1. Introduction
Colorectal cancer (CRC), one of the most common malignant tumors within the digestive system, primarily comprises two subtypes: colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ). CRC is the third most frequently diagnosed cancer and second most common cause of cancer-related death worldwide, with extremely high morbidity and mortality rates [
1]. In its early stages, CRC often presents no clinical symptoms, leading to most cases being diagnosed at advanced stages [
2]. Missing the optimal therapeutic window is a key factor contributing to the high mortality rate of CRC [
3]. Consequently, early diagnosis and treatment of CRC are crucial [
4].
UGP-glucose pyrophosphate synthase 2 (UGP2), an enzyme involved in the glycogen biosynthesis pathway, catalyzes the conversion of glucose-1-phosphate and UTP by UDP-glucose pyrophosphorylase into UDP-glucose and pyrophosphate [
5]. UGP2 plays a crucial role in carbohydrate metabolism and energy storage; its dysregulation has been linked to various metabolic disorders [
6]. Recent advances in cancer metabolism highlight the significant role of glucose metabolism in disease onset and progression [
7,
8]. UGP2 contributes to cancer biology, including cell proliferation, apoptosis, and metastasis [
9]. For instance, UGP2 expression is downregulated in hepatocellular carcinoma (HCC), where it is significantly associated with fatty acid metabolism, and its downregulation predicts poor prognosis in patients with HCC [
10]. Conversely, UGP2 is aberrantly overexpressed in glioblastoma multiforme (GBM) and positively correlates with pathological grading, making it a potential biomarker for predicting GBM prognosis [
9]. This dual role highlights the tissue-specific complexity of UGP2 in tumorigenesis and underscores the critical need to define its function within the specific metabolic landscape of CRC, where it remains incompletely characterized.
The aim of this study was to comprehensively investigate the expression, biological functions, and clinical significance of UGP2 in CRC, and to evaluate its potential as a diagnostic biomarker and therapeutic target. To achieve this, we employed an integrated strategy comprising three complementary phases: We first assessed UGP2 expression levels in CRC through proteomic screening, bioinformatics analysis of public datasets, and validation in our clinical cohort. Subsequently, bioinformatics analyses were conducted to examine its correlations with clinicopathological features, patient prognosis, DNA methylation status, and immune cell infiltration. Furthermore, functional validation assays were performed to determine its biological impact via in vitro knockdown and overexpression experiments on core cancer phenotypes. This multi-faceted approach was designed to systematically elucidate the role of UGP2 in CRC pathogenesis and clinical outcomes.
2. Materials and Methods
2.1. Clinical Samples
A total of 112 CRC tissue samples were collected from diagnosed patients, whereas 54 normal tissue samples were obtained from healthy individuals who underwent colonoscopy and were confirmed free of colorectal neoplasia or other significant pathologies. All samples, along with corresponding clinical data, were collected from the Department of Gastroenterology or surgical procedures at the Department of Gastrointestinal Surgery, Affiliated Hospital of Guangdong Medical University, between 2020 and 2024. All diagnoses were confirmed through clinical and pathological evaluations. A complete summary of patient demographics and clinicopathological characteristics, including age, gender, tumor location, size, differentiation grade, and TNM stage, is provided in
Supplementary File S1. Written informed consent was obtained from all participants before tissue collection and the study was conducted in strict compliance with the ethical standards established by the Ethics Committee of Affiliated Hospital of Guangdong Medical University (Ethics approval certificate No: PJKT2024-250).
2.2. Proteomic Analysis and Parallel Reaction Monitoring (PRM) Validation
Three CRC tissue samples and three normal colorectal mucosal tissue samples were selected for proteomic analysis (Shanghai Applied Protein Technology Co., Ltd., Shanghai, China). The analysis was performed using TMT (tandem mass tag) labeling coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). Proteins were extracted, digested, labeled with TMT reagents, fractionated, and analyzed by LC-MS/MS. Data were searched against a human protein database to identify and quantify proteins. From the proteomic data, we identified 10 differentially expressed proteins that have been minimally studied in CRC as target proteins. PRM was then conducted to validate and quantitatively analyze these selected proteins. Based on the PRM validation results, UGP2 was selected for further investigation due to its most significant and consistent dysregulation in CRC samples.
2.3. Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from CRC tissue samples or cellular samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), followed by reverse transcription into cDNA using the PrimeScript RT Master Mix Perfect Real Kit (Takara, Kusatsu, Japan). qRT-PCR was conducted using SYBR Premix Ex TaqTM II kits (Takara, Kusatsu, Japan) on a LightCycler 480 II (Roche, Basel, Switzerland). The relative expression level of UGP2 mRNA was calculated using the 2−ΔΔCt method, with GAPDH serving as the internal reference gene for normalization. Primers for amplification were synthesized by Tsingke Biotech (Guangzhou, China), with sequences as follows: UGP2 forward, 5′-CAGTAGGGGCTGCCATCAAA-3′; UGP2 reverse 5′-ACCAAGGGCACTGTAGGAAA-3′; GAPDH forward, 5′-GGGTGTGAACCATGAGAAGT-3′; GAPDH reverse, 5′-CAGTGATGGCATGGACTGTG-3′.
2.4. Western Blotting
Total protein from tissues or cells was lysed in RIPA buffer (Beyotime, Shanghai, China) supplemented with 1% phenylmethylsulfonyl fluoride. The protein concentrations were subsequently determined using the Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China). Protein samples (30 μg) were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transfer onto polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). Primary monoclonal antibodies against human UGP2, obtained from mice (Santa Cruz Biotechnology, Dallas, TX, USA), and GAPDH, obtained from rabbits (Affinity Biosciences, Longmont, CO, USA), were used. Horseradish peroxidase-labeled goat anti-mouse IgG (1:1000; Beyotime, Shanghai, China) was used as the secondary antibody. Optical images were captured using a 5200 imaging system (Tanon, Shanghai, China). The quantification of the integrated optical density of the protein bands was performed using ImageJ version 1.48v software (Bethesda, MD, USA). GAPDH was used as a loading control for normalization. GAPDH was used as a loading control for normalization.
2.5. Data Acquisition and Preprocessing
RNA sequencing data (in TPM format) and corresponding clinical information for CRC were obtained from The Cancer Genome Atlas (TCGA) for tumor samples, the Genotype-Tissue Expression (GTEx) database for normal tissue controls, and the Gene Expression Omnibus (GEO) for independent validation datasets (GSE39582, GSE17536, GSE44861). To establish a well-defined cohort, we included only samples with available RNA-seq data and complete clinical annotations pertinent to each specific analysis, while excluding duplicate samples based on unique IDs, samples lacking key clinical information required for the study, and, within the TCGA cohort, samples from non-CRC types. Subsequently, all expression values were converted to log2 (TPM + 1) for variance stabilization prior to downstream analysis.
2.6. Differential Expression Analysis of UGP2
We utilized R version 4.2.1 (R Foundation for Statistical Computing, Vienna, Austria) to conduct statistical analysis and data visualization of the datasets acquired from TCGA, GTEx, and GEO. Using the Wilcoxon rank-sum test, we assessed the expression differences of UGP2 in non-paired samples (comprising tumor and normal samples) for pan-cancer and CRC samples. Additionally, we evaluated the differences in UGP2 expression in paired samples (including tumor and adjacent non-tumor samples) for pan-cancer and CRC samples using the Wilcoxon signed-rank test. We set the significance threshold for the differential expression analysis as adjusted p-value < 0.05 and |Log2-fold change| > 1.
2.7. Functional Enrichment Analysis
The MSigDB website (
https://ngdc.cncb.ac.cn/databasecommons/, accessed on 30 October 2024) for the Gene Set Enrichment Analysis (GSEA) of UGP2-related genes was used. Gene Ontology (GO) terminology was used to explore the cellular components, molecular functions, and biological processes associated with UGP2. Additionally, pathway enrichment analysis was conducted using Kyoto Encyclopedia of Genes and Genomes (KEGG) to categorize various biological pathways involving UGP2-related genes. We set the significance threshold for enrichment at a false discovery rate (FDR) of <0.05,
q-value of <0.25, and nominal
p-value of <0.05.
2.8. Construction of the Interaction Network
We visualized the differentially expressed genes (DEGs) identified through screening and created a volcano plot using the threshold conditions |Log2 FC| > 1 and p-value of <0.05. The predicted protein–protein interaction (PPI) network for UGP2 was generated using the STRING database. The query was performed for Homo sapiens with a minimum interaction score of 0.400 and a maximum of 20 first-shell interactors to ensure a concise network. The resulting network was visualized in Cytoscape version 3.9.1 (Cytoscape Consortium, San Diego, CA, USA), where the MCODE plugin was applied solely for aesthetic layout. This network represents the top predicted interactors of UGP2 in a general biological context. To contextualize these findings in CRC, we subsequently conducted a co-expression analysis of the selected hub or top genes with UGP2 in the TCGA CRC dataset.
2.9. DNA Methylation Analysis
To systematically investigate the DNA methylation profile of
UGP2 in CRC, we employed a multi-step bioinformatic approach using established public platforms, each selected for its specific strengths. First, we used the UCSC Genome Browser (
http://genome.ucsc.edu, accessed on 18 October 2024) to visualize and obtain a preliminary assessment of CpG island distribution and methylation patterns within the
UGP2 promoter region, leveraging its comprehensive genomic annotation and user-friendly interface for initial exploration. Subsequently, we utilized the UALCAN database to quantitatively compare
UGP2 promoter methylation levels between CRC tumors and normal tissues, as this platform provides processed TCGA methylome data with integrated clinical information, enabling robust differential analysis. Finally, to evaluate the prognostic relevance of specific CpG sites, we employed the MethSurv tool, which is specifically designed for survival analysis based on CpG-site-level methylation data from TCGA. This complementary use of platforms allowed us to move from initial visualization to quantitative differential analysis and ultimately to clinical correlation.
2.10. Immune Cells Infiltration Analysis
We utilized the ssGSEA algorithm from the R-package GSVA [1.46.0] [
11] and referenced the markers of 24 immune cell types provided in the Immunity article [
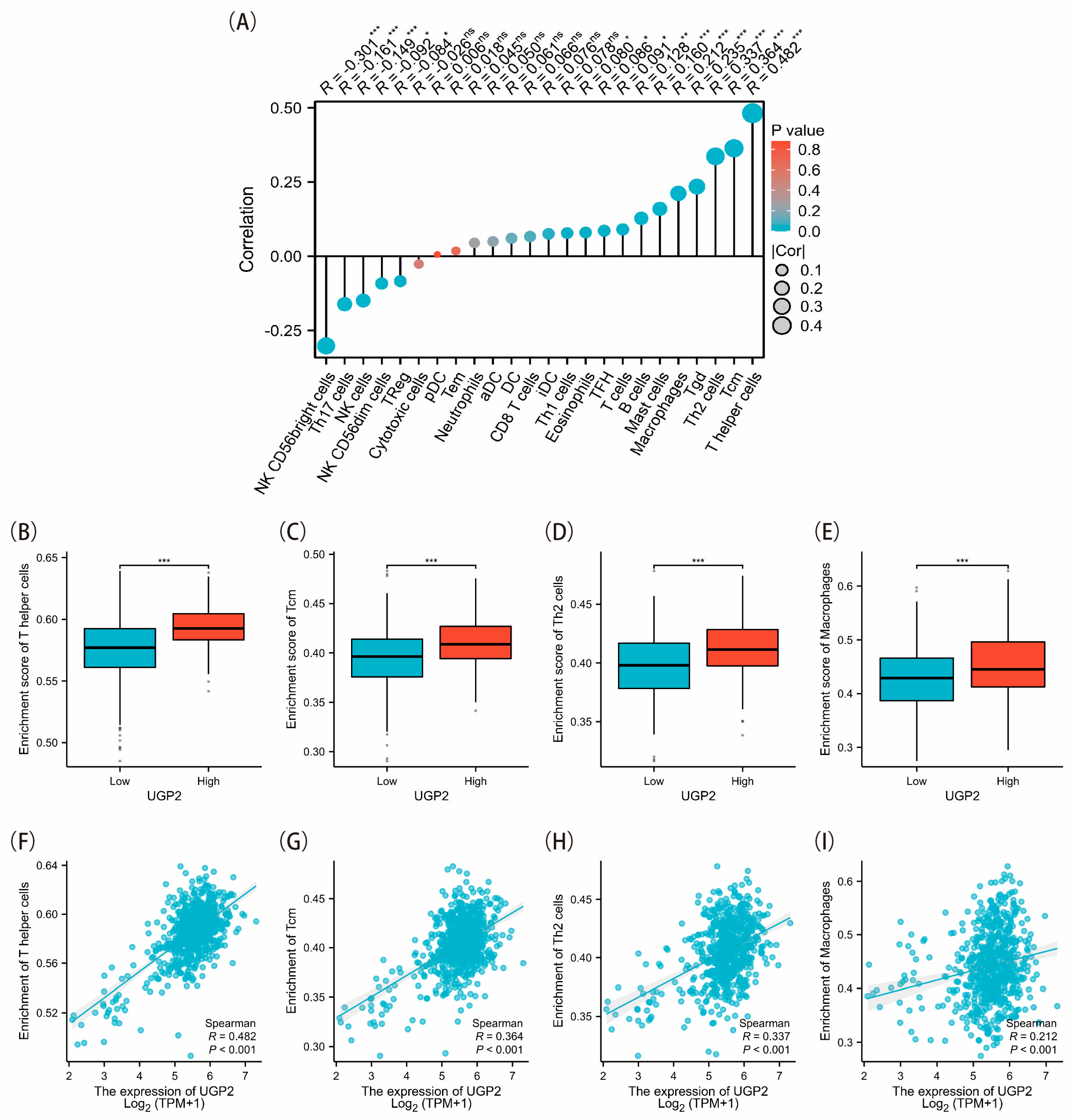
12]. Spearman’s rank correlation was applied to analyze RNA-seq data obtained and processed from TCGA database, along with the associated clinical data. The thresholds for high and low UGP2 expression were determined based on the median values obtained from our cohort analysis. Enrichment scores were then calculated to explore the differences in the infiltration levels of 24 immune cell types between the UGP2 high and low expression groups.
2.11. Diagnosis and Prognostic Evaluation
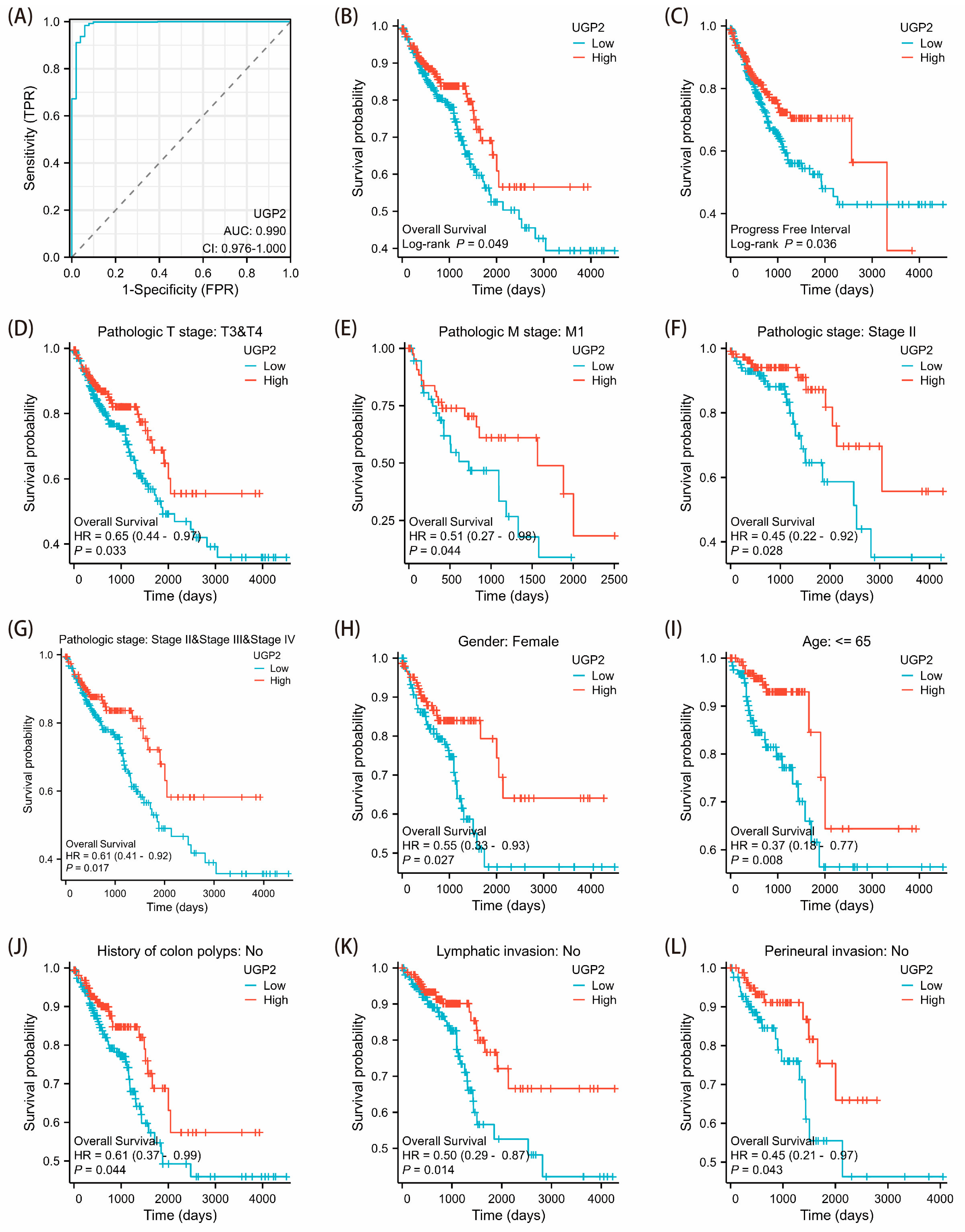
Based on the median expression value derived from the TCGA dataset, CRC patients were stratified into high and low UGP2 expression groups. The Wilcoxon rank-sum test was applied to assess the association between UGP2 expression levels and clinicopathological features. The Kaplan–Meier method, along with the log-rank test, was used to evaluate the relationship between UGP2 expression and clinical outcomes—including overall survival (OS) and progress free interval (PFI)—and to compute the corresponding p-values. Cox regression analysis was employed to examine the effect of UGP2 expression and other clinical variables on survival, with results expressed as hazard ratios (HR) and 95% confidence intervals (CI). Additionally, a receiver operating characteristic (ROC) analysis was conducted to assess the diagnostic value of UGP2 in predicting CRC outcomes. The performance was summarized using the area under the curve (AUC), where a value closer to 1 indicates stronger discriminatory power of the variable in predicting the outcome.
2.12. Cell Culture and Transfection
The human CRC cell lines HCT-116 and SW480 were obtained from the Cell Re-source Center of Chinese Academy of Sciences (Shanghai, China). Both cell lines were authenticated by short tandem repeat (STR) profiling and cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C with 5% CO2. UGP2 siRNA (target sequence: 5′-GAGCTAGAATTATCTGTGA-3′) and negative control siRNA (NC siRNA) (RiBoBio, Guangzhou, China) or UGP2 overexpression plasmid (UGP2 OE) and negative control overexpression plasmid (NC OE) (YuBo, Shanghai, China) were transfected into HCT-116 or SW480 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. Transfection efficiency was verified in each experiment by qRT-PCR analysis of UGP2 mRNA expression 24–48 h post-transfection.
2.13. Cell Proliferation, Migration Assay, and Apoptosis Assay
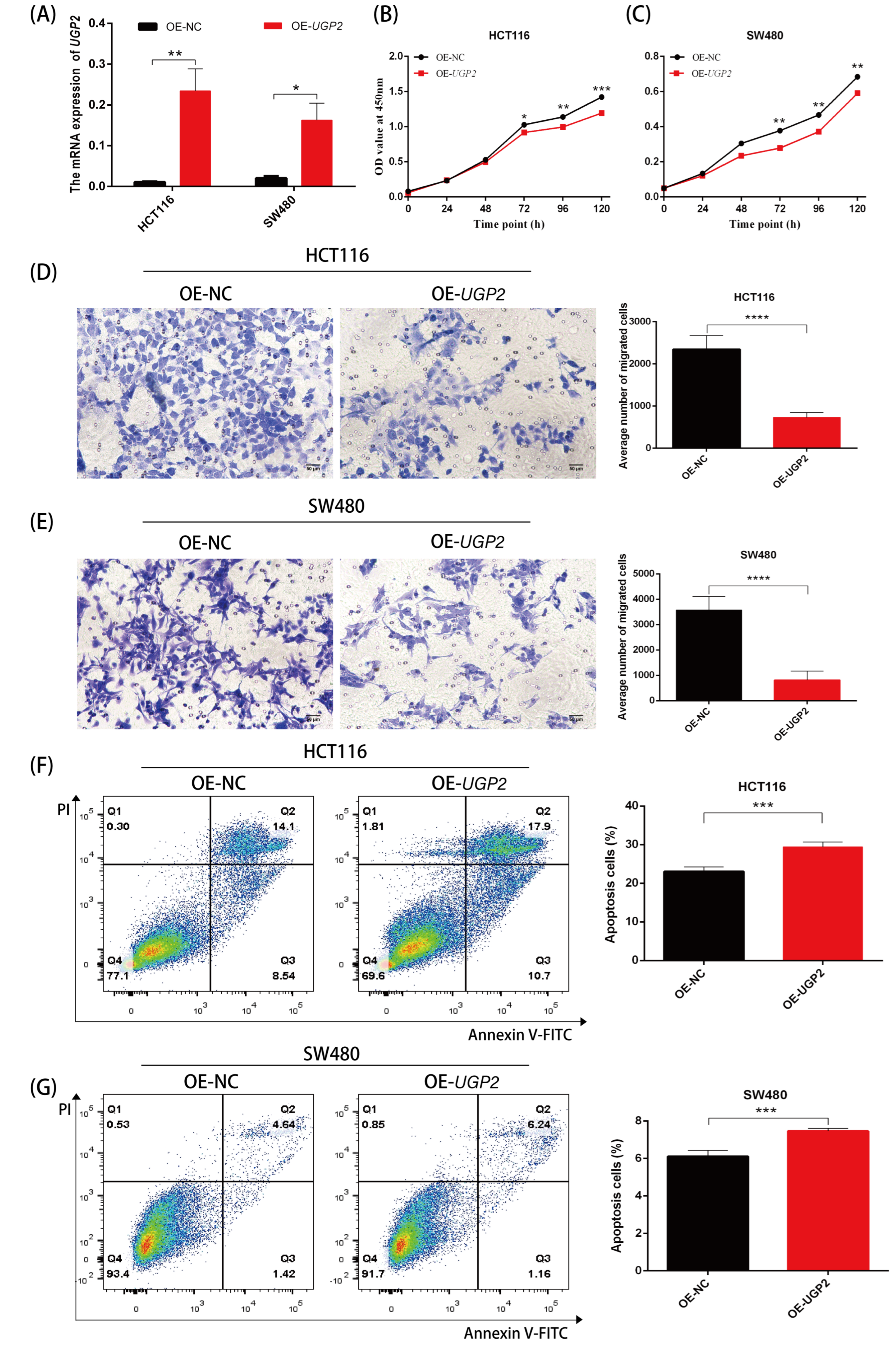
Suspensions of HCT-116 or SW480 cells were pre-cultured in 96-well plates for 24 h before being transfected with UGP2/NC siRNA or UGP2/NC OE. Cell proliferation was assessed 0, 24, 48, 72, 96, and 120 h after transfection using the Cell Counting Kit 8 (CCK-8; Dojindo, Kumamoto, Japan) per the manufacturer’s instructions. Absorbance at 450 nm was recorded as an indicator of cell viability. Cell migratory ability was assessed 24 h post-transfection using a migration chamber. Cells with weak migration ability on the upper surface of the chamber were carefully removed with a cotton swab, fixed with formalin, and stained with 0.1% crystal violet solution (Beyotime, Shanghai, China). Six random fields were examined for each insert under a microscope at ×100 magnification. Apoptosis was analyzed by flow cytometry using an Annexin V-FITC and propidium iodide (PI) detection kit (Beyotime, Shanghai, China). Cells were harvested 24 h or 48 h after transfection, stained according to the manufacturer’s protocol, and analyzed on a flow cytometer. The percentage of Annexin V-positive cells (early and late apoptotic populations) was quantified.
2.14. Statistical Analysis
All experimental data were analyzed using SPSS 19.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 6.01 (GraphPad Software, lnc., Boston, MA, USA), with results presented as means ± standard deviation (SD) based on at least three independent experiments. The expression levels of UGP2 in different tissue groups were assessed using a t-test. Two-way ANOVA was used for time-course proliferation assays. The significance threshold for all hypothesis testing was set at p < 0.05, unless otherwise specified for particular bioinformatics analyses (see details in the respective method sections). Specific statistical tests applied to different datasets are fully described in their corresponding methodological subsections.
4. Discussion
Our study revealed that UGP2 expression was significantly downregulated in CRC tissues compared with that in normal tissues. This was validated through PRM relative quantification analysis, data from the CPTAC database, qRT-PCR, and Western blotting. Additionally, analysis of the TCGA and GTEx databases demonstrated significant downregulation in 21 types of malignancies, including COAD and READ. UGP2 is aberrantly overexpressed and positively correlated with pathological grading in GBM in humans [
9]. This abnormal expression appears to be a rare occurrence and may be related to tissue specificity. The consistent downregulation of UGP2 across the majority of cancer types underscores its importance in maintaining normal cellular functions and suggests a broader involvement in tumorigenesis and progression.
Functional enrichment analysis of UGP2-associated DEGs revealed correlations with processes such as cellular detoxification, hydrogen peroxide metabolism, and, most notably, cholesterol and sterol homeostasis. Among these, cholesterol metabolism stands out. This bioinformatic finding prompts the hypothesis that the tumor-suppressive effect of UGP2 downregulation in CRC may be partially mediated through a metabolic rewiring that impacts lipid metabolism. This is highly relevant to CRC biology, as cholesterol metabolism is a well-established contributor to CRC pathogenesis, influencing tumor growth, invasion, and metastasis [
13,
14,
15]. For instance, the cholesterol transporter ABCA1, a key player in this pathway, has been identified as a marker of CRC invasion and survival [
16]. Furthermore, epidemiological studies have linked dyslipidemia to an increased risk of CRC [
17], and aberrant cholesterol metabolism has been implicated in promoting CRC liver metastasis [
18]. Therefore, our enrichment results do not establish causation but rather suggest a compelling link between the loss of UGP2 and perturbations in lipid metabolic pathways that are known to drive CRC progression. This provides a strong rationale for future mechanistic studies to investigate whether and how UGP2 directly regulates cholesterol metabolism in colorectal cells, which could uncover novel therapeutic avenues [
19,
20].
DNA methylation is a crucial epigenetic mechanism that regulates cell proliferation, apoptosis, differentiation, the cell cycle, and transformation while modulating gene expression and silencing [
21]. Aberrations in DNA methylation commonly occur in the promoter regions of transcription factors and play pivotal roles in tumorigenesis and progression [
22]. Tumor-specific methylation patterns are characterized by widespread hypomethylation and localized hypermethylation in specific gene promoter regions [
23]. Hypermethylation of tumor suppressor genes leads to their inactivation, which has emerged as a primary driver of tumor formation [
24]. Consequently, abnormal DNA methylation has gained attention as a potential biomarker for tumor development and prognostic assessment [
25]. For instance, diagnostic prediction models based on DNA methylation markers demonstrate high sensitivity for detecting early-stage lung cancer, supporting their use in noninvasive, blood-based diagnostics [
26]. Additionally, a study has identified six prognostic methylation genes by comparing the methylation states of head and neck squamous cell carcinoma (HNSCC) tissues with adjacent normal tissues. Validation confirmed that these six methylated genes could serve as independent prognostic markers for HNSCC [
27]. In the context of CRC, the methylation of the Septin9 gene has been extensively studied as a biomarker, with its methylation status showing significant potential for improving early diagnosis and prognosis of CRC [
28,
29,
30]. Moreover, the blood-based detection of Septin9 gene methylation and a detection reagent have received US FDA approval for risk assessment in individuals unwilling to undergo colonoscopy [
31]. In our study, data from the MethSurv database indicated that the
UGP2 promoter region was hypermethylated in COAD tissues compared with that in normal samples (
p < 0.01), likely resulting in the silencing of
UGP2 expression and contributing to tumorigenesis and progression. Patients with higher methylation levels exhibited significantly lower survival probabilities (
p < 0.05). Additionally, lower
UGP2 expression levels in patients with CRC were associated with more aggressive tumor behavior and poorer clinical outcomes, highlighting its potential as a prognostic marker. ROC curve analysis in our study demonstrated high accuracy in distinguishing patients with CRC, underscoring the value of UGP2 as a diagnostic marker.
The immune microenvironment is critical for the progression and prognosis of CRC [
32]. One of the two major subsets of peripheral T cells, Th cells, can recognize most tumor-specific antigens. Building on this, researchers have developed long-epitope vaccines for various cancers, utilizing Th cells as precise antitumor agonists [
33]. Tcm, which are crucial for long-term immune memory and rapid response upon antigen re-exposure, exhibit greater persistence and antitumor immunity than effector T cells (Teff). Researchers have identified the Tcm/Teff ratio as a predictive biomarker for the immune response to certain tumors [
34]. Th2 cells, which represent a unique subset of activated CD4 T cells, are capable of producing specific cytokines. These cytokines are crucial in promoting and coordinating immune defense responses against cancer cells and the tumor microenvironment (TME) [
35,
36]. Th2 cells produce interleukin-4 and exert strong anti-cancer effects, partly by promoting tumor stromal remodeling and tissue repair [
37]. Depending on their polarization state, macrophages can either promote or inhibit tumor progression, with M1 macrophages generally exerting antitumor effects and M2 macrophages supporting tumor growth and metastasis [
38]. Overall, immune cells can coordinate their actions within tumors through various immune responses, thereby regulating the TME and affecting tumor progression. Immune cell-mediated tissue-level immunity may represent a novel approach for TME-focused immunotherapy. Our study showed that UGP2 expression positively correlates with the infiltration levels of several immune cell types, such as Th cells, Tcm, Th2 cells, and macrophages. These immune cells are essential for orchestrating antitumor immune responses and their reduced infiltration in the UGP2 low expression group suggests a compromised immune surveillance mechanism. Th cells, particularly Th2 cells, are critical for modulating immune responses and activating other immune cells. The observed decrease in these immune cell populations in the low UGP2 expression group may contribute to a less effective antitumor immune response, facilitating tumor growth and progression. This underscores the potential of UGP2 as a biomarker of immune infiltration and its relevance to the immune landscape of CRC. Studying the interaction between UGP2 expression and immune cell infiltration could help us understand the mechanisms of immune evasion in CRC and highlight novel therapeutic targets for enhancing antitumor immunity.
The low expression of certain genes in tumors suggests their anti-cancer effects. Conversely, the reduced expression of tumor suppressor genes is commonly associated with poorer clinical outcomes in practice [
39]. Functionally, this may manifest as increased tumor cell proliferation and migration alongside reduced apoptosis. To confirm the clinical significance of UGP2 as a tumor suppressor gene in CRC, we first evaluated its diagnostic accuracy in identifying CRC. Subsequently, we analyzed the prognostic implications of UGP2 expression levels, including on OS, PFI, and survival rates, across various clinical subgroups. The ROC curve analysis demonstrated exceptionally high accuracy in distinguishing patients with CRC based on UGP2 expression, underscoring its value as a diagnostic marker. Furthermore, the KM curve analysis revealed that patients with low UGP2 expression exhibited poorer prognosis in terms of OS and PFI than those with high UGP2 expression. This was also evident across various clinical subgroups, including pathological T3 and T4 stages, pathological M1 stage, pathological stages II, III, and IV, female patients, those aged ≤ 65 years, and those with no history of colon polyps, no lymphatic invasion, and no perineural invasion. These results support the notion that UGP2 functions as a tumor suppressor in CRC and establish it as a potential prognostic marker. This could be used to stratify patients based on risk and guide the development of personalized treatment strategies. To further validate the function of UGP2 as a tumor suppressor gene on CRC at the cellular level, we conducted in vitro functional assays. Knockdown of UGP2 in HCT116 and SW480 cell lines enhanced cell proliferation and migration while reducing apoptosis; conversely, UGP2 overexpression reduced CRC cell proliferation and migration while enhancing apoptosis. This functional evidence reinforces the view that UGP2 is a tumor suppressor in CRC. Consequently, therapeutic approaches to restore UGP2 expression or function could inhibit CRC cell proliferation and migration, offering a promising strategy for combating CRC.
Our discovery that UGP2 acts as a tumor-suppressive factor in CRC indicates that its downregulation drives malignant progression and is associated with poor clinical outcomes, highlighting its potential as a dual-purpose diagnostic and prognostic candidate biomarker. A significant question that arises from this finding is how to reconcile this role with UGP2’s well-established function in glycogen synthesis. While UGP2 is widely recognized for its role in catalyzing the production of UDP-glucose for glycogen synthesis in normal physiology [
5,
6], our data strongly suggest a context-dependent, non-canonical tumor-suppressive role specifically within the tissue microenvironment of CRC. This observation is not merely an incidental finding but rather a key conclusion of our study. The fact that UGP2 is downregulated and functions as a tumor suppressor in CRC (and similarly in HCC [
10]) while being upregulated and acting as an oncogene in GBM [
9] indicates that its biological effects are not inherently pro- or anti-tumorigenic; instead, they are differentially regulated depending on the type of cancer. We propose that in CRC, the loss of UGP2 may provide a selective advantage to cancer cells not primarily by disrupting glycogen storage but possibly by altering the availability of UDP-glucose for other essential processes, such as the glycosylation of important membrane receptors or the hexosamine biosynthesis pathway. This change could interfere with normal cellular differentiation and promote a malignant phenotype. This tissue-specific functional divergence underscores the complexity of cancer metabolism and positions UGP2 not just as a metabolic housekeeping gene but as a context-dependent metabolic regulator with opposing roles in different cancers.
There are some limitations to this study. First, although this study provides a comprehensive analysis of UGP2’s differential expression and its functional impact in CRC, the exploration of underlying mechanisms remains primarily at the level of prediction using public databases, lacking direct experimental validation. For instance, the association between UGP2 promoter hypermethylation and favorable prognosis was identified through bioinformatic mining of public datasets; this correlation has not been confirmed by experimental techniques such as methylation-specific PCR (MSP) or pyrosequencing in our clinical cohort. Future investigations employing both in vitro and in vivo models are warranted to mechanistically dissect how UGP2 influences CRC pathogenesis. Second, the sample size, although substantial, might still be considered limited, potentially affecting the generalizability of the findings. The study included 112 CRC tissues and 54 normal tissues, which, although informative, may not capture the full heterogeneity of CRC. Finally, the lack of extensive clinical validation analyses means that the clinical applicability of UGP2 as a promising candidate biomarker or therapeutic target remains to be established. Its clinical applicability requires future confirmation in larger, prospective studies and in vivo experiments.