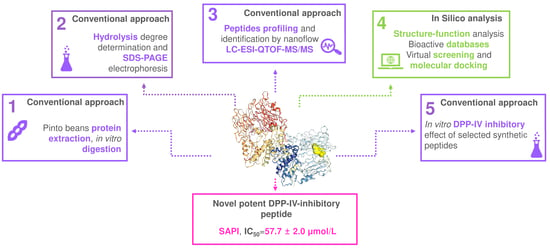
Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of the Pinto Bean Protein Extract
2.3. In Vitro Gastrointestinal Digestion
2.4. Assessment of Protein Hydrolysis during In Vitro Gastrointestinal Digestion
2.5. Sodium Dodecyl Sulphate Poly-Acrylamide Gel Electrophoresis (SDS-PAGE)
2.6. Peptidomics Analysis of Low Molecular Weight Peptides by Nanoflow LC-ESI-QTOF-MS/MS
2.7. In Silico Analysis
2.7.1. Identification of Previously Reported Bioactive Peptides
2.7.2. Identification of Novel DPP-IV-Inhibitory Peptides
2.8. Statistical Analysis
3. Results and Discussion
3.1. Assessment of Protein Hydrolysis during In Vitro Gastrointestinal Digestion
3.2. Peptidomics Profiles of Low-Molecular Weight Peptides after In Vitro Gastrointestinal Digestion and Identification of Previously Known Bioactive Peptides
3.3. Identification of Novel Potential DPP-IV-Inhibitory Peptides by In Silico Approach and Structure–Activity Relationship Modelling
3.4. In Vitro DPP-IV Inhibitory Activity of Synthetic Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Montone, C.M.; Zenezini Chiozzi, R.; Laganà, A. Recent trends and analytical challenges in plant bioactive peptide separation, identification and validation. Anal. Bioanal. Chem. 2018, 410, 3425–3444. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rivera, L.; Martínez-Maqueda, D.; Cruz-Huerta, E.; Miralles, B.; Recio, I. Peptidomics for discovery, bioavailability and monitoring of dairy bioactive peptides. Food Res. Int. 2014, 63, 170–181. [Google Scholar] [CrossRef]
- Arroume, N.; Froidevaux, R.; Kapel, R.; Cudennec, B.; Ravallec, R.; Flahaut, C.; Bazinet, L.; Jacques, P.; Dhulster, P. Food peptides: Purification, identification and role in the metabolism. Curr. Opin. Food Sci. 2016, 7, 101–107. [Google Scholar] [CrossRef]
- Cicero, A.F.G.; Fogacci, F.; Colletti, A. Potential role of bioactive peptides in prevention and treatment of chronic diseases: A narrative review. Br. J. Pharm. 2017, 174, 1378–1394. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Tagliazucchi, D.; Babini, E.; Rutella, G.S.; Taneyo Saa, D.L.; Gianotti, A. Bioactive peptides from vegetable food matrices: Research trends and novel biotechnologies for synthesis and recovery. J. Funct. Foods 2016, 27, 549–569. [Google Scholar] [CrossRef]
- Tagliazucchi, D.; Martini, S.; Solieri, L. Bioprospecting for bioactive peptide production by lactic acid bacteria isolated from fermented dairy food. Fermentation 2019, 5, 96. [Google Scholar] [CrossRef] [Green Version]
- Xing, L.; Liu, R.; Cao, S.; Zhang, W.; Guanghong, Z. Meat protein based bioactive peptides and their potential functional activity: A review. Int. J. Food Sci. Technol. 2019, 54, 1956–1966. [Google Scholar] [CrossRef] [Green Version]
- Tulipano, G. Role of bioactive peptide sequences in the potential impact of dairy protein intake on metabolic health. Int. J. Mol. Sci 2020, 21, 8881. [Google Scholar] [CrossRef]
- Fernández-Tomé, S.; Hernández-Ledesma, B. Gastrointestinal digestion of food proteins under the effects of released bioactive peptides on digestive health. Mol. Nutr. Food Res. 2020, 64, 2000401. [Google Scholar] [CrossRef]
- Guha, S.; Sharma, H.; Deshwal, G.K.; Rao, P.S. A comprehensive review on bioactive peptides derived from milk and milk products of minor dairy species. Food Prod. Process. Nutr. 2021, 3, 2. [Google Scholar] [CrossRef]
- Martini, S.; Conte, A.; Tagliazucchi, D. Comparative peptidomic profile and bioactivities of cooked beef, pork, chicken and turkey meat after in vitro gastro-intestinal digestion. J. Proteom. 2019, 208, 103500. [Google Scholar] [CrossRef] [PubMed]
- Malaguti, M.; Dinelli, G.; Leoncini, E.; Bregola, V.; Bosi, S.; Cicero, A.F.G.; Hrelia, S. Bioactive peptides in cereals and legumes: Agronomical, biochemical and clinical aspects. Int. J. Mol. Sci. 2014, 15, 21120–21135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Valdespino, C.A.; Luna-Vital, D.; Camacho-Ruiz, R.M.; Mojica, L. Bioactive proteins and phytochemicals from legumes: Mechanisms of action preventing obesity and type-2 diabetes. Food Res. Int. 2020, 130, 108905. [Google Scholar] [CrossRef]
- Ngoh, Y.Y.; Gan, C.Y. Identification of Pinto bean peptides with inhibitory effects on α-amylase and angiotensin converting enzyme (ACE) activities using an integrated bioinformatics-assisted approach. Food Chem. 2018, 267, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Tagliazucchi, D.; Martini, S.; Bellesia, A.; Conte, A. Identification of ACE-inhibitory peptides from Phaseolus vulgaris after in vitro gastrointestinal digestion. Int. J. Food Sci. Nutr. 2015, 66, 774–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojica, L.; Luna-Vital, D.A.; González de Mejía, E. Characterization of peptides from common bean protein isolates and their potential to inhibit markers of type-2 diabetes, hypertension and oxidative stress. J. Sci. Food Agric. 2017, 97, 2401–2410. [Google Scholar] [CrossRef]
- Mojica, L.; González de Mejía, E. Optimization of enzymatic production of anti-diabetic peptides from black bean (Phaseolus vulgaris L.) proteins, their characterization and biological potential. Food Funct. 2016, 7, 713–727. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.J.; Cermeño, M.; Khalesi, M.; Kleekayai, T.; Amigo-Benavent, M. Application of in silico approaches for the generation of milk protein-derived bioactive peptides. J. Funct. Foods 2020, 64, 103636. [Google Scholar] [CrossRef]
- Martini, S.; Solieri, L.; Cattivelli, A.; Pizzamiglio, V.; Tagliazucchi, D. An integrated peptidomics and in silico approach to identify novel anti-diabetic peptides in Parmigiano-Reggiano cheese. Biology 2021, 10, 563. [Google Scholar] [CrossRef]
- Martini, S.; Solieri, L.; Tagliazucchi, D. Peptidomics: New trends in food science. Curr. Opin. Food Sci. 2021, 39, 51–59. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, 45012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabuco, L.G.; Lise, S.; Petsalaki, E.; Russell, R.B. PepSite: Prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012, 40, W423–W427. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Jacinto-Hernández, C.; Alaiz, M.; Girón-Calle, J.; Vioque, J.; Dávila-Ortiz, G. Antioxidant and metal chelating activities of Phaseolus vulgaris L. var. Jamapa protein isolates, phaseolin and lectin hydrolysates. Food Chem. 2012, 131, 1157–1164. [Google Scholar]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzensulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Tagliazucchi, D.; Shamsia, S.; Helal, A.; Conte, A. Angiotensin-converting enzyme inhibitory peptides from goats’ milk released by in vitro gastrointestinal digestion. Int. Dairy J. 2017, 71, 6–16. [Google Scholar] [CrossRef]
- Tagliazucchi, D.; Martini, S.; Shamsia, S.; Helal, A.; Conte, A. Biological activity and peptidomic profile of in vitro digested cow, camel, goat and sheep milk. Int. Dairy J. 2018, 81, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Montoya, C.A.; Leterme, P.; Victoria, N.F.; Toro, O.; Souffrant, W.B.; Beebe, S.; Lallès, J.P. Susceptibility of phaseolin to in vitro proteolysis is highly variable across common bean varieties (Phaseolus vulgaris). J. Agric. Food Chem. 2008, 56, 2183–2191. [Google Scholar] [CrossRef]
- Luna-Vital, D.A.; Mojica, L.; González de Mejía, E.; Mendoza, S.; Loarca-Piña, G. Biological potential of protein hydrolysates and peptides from common bean (Phaseolus vulgaris L.): A review. Food Res. Int. 2015, 76, 39–50. [Google Scholar] [CrossRef]
- Iroyukifujita, H.; Eiichiyokoyama, K.; Yoshikawa, M. Classification and antihypertensive activity of angiotensin I-converting enzyme inhibitory peptides derived from food proteins. J. Food Sci. 2000, 65, 564–569. [Google Scholar] [CrossRef]
- He, R.; Malomo, S.A.; Alashi, A.; Girgih, A.T.; Ju, X.; Aluko, R.E. Purification and hypotensive activity of rapeseed protein-derived renin and angiotensin converting enzyme inhibitory peptides. J. Funct. Foods 2013, 5, 781–789. [Google Scholar] [CrossRef]
- Vercruysse, L.; Van Camp, J.; Morel, N.; Rougè, P.; Herregods, G.; Smagghe, G. Ala-Val-Phe and Val-Phe: ACE inhibitory peptides derived from insect protein with antihypertensive activity in spontaneously hypertensive rats. Peptides 2010, 31, 482–488. [Google Scholar] [CrossRef]
- Suetsuna, K.; Maekawa, K.; Chen, J.R. Antihypertensive effects of Undaria pinnatifida (wakame) peptide on blood pressure in spontaneously hypertensive rats. J. Nutr. Biochem. 2004, 15, 267–272. [Google Scholar] [CrossRef]
- Van Platerink, C.J.; Janssen, H.G.M.; Horsten, R.; Haverkamp, J. Quantification of ACE inhibiting peptides in human plasma using high performance liquid chromatography–mass spectrometry. J. Chromatogr. B 2006, 830, 151–157. [Google Scholar] [CrossRef] [Green Version]
- Foltz, M.; Meynen, E.E.; Bianco, V.; Van Platerink, C.J.; Koning, T.M.M.G.; Kloek, J. Angiotensin converting enzyme inhibitory peptides from a lactotripeptide-enriched milk beverage are absorbed intact into the circulation. J. Nutr. 2007, 137, 953–958. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.; FitzGerald, R.J. Structure activity relationship modelling of milk protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Peptides 2016, 79, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.; FitzGerald, R.J. Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [Green Version]
- Iwaniak, A.; Mogut, D. Metabolic syndrome-preventive peptides derived from milk proteins and their presence in cheeses: A review. Appl. Sci. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Deacon, C.F. Dipeptidyl peptidase 4 inhibitors in the treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2020, 16, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Tulipano, G.; Faggi, L.; Nardone, A.; Cocchi, D.; Caroli, A.M. Characterisation of the potential of β-lactoglobulin and α-lactalbumin as sources of bioactive peptides affecting incretin function: In silico and in vitro comparative studies. Int. Dairy J. 2015, 48, 66–72. [Google Scholar] [CrossRef]
- Power, O.; Nongonierma, A.B.; Jakeman, P.; FitzGerald, R.J. Food protein hydrolysates as a source of dipeptidyl peptidase IV inhibitory peptides for the management of type 2 diabetes. Proc. Nutr. Soc. 2014, 73, 34–46. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabeno, M.; Akahoshi, F.; Kishida, H.; Miyaguchi, I.; Tanaka, Y.; Ishii, S.; Kadowaki, T. A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site. Biochem. Biophys. Res. Commun. 2013, 434, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Torres-Fuentes, C.; Contreras, M.M.; Recio, I.; Alaiz, M.; Vioque, J. Identification and characterization of antioxidant peptides from chickpea protein hydrolysates. Food Chem. 2015, 180, 194–202. [Google Scholar] [CrossRef] [Green Version]
- Samaei, S.P.; Ghorbani, M.; Tagliazucchi, D.; Martini, S.; Gotti, R.; Themelis, T.; Tesini, F.; Gianotti, A.; Gallina Toschi, T.; Babini, E. Functional, nutritional, antioxidant, sensory properties and comparative peptidomic profile of faba bean (Vicia faba, L.) seed protein hydrolysates and fortified apple juice. Food Chem. 2020, 330, 127120. [Google Scholar] [CrossRef] [PubMed]


| Peptide Sequence | Protein Precursor | Bioactivity 1 |
|---|---|---|
| AI | Various proteins | ACE-inhibition |
| AL | Various proteins | DPP-IV-inhibition |
| EI | Various proteins | ACE-inhibition DPP-IV-inhibition |
| EL | Various proteins | Antioxidant |
| EY | Various proteins | ACE-inhibition DPP-IV-inhibition |
| FVPH | α and β subunits of phaseolins | Antioxidant |
| GI | Various proteins | ACE-inhibition DPP-IV-inhibition |
| GL | Various proteins | ACE-inhibition DPP-IV-inhibition |
| IA | Various proteins | ACE-inhibition DPP-IV-inhibition |
| IE | Various proteins | ACE-inhibition |
| IH | Various proteins | DPP-IV-inhibition |
| II | Various proteins | DPP-IV-inhibition |
| IL | Various proteins | ACE-inhibition DPP-IV-inhibition |
| IP | Various proteins | ACE-inhibition DPP-IV-inhibition |
| IY | Various proteins | ACE-inhibition |
| LA | Various proteins | ACE-inhibition DPP-IV-inhibition |
| LH | Various proteins | DPP-IV-inhibition |
| LI | Various proteins | DPP-IV-inhibition |
| LKA | Various proteins | ACE-inhibition |
| LL | Various proteins | ACE-inhibition DPP-IV-inhibition |
| LP | Various proteins | DPP-IV-inhibition |
| LPQ | α and β subunits of phaseolins | DPP-IV-inhibition |
| LT | Various proteins | DPP-IV-inhibition |
| LY | Various proteins | ACE-inhibition |
| MI | Various proteins | DPP-IV-inhibition |
| ML | Various proteins | DPP-IV-inhibition |
| PR | Various proteins | ACE-inhibition |
| TI | Various proteins | DPP-IV-inhibition |
| TL | Various proteins | DPP-IV-inhibition |
| VAV | Various proteins | ACE-inhibition |
| VF | Various proteins | ACE-inhibition DPP-IV-inhibition |
| VI | Various proteins | DPP-IV-inhibition |
| VL | Various proteins | DPP-IV-inhibition |
| VM | Various proteins | DPP-IV-inhibition |
| VR | Various proteins | ACE-inhibition DPP-IV-inhibition |
| VV | Various proteins | DPP-IV-inhibition |
| YR | Various proteins | DPP-IV-inhibition |
| Sequence | PepSite2 p-Value for DPPIV (pdb: 1NU6) | SAR |
|---|---|---|
| PR | 1.45 × 10−4 | |
| IP * | 1.98 × 10−4 | |
| LP * | 2.07 × 10−4 | |
| LPQ * | 4.91 × 10−4 | |
| FT | 1.49 × 10−3 | |
| SIPR | 1.77 × 10−3 | R at the C-terminus and p in penultimate position |
| GI * | 2.07 × 10−3 | |
| MI * | 2.12 × 10−3 | |
| SAPI | 2.23 × 10−3 | p in penultimate position and A in second position |
| ML * | 2.25 × 10−3 | |
| GL * | 3.53 × 10−3 | |
| FVPH | 3.56 × 10−3 | F at the N-terminus and p in penultimate position |
| VF * | 3.63 × 10−3 | |
| FV | 3.63 × 10−3 | |
| AL * | 5.87 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martini, S.; Cattivelli, A.; Conte, A.; Tagliazucchi, D. Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract. Curr. Issues Mol. Biol. 2022, 44, 139-151. https://doi.org/10.3390/cimb44010011
Martini S, Cattivelli A, Conte A, Tagliazucchi D. Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract. Current Issues in Molecular Biology. 2022; 44(1):139-151. https://doi.org/10.3390/cimb44010011
Chicago/Turabian StyleMartini, Serena, Alice Cattivelli, Angela Conte, and Davide Tagliazucchi. 2022. "Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract" Current Issues in Molecular Biology 44, no. 1: 139-151. https://doi.org/10.3390/cimb44010011
APA StyleMartini, S., Cattivelli, A., Conte, A., & Tagliazucchi, D. (2022). Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract. Current Issues in Molecular Biology, 44(1), 139-151. https://doi.org/10.3390/cimb44010011