Koskela, H.L.; Eldfors, S.; Ellonen, P.; van Adrichem, A.J.; Kuusanmäki, H.; Andersson, E.I.; Lagström, S.; Clemente, M.J.; Olson, T.; Jalkanen, S.E.; et al. Somatic STAT3 Mutations in Large Granular Lymphocytic Leukemia. N. Engl. J. Med. 2012, 366, 1905–1913. [Google Scholar] [CrossRef]
- Savola, P.; Bhattacharya, D.; Huuhtanen, J. The spectrum of somatic mutations in large granular lymphocyte leukemia, rheumatoid arthritis, and Felty’s syndrome. Semin. Hematol. 2022, 59, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Sokol, L.; Agrawal, D.; Loughran, T.P. Characterization of HTLV envelope seroreactivity in large granular lymphocyte leukemia. Leuk. Res. 2005, 29, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Hadlock, K.G.; Goh, C.J.; Bradshaw, P.A.; Perkins, S.; Lo, J.; Kaplan, J.E.; Khabbaz, R.; Foung, S.K. Delineation of an immunodominant and human T-cell lymphotropic virus (HTLV)-specific epitope within the HTLV-I transmembrane glycoprotein. Blood 1995, 86, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Lamy, T.; Liu, J.H.; Landowski, T.H.; Dalton, W.S.; Loughran, T.P. Dysregulation of CD95/CD95 ligand-apoptotic pathway in CD3(+) large granular lymphocyte leukemia. Blood 1998, 92, 4771–4777. [Google Scholar] [CrossRef]
- Steinway, S.N.; LeBlanc, F.; Loughran, T.P., Jr. The pathogenesis and treatment of large granular lymphocyte leukemia. Blood Rev. 2014, 28, 87–94. [Google Scholar] [CrossRef]
- Wlodarski, M.W.; Nearman, Z.; Jiang, Y.; Lichtin, A.; Maciejewski, J.P. Clonal predominance of CD8(+) T cells in patients with unexplained neutropenia. Exp. Hematol. 2008, 36, 293–300. [Google Scholar] [CrossRef]
- Maciejewski, J.P.; O’Keefe, C.; Gondek, L.; Tiu, R. Immune-mediated bone marrow failure syndromes of progenitor and stem cells: Molecular analysis of cytotoxic T cell clones. Folia Histochem. Cytobiol. 2007, 45, 5–14. [Google Scholar]
- Mavroudi, I.; Papadaki, H.A. Genetic associations in acquired immune-mediated bone marrow failure syndromes: Insights in aplastic anemia and chronic idiopathic neutropenia. Clin. Dev. Immunol. 2012, 2012, 123789. [Google Scholar] [CrossRef]
- Papadaki, H.A.; Pontikoglou, C. Pathophysiologic mechanisms, clinical features and treatment of idiopathic neutropenia. Expert. Rev. Hematol. 2008, 1, 217–229. [Google Scholar] [CrossRef]
- Papadaki, H.A.; Palmblad, J.; Eliopoulos, G.D. Non-immune chronic idiopathic neutropenia of adult: An overview. Eur. J. Haematol. 2001, 67, 35–44. [Google Scholar] [CrossRef]
- Papadaki, H.A.; Eliopoulos, G.D.; Coulocheri, S.A.; Spyropoulou, M.; Stavropoulos-Giokas, C. Increased frequency of HLA-DRB1*1302 haplotype in patients with nonimmune chronic idiopathic neutropenia of adults. Blood 2001, 97, 580–581. [Google Scholar] [CrossRef] [PubMed]
- Palmblad, J.; Papadaki, H.A. Chronic idiopathic neutropenias and severe congenital neutropenia. Curr. Opin. Hematol. 2008, 15, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Dale, D.C.; Bolyard, A.A. An update on the diagnosis and treatment of chronic idiopathic neutropenia. Curr. Opin. Hematol. 2017, 24, 46–53. [Google Scholar] [CrossRef]
- Papadaki, H.A.; Stamatopoulos, K.; Damianaki, A.; Gemetzi, C.; Anagnostopoulos, A.; Papadaki, T.; Eliopoulos, A.G.; Eliopoulos, G.D. Activated T-lymphocytes with myelosuppressive properties in patients with chronic idiopathic neutropenia. Br. J. Haematol. 2005, 128, 863–876. [Google Scholar] [CrossRef]
- Rajala, H.L.; Olson, T.; Clemente, M.J.; Lagström, S.; Ellonen, P.; Lundan, T.; Hamm, D.E.; Zaman, S.A.; Lopez Marti, J.M.; Andersson, E.I.; et al. The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia. Haematologica 2015, 100, 91–99. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jerez, A.; Clemente, M.J.; Makishima, H.; Rajala, H.; Gómez-Seguí, I.; Olson, T.; McGraw, K.; Przychodzen, B.; Kulasekararaj, A.; Afable, M.; et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood 2013, 122, 2453–2459. [Google Scholar] [CrossRef] [PubMed]
- Spanoudakis, M.; Koutala, H.; Ximeri, M.; Pyrovolaki, K.; Stamatopoulos, K.; Papadaki, H.A. T-cell receptor Vβ repertoire analysis in patients with chronic idiopathic neutropenia demonstrates the presence of aberrant T-cell expansions. Clin. Immunol. 2010, 137, 384–395. [Google Scholar] [CrossRef]
- Mastrodemou, S.; Stalika, E.; Vardi, A.; Gemenetzi, K.; Spanoudakis, M.; Karypidou, M.; Mavroudi, I.; Hadzidimitriou, A.; Stavropoulos-Giokas, C.; Papadaki, H.A.; et al. Cytotoxic T cells in chronic idiopathic neutropenia express restricted antigen receptors. Leuk. Lymphoma 2017, 58, 2926–2933. [Google Scholar] [CrossRef]
- Uderzo, C.; Corti, P.; Pappalettera, M.; Baldini, V.; Lucchini, G.; Meani, D.; Rovelli, A. Life Satisfaction in Young Adults 10 or More Years after Hematopoietic Stem Cell Transplantation for Childhood Malignant and Nonmalignant Diseases Does Not Show Significant Impairment Compared with Healthy Controls: A Case-Matched Study. Biol. Blood Marrow Transplant. 2012, 18, 1759–1764. [Google Scholar] [CrossRef]
- Papadaki, T.; Stamatopoulos, K.; Anagnostopoulos, A.; Fassas, A. Rituximab-associated immune myelopathy. Blood 2003, 102, 1557–1558. [Google Scholar] [CrossRef][Green Version]
- Werner, L.; Dor, C.; Salamon, N.; Nagar, M.; Shouval, D.S. T and B Cell Receptor Immune Repertoire Analysis using Next-generation Sequencing. J. Vis. Exp. 2021, 167, e61792. [Google Scholar] [CrossRef] [PubMed]
- de Masson, A.; O’Malley, J.T.; Elco, C.P.; Garcia, S.S.; Divito, S.J.; Lowry, E.L.; Tawa, M.; Fisher, D.C.; Devlin, P.M.; Teague, J.E.; et al. High-throughput sequencing of the T cell receptor beta gene identifies aggressive early-stage mycosis fungoides. Sci. Transl. Med. 2018, 10, eaar5894. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, T.-Y. Revealing the Immune Heterogeneity between Systemic Lupus Erythematosus and Rheumatoid Arthritis Based on Multi-Omics Data Analysis. Int. J. Mol. Sci. 2022, 23, 5166. [Google Scholar] [CrossRef]
- Gkazi, A.S.; Margetts, B.K.; Attenborough, T.; Mhaldien, L.; Standing, J.F.; Oakes, T.; Heather, J.M.; Booth, J.; Pasquet, M.; Chiesa, R.; et al. Clinical T Cell Receptor Repertoire Deep Sequencing and Analysis: An Application to Monitor Immune Reconstitution Following Cord Blood Transplantation. Front. Immunol. 2018, 9, 2547. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, Y.; Zheng, B.; Luo, L.; Su, Z. Human TCR repertoire in cancer. Cancer Med. 2024, 13, e70164. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Keane, C.; Gould, C.; Jones, K.; Hamm, D.; Talaulikar, D.; Ellis, J. The T-cell receptor repertoire influences the tumor microenvironment and is associated with survival in aggressive b-cell lymphoma. Clin. Cancer Res. 2017, 23, 1820–1828. [Google Scholar] [CrossRef]
- Olschewski, V.; Witte, H.M.; Bernard, V.; Steinestel, K.; Peter, W.; Merz, H.; Rieken, J.; Biersack, H.; von Bubnoff, N.; Feller, A.C.; et al. Systemic inflammation and tumour-infiltrating T-cell receptor repertoire diversity are predictive of clinical outcome in high-grade b-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements. Cancers 2021, 13, 887. [Google Scholar] [CrossRef]
- Assmann, J.L.J.C.; Vlachonikola, E.; Kolijn, P.M.; Agathangelidis, A.; Pechlivanis, N.; Papalexandri, A.; Stamatopoulos, K.; Chatzidimitriou, A.; Langerak, A.W. Context-dependent T-cell Receptor Gene Repertoire Profiles in Proliferations of T Large Granular Lymphocytes. Hemasphere 2023, 7, e929. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stalika, E.; Papalexandri, A.; Kannelis, G.; Batsis, I.; Papadaki, T.; Anagnostopoulos, A.; Stamatopoulos, K. Transient monoclonal CD3+ T large granular lymphocyte proliferation in a case of mantle cell lymphoma with Rituximab-associated late onset neutropenia. Hematol. Oncol. 2011, 29, 144–146. [Google Scholar] [CrossRef]
- Sumi, M.; Watanabe, M.; Sato, K.; Shimizu, I.; Ueki, T.; Akahane, D.; Ueno, M.; Ichikawa, N.; Asano, N.; Kobayashi, H. Epstein-Barr virus-associated lymphoproliferative disorder developed after anti-thymocyte globulin therapy in a patient with bone marrow failure associated with T-cell large granular lymphocytic leukemia. [Rinsho Ketsueki] Jpn. J. Clin. Hematol. 2011, 52, 1782–1787. [Google Scholar]
- Nann-Rütti, S.; Tzankov, A.; Cantoni, N.; Halter, J.; Heim, D.; Tsakiris, D.; Arber, C.; Buser, A.; Gratwohl, A.; Tichelli, A.; et al. Large Granular Lymphocyte Expansion after Allogeneic Hematopoietic Stem Cell Transplant Is Associated with a Cytomegalovirus Reactivation and Shows an Indolent Outcome. Biol. Blood Marrow Transplant. 2012, 18, 1765–1770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poch Martell, M.; Hamad, N.; Shin, E.; Moon, J.H.; Sohn, S.K.; Uhm, J.; Fotios, V.M.; Auro, V.; Jeffrey, H.L.; Hans, A.M.; et al. Distinctive clinical characteristics and favorable outcomes in patients with large granular lymphocytosis after allo-HCT: 12-year follow-up data. Eur. J. Haematol. 2017, 99, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Kallemeijn, M.J.; de Ridder, D.; Schilperoord-Vermeulen, J.; van der Klift, M.Y.; Sandberg, Y.; van Dongen, J.J.M.; Langerak, A.W. Dysregulated signaling, proliferation and apoptosis impact on the pathogenesis of TCRγδ+ T cell large granular lymphocyte leukemia. Lee, S.-G, editor. PLoS ONE 2017, 12, e0175670. [Google Scholar] [CrossRef]
- Camagna, A.; Cedrone, L.; Caré, A.; Samoggia, P.; De Marco, M.C.; Del Duca, P.; De Martinis, C.; Testa, U. Polyclonal expansion of CD3(+)/CD4(+)/CD56(+) large granular lymphocytes and autoimmunity associated with dysregulation of Fas/FasL apoptotic pathway. Br. J. Haematol. 2001, 112, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Epling-Burnette, P.K.; Painter, J.S.; Zou, J.; Bai, F.; Wei, S.; Loughran, T.P., Jr. Antigen activation and impaired Fas-induced death-inducing signaling complex formation in T-large-granular lymphocyte leukemia. Blood 2007, 111, 1610–1616. [Google Scholar] [CrossRef]
- Zhang, D.; Loughran, T.P., Jr. Large granular lymphocytic leukemia: Molecular pathogenesis, clinical manifestations, and treatment. Hematol. Am. Soc. Hematol. Educ. Program 2012, 2012, 652–659. [Google Scholar] [CrossRef]
- Oshimi, K. Clinical Features, Pathogenesis, and Treatment of Large Granular Lymphocyte Leukemias. Intern. Med. 2017, 56, 1759–1769. [Google Scholar] [CrossRef]
- Shah, M.V.; Zhang, R.; Loughran, T.P. Never Say Die: Survival Signaling in Large Granular Lymphocyte Leukemia. Clin. Lymphoma Myeloma 2009, 9, S244–S253. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, B.; Gao, L.; Liu, J.; Chen, X.; Huang, H.; Zhao, Z. Next generation sequencing reveals changes of the γδ T cell receptor repertoires in patients with pulmonary tuberculosis. Sci. Rep. 2018, 8, 3956. [Google Scholar] [CrossRef]
- Dziubianau, M.; Hecht, J.; Kuchenbecker, L.; Sattler, A.; Stervbo, U.; Rödelsperger, C.; Nickel, P.; Neumann, A.U.; Robinson, P.N.; Mundlos, S.; et al. TCR Repertoire Analysis by Next Generation Sequencing Allows Complex Differential Diagnosis of T Cell-Related Pathology. Am. J. Transplant. 2013, 13, 2842–2854. [Google Scholar] [CrossRef]
- O′Connell, A.E.; Volpi, S.; Dobbs, K.; Fiorini, C.; Tsitsikov, E.; de Boer, H.; Barlan, I.B.; Despotovic, J.M.; Espinosa-Rosales, F.J.; Hanson, I.C.; et al. Next Generation Sequencing Reveals Skewing of the T and B Cell Receptor Repertoires in Patients with wiskott-Aldrich Syndrome. Front. Immunol. 2014, 5, 340. [Google Scholar] [PubMed]
- Fang, H.; Yamaguchi, R.; Liu, X.; Daigo, Y.; Yew, P.Y.; Tanikawa, C.; Matsuda, K.; Imoto, S.; Miyano, S.; Nakamura, Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next-generation sequencing (NGS). OncoImmunology 2014, 3, e968467. [Google Scholar] [CrossRef] [PubMed]
- Thapa, D.R.; Tonikian, R.; Sun, C.; Liu, M.; Dearth, A.; Petri, M.; Pepin, F.; Emerson, R.O.; Ranger, A. Longitudinal analysis of peripheral blood T cell receptor diversity in patients with systemic lupus erythematosus by next-generation sequencing. Arthritis Res. Ther. 2015, 17, 132. [Google Scholar] [CrossRef]
- Sufficool, K.E.; Lockwood, C.M.; Abel, H.J.; Hagemann, I.S.; Schumacher, J.A.; Kelley, T.W.; Duncavage, E.J. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J. Am. Acad. Dermatol. 2015, 73, 228–236.e2. [Google Scholar] [CrossRef]
- Kitaura, K.; Shini, T.; Matsutani, T.; Suzuki, R. A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and β repertoires and identifying potential new invariant TCR α chains. BMC Immunol. 2016, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Langerak, A.W.; Brüggemann, M.; Davi, F.; Darzentas, N.; van Dongen, J.J.M.; Gonzalez, D.; Cazzaniga, G.; Giudicelli, V.; Lefranc, M.P.; Giraud, M.; et al. High-Throughput Immunogenetics for Clinical and Research Applications in Immunohematology: Potential and Challenges. J. Immunol. 2017, 198, 3765–3774. [Google Scholar]
- Papalexandri, A.; Stalika, E.; Iskas, M.; Karypidou, M.; Zerva, P.; Touloumenidou, T.; Tachynopoulou, V.; Batsis, I.; Papadaki, T.; Sakellari, I.; et al. Molecular evidence for repertoire skewing of T large granular lymphocyte proliferation after allogeneic hematopoietic SCT: Report of two cases. Bone Marrow Transplant. 2013, 48, 1260–1261. [Google Scholar] [CrossRef]
- Vardi, A.; Agathangelidis, A.; Stalika, E.; Karypidou, M.; Siorenta, A.; Anagnostopoulos, A.; Rosenquist, R.; Hadzidimitriou, A.; Ghia, P.; Sutton, L.A.; et al. Antigen selection shapes the T-cell repertoire in chronic lymphocytic leukemia. Clin. Cancer Res. 2016, 22, 167–174. [Google Scholar] [CrossRef]
- Boeckx, N.; Uyttebroeck, A.; Langerak, A.W.; Brusselmans, C.; Goossens, W.; Bossuyt, X. Clonal proliferation of T-Cell large granular lymphocytes. Pediatr. Blood Cancer 2004, 42, 275–277. [Google Scholar] [CrossRef]
- Wlodarski, M.W.; Schade, A.E.; Maciejewski, J.P. T-large granular lymphocyte leukemia: Current molecular concepts. Hematology 2006, 11, 245–256. [Google Scholar] [CrossRef]
- Wlodarski, M.W.; Gondek, L.P.; Nearman, Z.P.; Plasilova, M.; Kalaycio, M.; His, E.D.; Maciejewski, J.P. Molecular strategies for detection and quantitation of clonal cytotoxic T-cell responses in aplastic anemia and myelodysplastic syndrome. Blood 2006, 108, 2632–2641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gill, H.; Ip, A.H.W.; Leung, R.; So, J.C.C.; Pang, A.W.K.; Tse, E.; Leung, A.Y.; Lie, A.K.; Kwong, Y.L. Indolent T-cell large granular lymphocyte leukaemia after haematopoietic SCT: A clinicopathologic and molecular analysis. Bone Marrow Transplant. 2012, 47, 952–956. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clemente, M.J.; Wlodarski, M.W.; Makishima, H.; Viny, A.D.; Bretschneider, I.; Shaik, M.; Bejanyan, N.; Lichtin, A.E.; His, E.D.; Paquette, R.L.; et al. Clonal drift demonstrates unexpected dynamics of the T-cell repertoire in T-large granular lymphocyte leukemia. Blood 2011, 118, 4384–4393. [Google Scholar] [CrossRef]
- Clemente, M.J.; Przychodzen, B.; Jerez, A.; Dienes, B.E.; Afable, M.G.; Husseinzadeh, H.; Rajala, H.L.; Wlodarski, M.W.; Mustjoki, S.; Maciejewski, J.P. Deep sequencing of the T-cell receptor repertoire in CD8+ T-large granular lymphocyte leukemia identifies signature landscapes. Blood 2013, 122, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Almeida, J.; Santos, A.H.; dos Anjos Teixeira, M.; Alguero, M.C.; Queirós, M.L.; Balanzategui, A.; Justiça, B.; Gonzalez, M.; San Miguel, J.F.; et al. Immunophenotypic analysis of the TCR-Vbeta repertoire in 98 persistent expansions of CD3(+)/TCR-alphabeta(+) large granular lymphocytes: Utility in assessing clonality and insights into the pathogenesis of the disease. Am. J. Pathol. 2001, 159, 1861–1868. [Google Scholar] [CrossRef]
- Sandberg, Y.; Kallemeijn, M.J.; Dik, W.A.; Tielemans, D.; Wolvers-Tettero, I.L.M.; van Gastel-Mol, E.J.; Szczepanski, T.; Pol, Y.; Darzentas, N.; van Dongen, J.J.; et al. Lack of common TCRA and TCRB clonotypes in CD8(+)/TCRαβ(+) T-cell large granular lymphocyte leukemia: A review on the role of antigenic selection in the immunopathogenesis of CD8(+) T-LGL. Blood Cancer J. 2014, 4, e172. [Google Scholar] [CrossRef] [PubMed]
- Zambello, R.; Semenzato, G. Large granular lymphocyte disorders: New etiopathogenetic clues as a rationale for innovative therapeutic approaches. Haematologica 2009, 94, 1341–1345. [Google Scholar] [CrossRef]
- Halapi, E.; Jeddi-Tehrani, M.; Blücher, Å.; Andersson, R.; Rossi, P.; Wigzell, H.; Grunewald, J. Diverse T-cell receptor CDR3 length patterns in human CD4+ and CD8+T lymphocytes from newborns and adults. Scand. J. Immunol. 1999, 49, 149–154. [Google Scholar] [CrossRef][Green Version]
- Costello, P.J.; Winchester, R.J.; Curran S a Peterson, K.S.; Kane, D.J.; Bresnihan, B.; FitzGerald, O.M. Psoriatic arthritis joint fluids are characterized by CD8 and CD4 T cell clonal expansions appear antigen driven. J. Immunol. 2001, 166, 2878–2886. [Google Scholar] [CrossRef]
- Venturi, V.; Kedzierska, K.; Price, D.A.; Doherty, P.C.; Douek, D.C.; Turner, S.J.; Davenport, M.P. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc. Natl. Acad. Sci. USA 2006, 103, 18691–18696. [Google Scholar] [CrossRef]
- Madi, A.; Shifrut, E.; Reich-Zeliger, S.; Gal, H.; Best, K.; Ndifon, W.; Chain, B.; Cohen, I.R.; Friedman, N. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res. 2014, 24, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, J.D.; Zeng, T.; Choe, G.; Munro, S.; Moore, R.; Webb, J.R.; Holt, R.A. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011, 21, 790–797. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stalika, E.; Stavroyianni, N.; Anagnostopoulos, A.; Papadaki, H.; Papadaki, T.; Stamatopoulos, K. P083 A father and his son presenting with cytopenias associated with CD3 T large granular lymphocyte proliferation. Leuk. Res. 2009, 33 (Suppl. S1), S106. [Google Scholar] [CrossRef]
- Stalika, E.; Papalexandri, A.; Iskas, M.; Stavroyianni, N.; Kanellis, G.; Kotta, K.; Pontikoglou, C.; Siorenta, A.; Anagnostopoulos, A.; Papadaki, H.; et al. Familial CD3+T large granular lymphocyte leukemia: Evidence that genetic predisposition and antigen selection promote clonal cytotoxic T-cell responses. Leuk. Lymphoma 2014, 55, 1781–1787. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.; Massarotti, E. Rituximab. Drugs Today 2005, 41, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Rajala, H.L.; Porkka, K.; Maciejewski, J.P.; Loughran, T.P., Jr.; Mustjoki, S. Uncovering the pathogenesis of large granular lymphocytic leukemia-novel STAT3 and STAT5b mutations. Ann. Med. 2014, 46, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Lee, Y.J.; Kim, Y.; Kim, H.K.; Lim, J.H.; Jo, J.C. T-large granular lymphocytic leukemia. Blood Res. 2023, 58 (Suppl. S1), S52–S57. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ullah, F.; Markouli, M.; Orland, M.; Ogbue, O.; Dima, D.; Omar, N.; Mustafa Ali, M.K. Large Granular Lymphocytic Leukemia: Clinical Features, Molecular Pathogenesis, Diagnosis and Treatment. Cancers 2024, 16, 1307. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dufour, C.; Corcione, A.; Svahn, J.; Haupt, R.; Poggi, V.; Béka’ssy, A.N.; Scimè, R.; Pistorio, A.; Pistoia, V. TNF-αandIFN-γare overexpressed in the bone marrow of Fanconi anemia patients and TNF-α suppresses erythropoiesis in vitro. Blood 2003, 102, 2053–2059. [Google Scholar] [CrossRef]
- Tsamesidis, I.; Pantaleo, A.; Pekou, A.; Gusani, A.; Iliadis, S.; Makedou, K.; Manca, A.; Carruale, A.; Lymperaki, E.; Fozza, C. Correlation of Oxidative Stress Biomarkers and Hematological Parameters in Blood Cancer Patients from Sardinia, Italy. Int. J. Hematol. Oncol. Stem Cell Res. 2019, 13, 49–57. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lian, J.; Yue, Y.; Yu, W.; Zhang, Y. Immunosenescence: A key player in cancer development. J. Hematol. Oncol. 2020, 13, 151, Erratum in J. Hematol. Oncol. 2025, 18, 25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Clinical Entity | Conclusions | Reference |
|---|---|---|
| Cutaneous T cell lymphomas (CTCLs)—Mycosis fungoides | 1. An increased proportion of malignant T cell clones found in the skin is notably linked to reduced progression-free survival and overall survival rates in patients diagnosed with CTCL. 2. High-throughput DNA sequencing of the TCRβ gene revealed that a tumor clone frequency greater than 25% serves as a significant predictor of disease progression and unfavorable survival outcomes for MF patients with localized skin disease. | [52] |
| Systemic Lupus Erythematosus (SLE) and Rheumatoid Arthritis (RA) | 1. Autoimmune diseases could lead to an increase in specific CDR3 amino acid sequences. 2. Significant changes in TRBV, TRBJ, IGHV and IGHJ genes. 3. Improve the understanding of TCR and BCR repertoires’ features and suggest an immune response to the common autoantigens in SLE or RA patients, which could assist in the development of targeted biotherapy and the diagnosis of SLE and RA. | [53] |
| Cord blood transplantation | Observation of clonal expansions in cord blood transplantation group by pinpointing antigen “specific” sequences in samples through VDJdb. The presence of clonally expanded sequences that are “specific” to HIV-1 in clinically confirmed HIV seronegative samples reinforces the notion of significant TCR cross-reactivity throughout the repertoire. | [54] |
| TCR repertoire in cancer | 1. Evaluate immune diversity, assisting with early-stage cancer diagnosis, treatment selection, and prognosis prediction. 2. Dynamic TCR repertoire analysis may serve as a useful indicator of cancer development and guide immunotherapy. 3. In gastric cancer, low diversity of the TCR repertoire within the tumor-adjacent mucosal tissue is associated with a poor clinical prognosis in patients. 4. In lung cancer, a higher TCR repertoire diversity in tumor tissues is associated with worse cancer outcomes, while patients with a higher TCR diversity in peripheral blood show longer progression-free survival (PFS). | [55] |
| In B-cell lymphomas (BCL)/Diffuse Large B cell Lymphomas (DLBCL) | In BCL, a restricted TR repertoire is found to be associated with a poor outcome in DLBCL treated without immune-checkpoint inhibition and in high-grade B-cell lymphomas. | [56,57] |
| T-LGL lymphoproliferative disorders | 1. The TRB gene repertoire in patients with T-LGL lymphoproliferations is highly dependent on context. 2. Different patterns of clonality across various disease scenarios. 3. Significant temporal clonal changes, suggesting that T-LGL lymphoproliferations could serve as an epiphenomenon in the presence of other cancers, potentially reacting to tumor antigens, for instance. | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stalika, E.; Tsamesidis, I. T Cell Repertoire Analysis as a Molecular Signature of the Spectrum of T-LGL Lymphoproliferative Disorders: Tracing the Literature. Curr. Issues Mol. Biol. 2025, 47, 264. https://doi.org/10.3390/cimb47040264
Stalika E, Tsamesidis I. T Cell Repertoire Analysis as a Molecular Signature of the Spectrum of T-LGL Lymphoproliferative Disorders: Tracing the Literature. Current Issues in Molecular Biology. 2025; 47(4):264. https://doi.org/10.3390/cimb47040264
Chicago/Turabian StyleStalika, Evangelia, and Ioannis Tsamesidis. 2025. "T Cell Repertoire Analysis as a Molecular Signature of the Spectrum of T-LGL Lymphoproliferative Disorders: Tracing the Literature" Current Issues in Molecular Biology 47, no. 4: 264. https://doi.org/10.3390/cimb47040264
APA StyleStalika, E., & Tsamesidis, I. (2025). T Cell Repertoire Analysis as a Molecular Signature of the Spectrum of T-LGL Lymphoproliferative Disorders: Tracing the Literature. Current Issues in Molecular Biology, 47(4), 264. https://doi.org/10.3390/cimb47040264







