Multiplex Microarrays in 96-Well Plates Photoactivated with 4-Azidotetrafluorobenzaldehyde for the Identification and Quantification of β-Lactamase Genes and Their RNA Transcripts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Bacterial Strains and DNA and RNA Samples
2.3. Photochemical Activation of Polystyrene Surface
2.4. Oligonucleotide Microarray Fabrication
2.5. Microarray Hybridization and Detection
3. Results
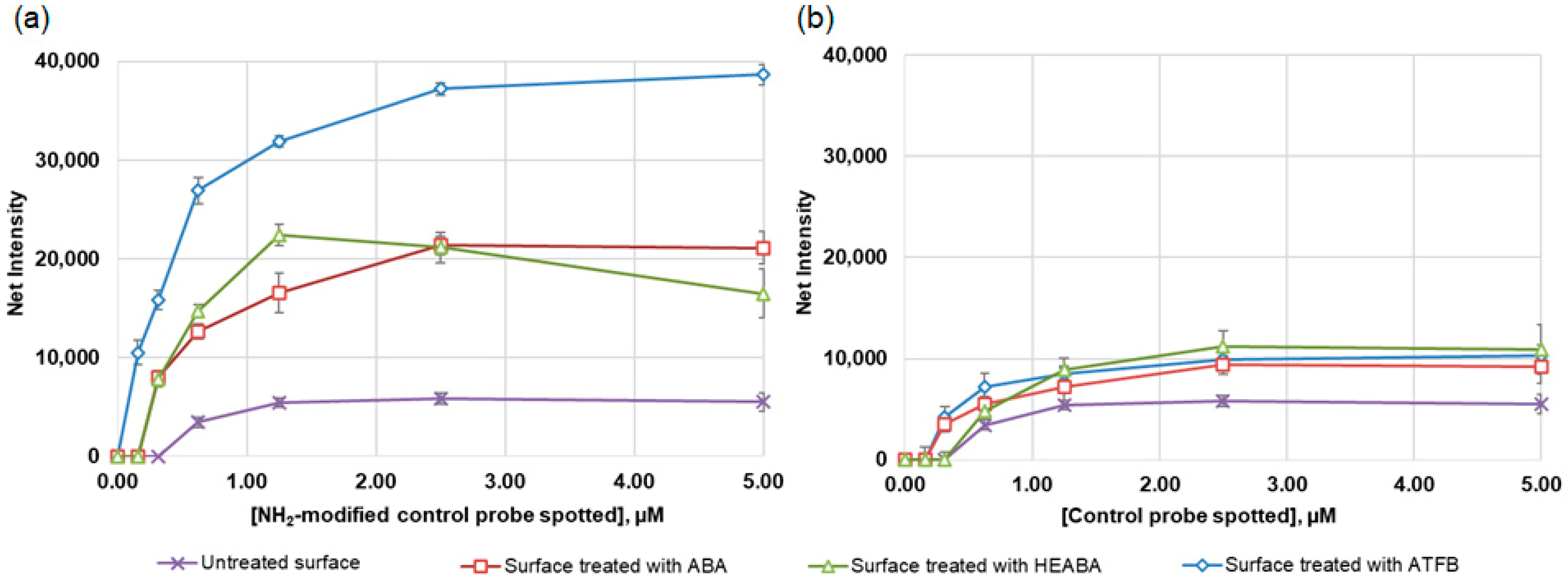
3.1. Photoactivation of 96-Well Polystyrene Plates with Different Bifunctional Linkers
3.2. Sensitivity of Hybridization Analysis on Photoactivated 96-Well Polystyrene Plates
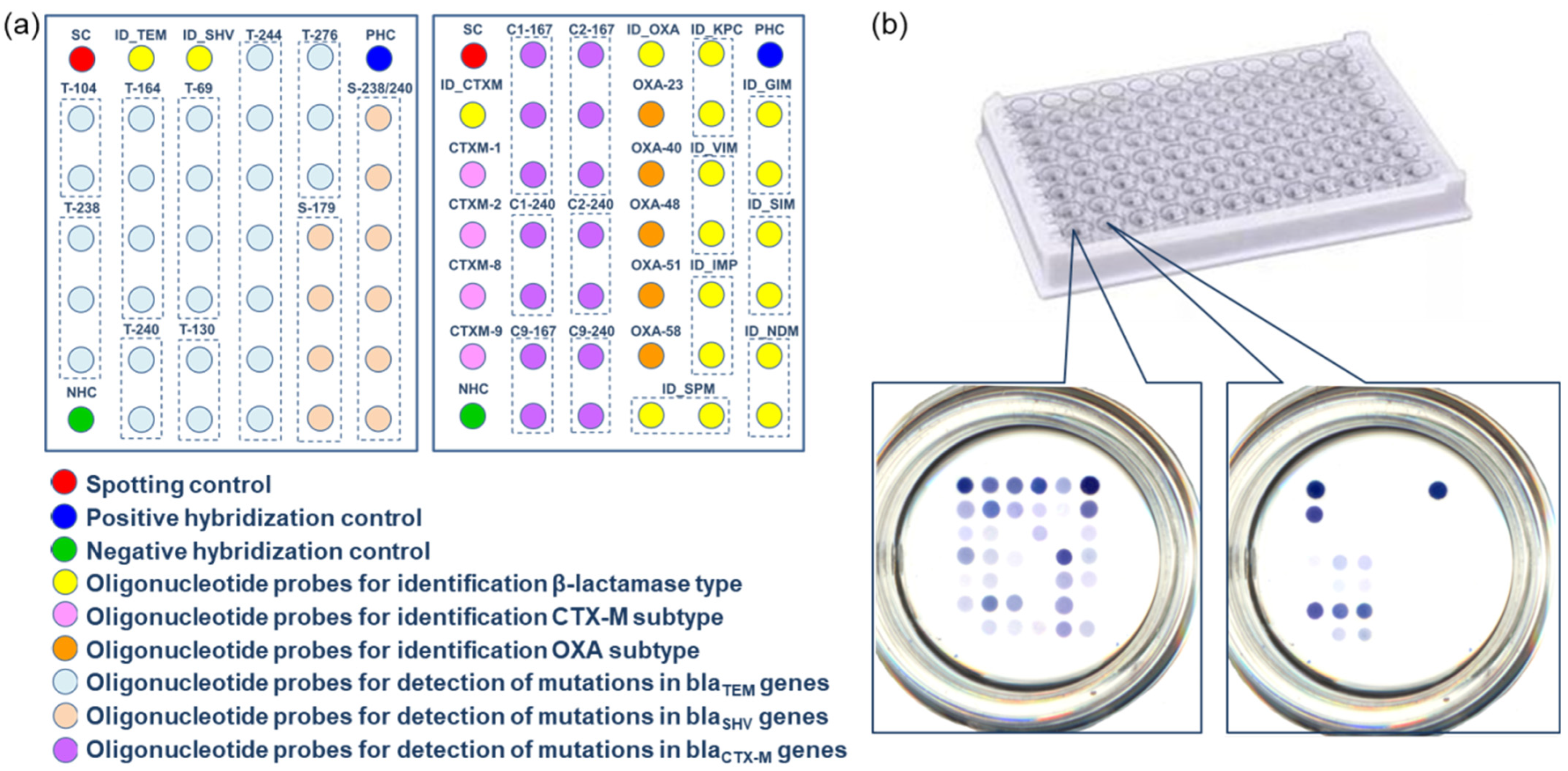
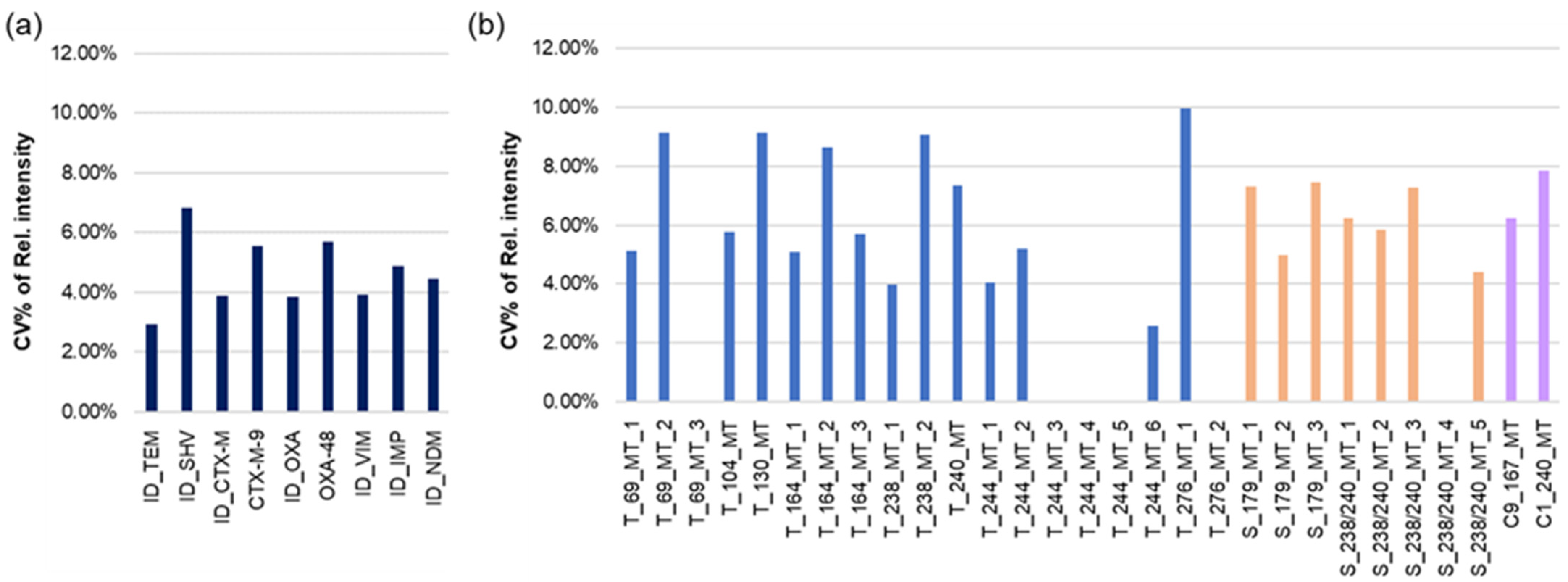
3.3. Microarrays in the Wells of the Microtiter Plate for the Identification of ESBLs and Carbapenemases
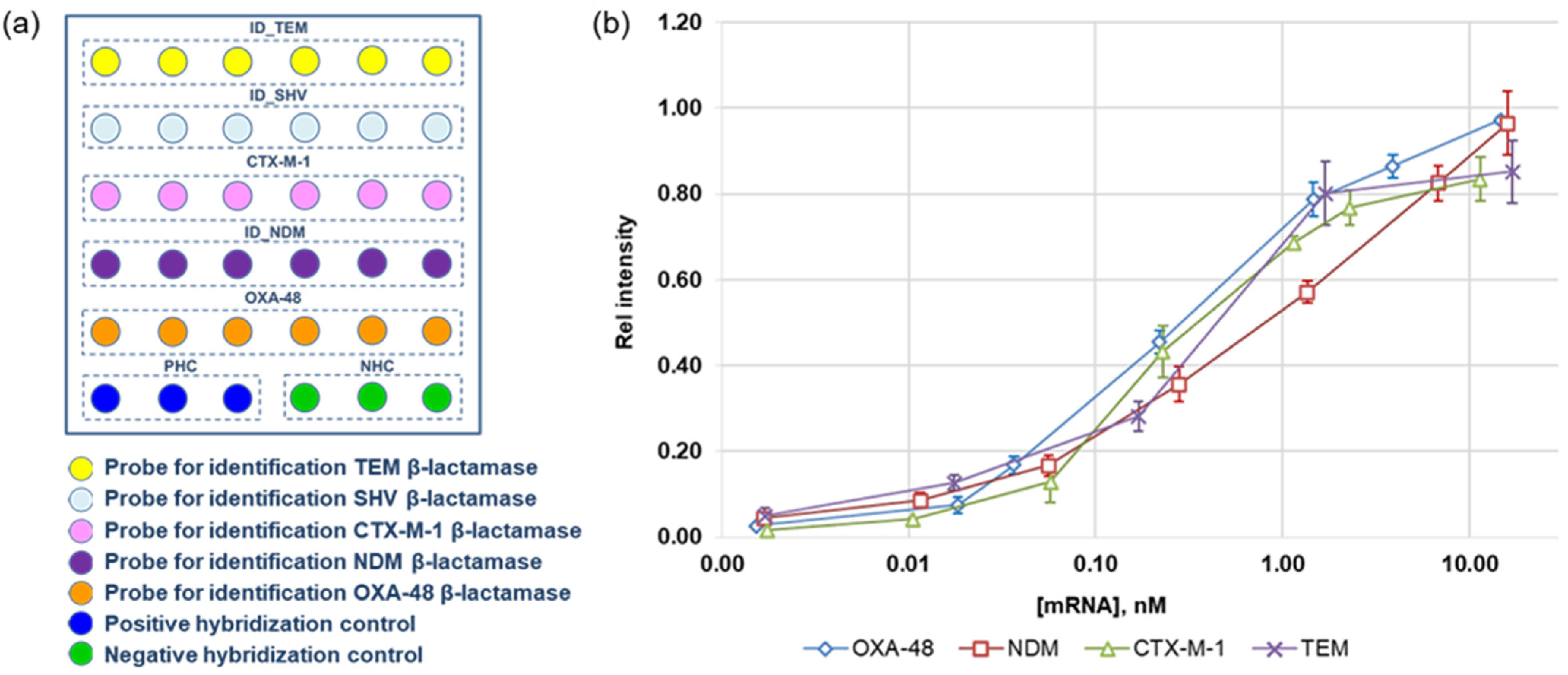
3.4. Microarrays in the Wells of the Microtiter Plate for the Quantification of mRNAs of β-Lactamases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersson, D.I.; Balaban, N.Q.; Baquero, F.; Courvalin, P.; Glaser, P.; Gophna, U.; Kishony, R.; Molin, S.; Tonjum, T. Antibiotic resistance: Turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 2020, 44, 171–188. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 21 November 2023).
- Van Belkum, A.; Burnham, C.A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.; Gonzalez-Zorn, A.B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Rood, I.G.H.; Li, Q.G. Molecular Detection of Extended-Spectrum-ß-Lactamase- and Carbapenemase-Producing Enterobacteriaceae in a Clinical Setting. Diagn. Microbiol. Infect.Dis. 2017, 89, 245–250. [Google Scholar] [CrossRef]
- Comini, S.; Bianco, G.; Boattini, M.; Banche, G.; Ricciardelli, G.; Allizond, V.; Cavallo, R.; Costa, C. Evaluation of a diagnostic algorithm for rapid identification of Gram-negative species and detection of extended-spectrum β-lactamase and carbapenemase directly from blood cultures. J. Antimicrob. Chemother. 2022, 77, 2632–2641. [Google Scholar] [CrossRef]
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar] [CrossRef]
- Wallace, R.B.; Shaffer, J.; Murphy, R.F.; Bonner, J.; Hirose, T.; Itakura, K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: The effect of single base pair mismatch. Nucleic Acids Res. 1979, 6, 3543–3557. [Google Scholar] [CrossRef]
- Bañuls, M.J.; Jiménez-Meneses, P.; Meyer, A.; Vasseur, J.J.; Morvan, F.; Escorihuela, J.; Puchades, R.; Maquieira, Á. Improved Performance of DNA Microarray Multiplex Hybridization Using Probes Anchored at Several Points by Thiol-Ene or Thiol-Yne Coupling Chemistry. Bioconjug. Chem. 2017, 28, 496–506. [Google Scholar] [CrossRef]
- Sabourin, D.; Petersen, J.; Snakenborg, D.; Brivio, M.; Gudnadson, H.; Wolff, A.; Dufva, M. Microfluidic DNA microarrays in PMMA chips: Streamlined fabrication via simultaneous DNA immobilization and bonding activation by brief UV exposure. Biomed. Microdevices 2010, 12, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Tsougeni, K.; Koukouvinos, G.; Petrou, P.S.; Tserepi, A.; Kakabakos, S.E.; Gogolides, E. High-capacity and high-intensity DNA microarray spots using oxygen-plasma nanotextured polystyrene slides. Anal. Bioanal. Chem. 2012, 403, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Bilitewski, U. DNA microarrays: An introduction to the technology. Methods Mol Biol. 2009, 509, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, S.A.; Gardner, C.; Das, S. Diagnosis and Management of Bloodstream Infections with Rapid, Multiplexed Molecular Assays. Front. Cell Infect. Microbiol. 2022, 12, 859935. [Google Scholar] [CrossRef] [PubMed]
- Shaskolskiy, B.; Kandinov, I.; Kravtsov, D.; Vinokurova, A.; Gorshkova, S.; Filippova, M.; Kubanov, A.; Solomka, V.; Deryabin, D.; Dementieva, E.; et al. Hydrogel Droplet Microarray for Genotyping Antimicrobial Resistance Determinants in Neisseria gonorrhoeae Isolates. Polymers 2021, 13, 3889. [Google Scholar] [CrossRef] [PubMed]
- Charnock, C.; Samuelsen, Ø.; Nordlie, A.-L.; Hjeltnes, B. Use of a Commercially Available Microarray to Characterize Antibiotic-Resistant Clinical Isolates of Klebsiella pneumoniae. Curr. Microbiol. 2017, 75, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Leinberger, D.M.; Grimm, V.; Rubtsova, M.; Weile, Y.; Schröppe, K.; Wichelhaus, T.A.; Knabbe, C.; Schmid, R.D.; Bachmann, T.T. Integrated detection of extended-spectrum-beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J. Clin. MIcrobiol. 2010, 48, 460–471. [Google Scholar] [CrossRef]
- Uddin, F.; McHugh, T.D.; Roulston, K.; Platt, G.; Khan, T.A.; Sohail, M. Detection of carbapenemases, AmpC and ESBL genes in Acinetobacter isolates from ICUs by DNA microarray. J. Microbiol. Meth. 2018, 155, 19–23. [Google Scholar] [CrossRef]
- Chiodi, E.; Marn, A.M.; Geib, M.T.; Unlu, M.S. The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview. Polymers 2021, 13, 1026. [Google Scholar] [CrossRef]
- Yan, M.; Cai, S.X.; Wybourne, M.N.; Keana, F.W. Photochemical functionalization polymer surfaces and the production of biomolecule-carrying micrometer-scale structures by UV lithograph using 4-substituted perfluoropherazides. J. Am. Chem. Soc. 1993, 115, 814–816. [Google Scholar] [CrossRef]
- Rubtsova, M.Y.; Samsonova, J.V.; Egorov, A.M.; Schmid, R.D. Simultaneous determination of several pesticides with chemiluminescent immunoassay on a multi-spot membrane strip. Food Agric. Immunol. 1998, 10, 223–235. [Google Scholar] [CrossRef]
- Ulyashova, M.M.; Khalilova, Y.I.; Rubtsova, M.Y.; Edelstein, M.V.; Alexandrova, I.M.; Egorov, A.M. Oligonucleotide microarray for the identification of carbapenemase genes of molecular classes A, B, and D. Acta Naturae 2010, 2, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Rubtsova, M.Y.; Ulyashova, M.M.; Pobolelova, Y.I.; Presnova, G.V.; Egorov, A.M. Biochip for simultaneous identification of beta-lactamase and carbapenemase genes conferring the bacterial resistance to beta-lactam antibiotics. Appl. Biochem. Microbiol. 2020, 56, 130–140. [Google Scholar] [CrossRef]
- Dankbar, D.M.; Gauglitz, G. A study on photolinkers used for biomolecule attachment to polymer surfaces. Anal. Bioanal. Chem. 2006, 386, 1967–1974. [Google Scholar] [CrossRef]
- Kannoujia, D.K.; Ali, S.; Nahar, P. Single-step covalent immobilization of oligonucleotides onto solid surface. Anal. Methods 2010, 2, 212. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, D.; Ahirwar, R.; Nahar, P. Exploring the flexible chemistry of 4-fluoro-3-nitrophenyl azide for biomolecule immobilization and bioconjugation. Anal. Bioanal. Chem. 2016, 408, 6945–6956. [Google Scholar] [CrossRef]
- Hermanson, G.T. Introduction to bioconjugation. In Bioconjugate Techniques, 3rd ed.; Academic Press: Boston, MA, USA, 2013; pp. 1–125. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulyashova, M.M.; Presnova, G.V.; Filippova, A.A.; Grigorenko, V.G.; Egorov, A.M.; Rubtsova, M.Y. Multiplex Microarrays in 96-Well Plates Photoactivated with 4-Azidotetrafluorobenzaldehyde for the Identification and Quantification of β-Lactamase Genes and Their RNA Transcripts. Curr. Issues Mol. Biol. 2024, 46, 53-66. https://doi.org/10.3390/cimb46010005
Ulyashova MM, Presnova GV, Filippova AA, Grigorenko VG, Egorov AM, Rubtsova MY. Multiplex Microarrays in 96-Well Plates Photoactivated with 4-Azidotetrafluorobenzaldehyde for the Identification and Quantification of β-Lactamase Genes and Their RNA Transcripts. Current Issues in Molecular Biology. 2024; 46(1):53-66. https://doi.org/10.3390/cimb46010005
Chicago/Turabian StyleUlyashova, Mariya M., Galina V. Presnova, Anna A. Filippova, Vitaly G. Grigorenko, Alexey M. Egorov, and Maya Yu. Rubtsova. 2024. "Multiplex Microarrays in 96-Well Plates Photoactivated with 4-Azidotetrafluorobenzaldehyde for the Identification and Quantification of β-Lactamase Genes and Their RNA Transcripts" Current Issues in Molecular Biology 46, no. 1: 53-66. https://doi.org/10.3390/cimb46010005
APA StyleUlyashova, M. M., Presnova, G. V., Filippova, A. A., Grigorenko, V. G., Egorov, A. M., & Rubtsova, M. Y. (2024). Multiplex Microarrays in 96-Well Plates Photoactivated with 4-Azidotetrafluorobenzaldehyde for the Identification and Quantification of β-Lactamase Genes and Their RNA Transcripts. Current Issues in Molecular Biology, 46(1), 53-66. https://doi.org/10.3390/cimb46010005





