Computer-Assisted Discovery of Alkaloids with Schistosomicidal Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. VolSurf+ Descriptors
2.3. Random Forest Model
2.4. MuDRA Model
2.5. Principal Component Analysis
2.6. Molecular Docking
2.7. Metabolic Prediction
2.8. Toxicity and Drug-Likeness Assessment
3. Results and Discussion
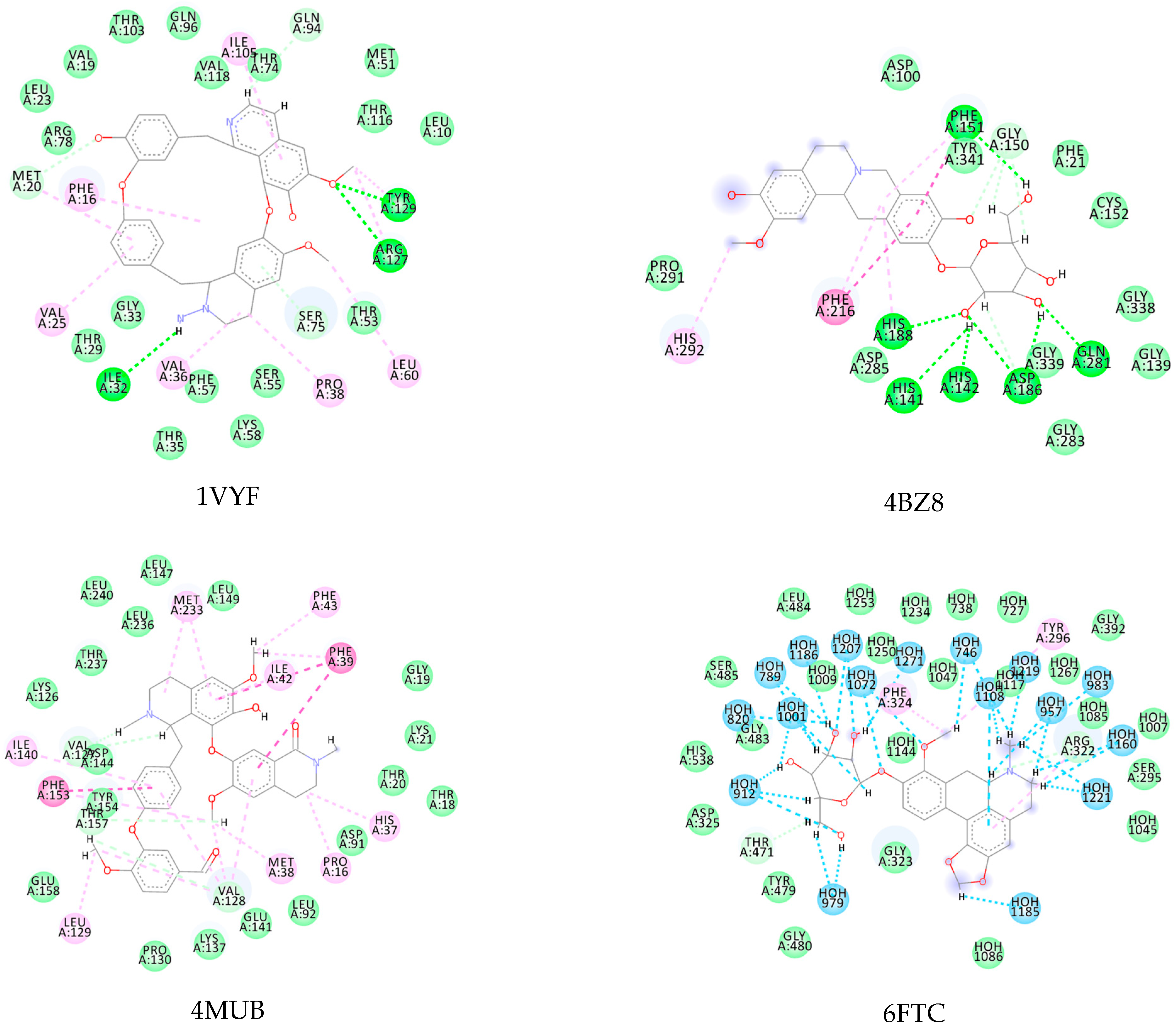
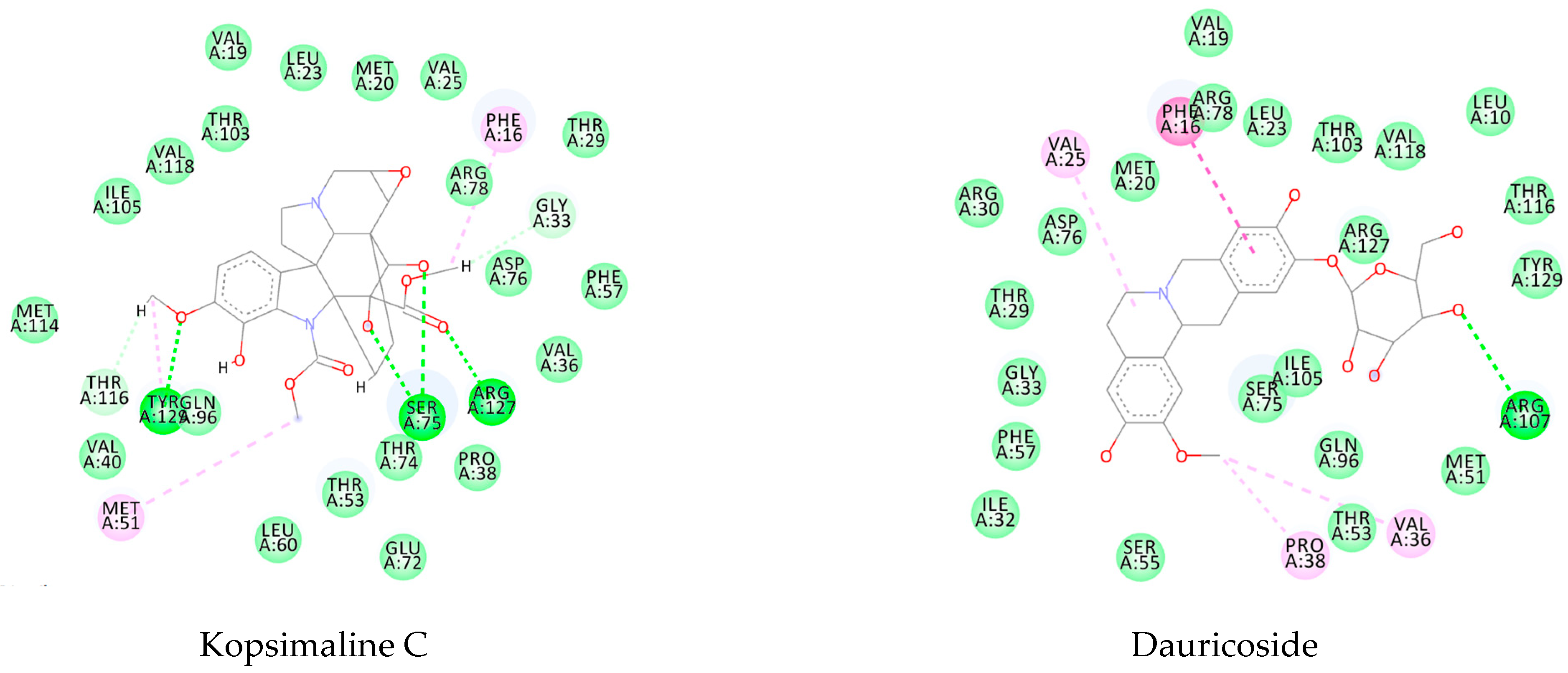
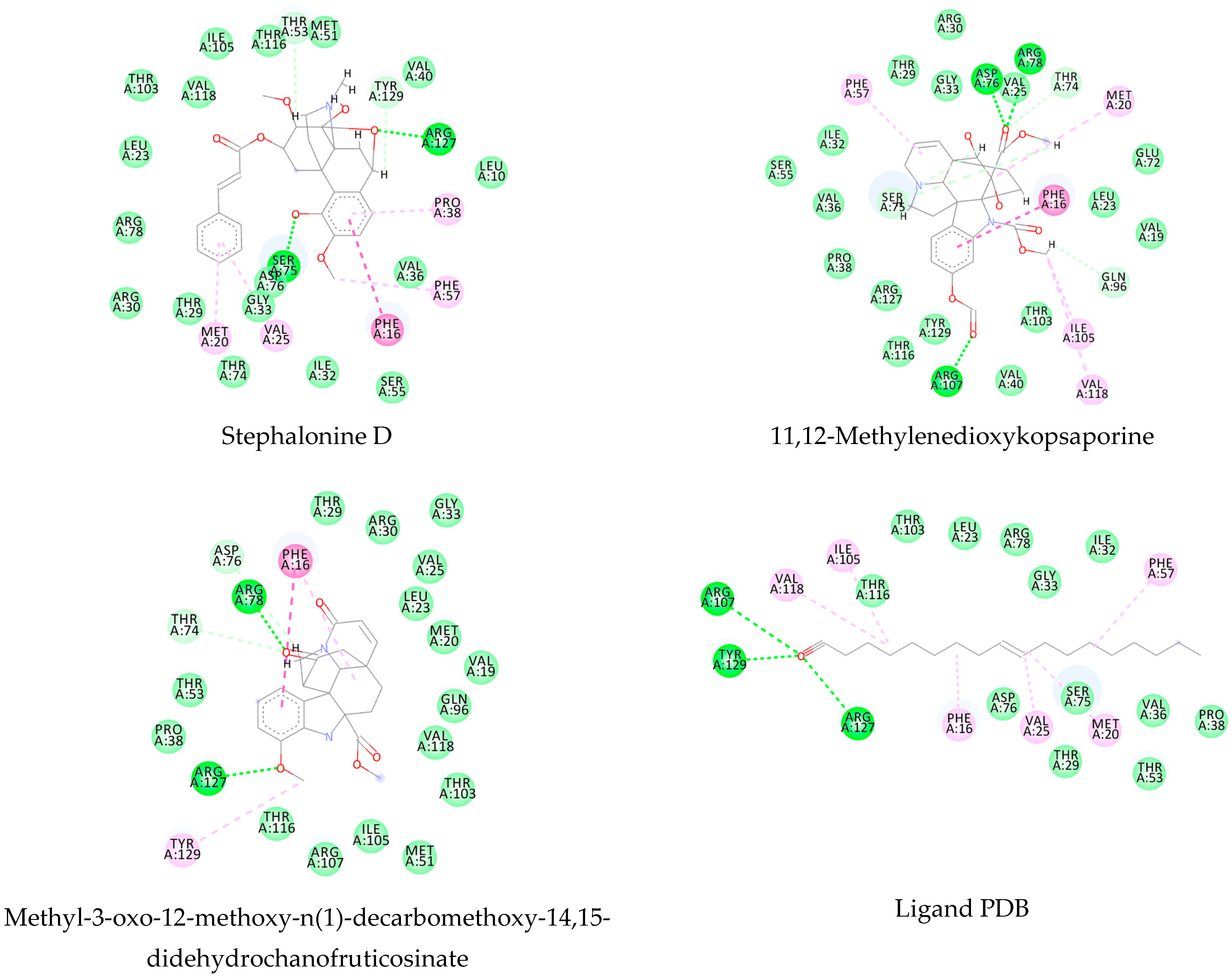
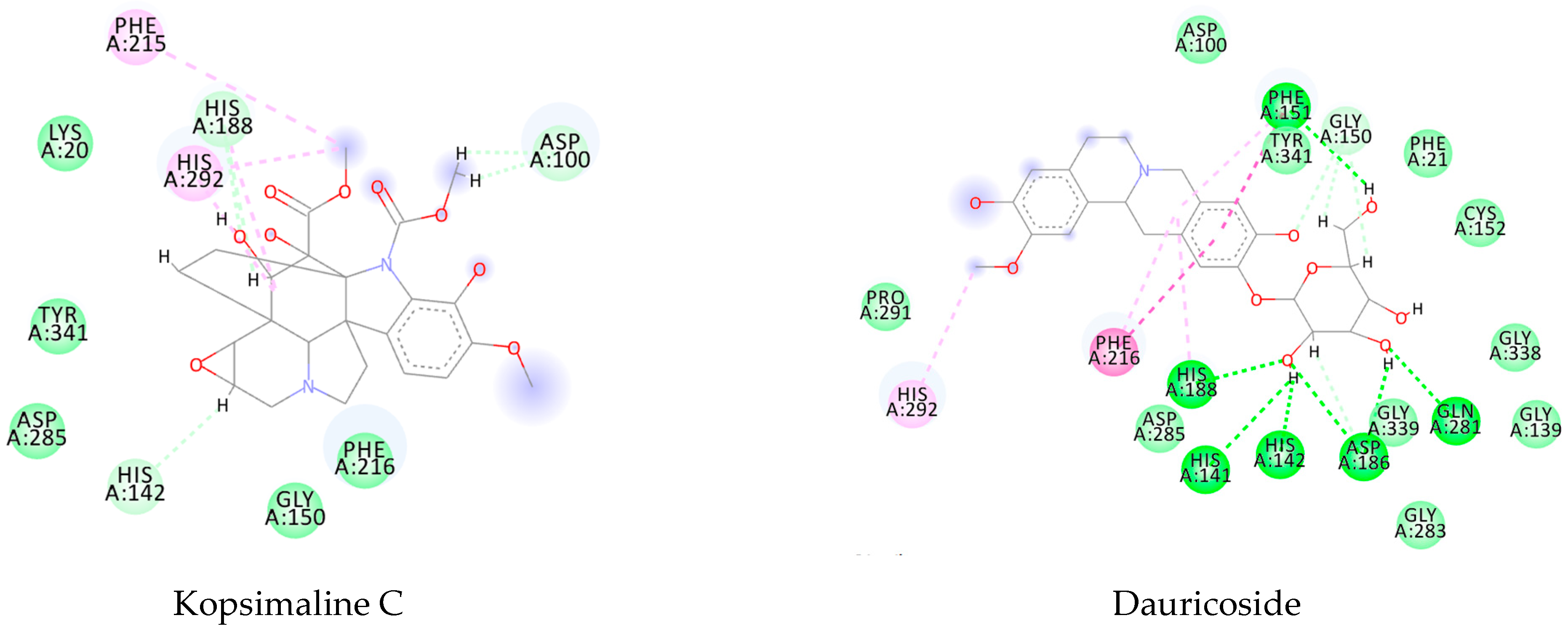
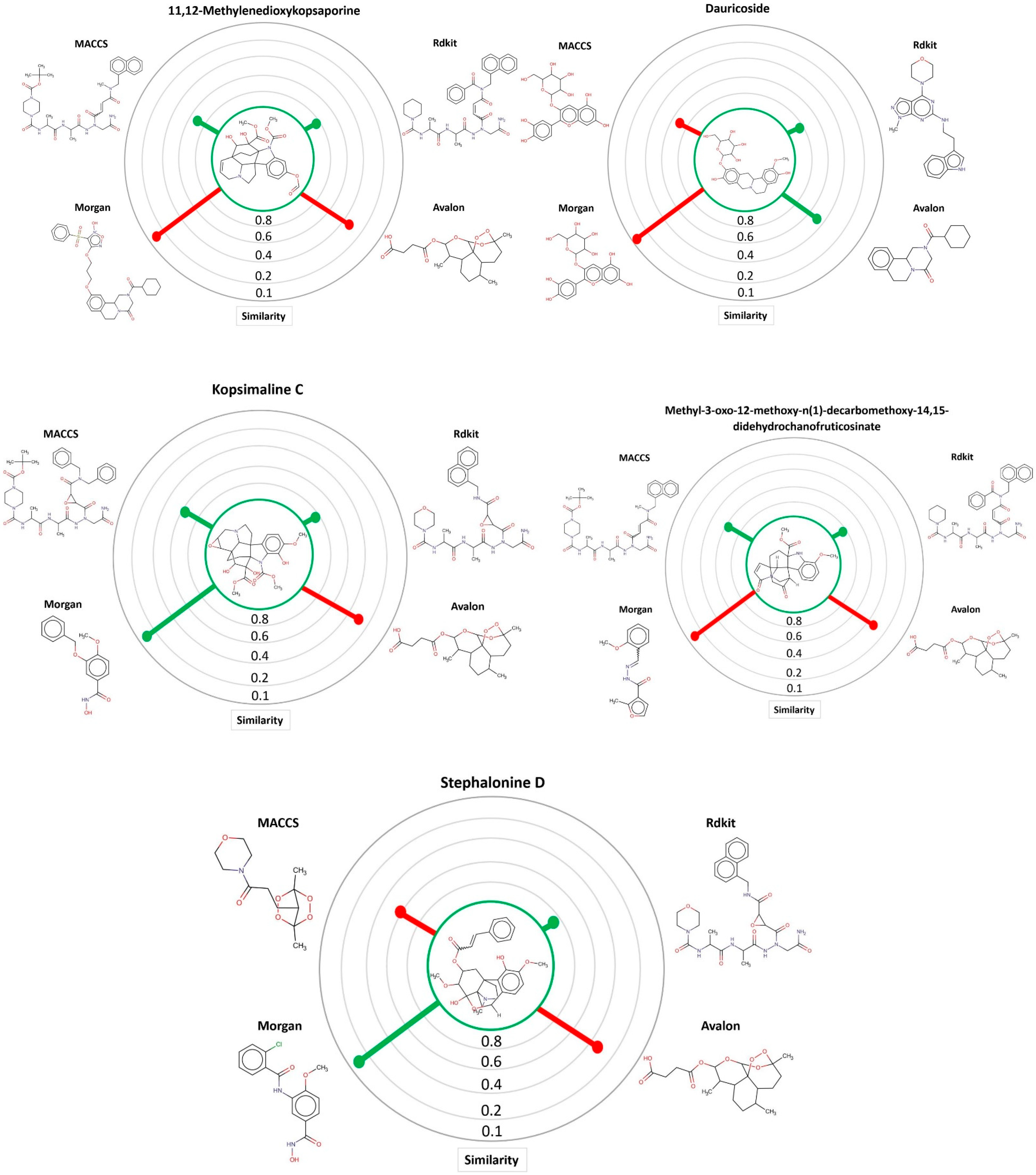
3.1. Ligand-Based Virtual Screening
3.2. Structure-Based Virtual Screening
3.3. Consensus Analysis
3.4. Prediction of Metabolism of Selected Alkaloids
3.5. Drug-like and Toxicity Analyzes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Compound_Name | 1VYF | 4BZ8 | 4MUB | 6FTC |
|---|---|---|---|---|
| Bisaknadinine | −11.72 | −82.27 | −66.68 | −63.46 |
| Nitaphylline | −93.64 | −119.96 | 111.63 | 4.72 |
| Leucolusine | −91.23 | −45.91 | −75 | −81.92 |
| Stesakine 9-O-b-D-glucoside | −98.81 | −128.85 | −136.44 | −125.82 |
| Bisinomenine | −109.73 | 29.43 | −90.57 | 20.6 |
| Kopsimaline C | −119.07 | −108.56 | −129.61 | −88.98 |
| Dauricoside | −121.12 | −167.78 | −130.14 | −111.4 |
| Kopsimaline F | −47.47 | −99.61 | −118.64 | −88.49 |
| Mersilongine | −51.53 | −92.85 | −97.84 | −41.96 |
| Mersilfoline B | −55.62 | −70.08 | −78.49 | −37.56 |
| Stephalonine D | −120.18 | −147.16 | −130.34 | −88.71 |
| Kopsifoline C | −92.87 | −82.48 | −131.52 | −63.74 |
| Kopsiloscine D | −108.95 | −96.57 | −110.53 | −96.6 |
| Kopsingine | −91.06 | −90.3 | −100.04 | −44.05 |
| 11,12-Methylenedioxykopsaporine | −97.19 | −140.91 | −106.58 | −89.29 |
| Voachalotine oxindole | −71.4 | −75.38 | −104.95 | −66.48 |
| 11,12-Methylenedioxykopsinol | −100.82 | −110.79 | −128.9 | −74.89 |
| Kopsidasine n-oxide | −49.93 | −54.62 | −64.24 | −15.24 |
| Stephalonine E | −95.68 | −108.73 | −96.83 | −71.08 |
| Kopsifoline b | −89.52 | −75.85 | −121.72 | −60.65 |
| Affinine | −99.97 | −98.56 | −109.07 | −106.94 |
| Jerantinine c | −96.02 | −108.01 | −112.42 | −81.91 |
| 12-Demethoxykopsingine | −93.37 | −110.82 | −93.47 | −50.82 |
| Kopsimaline a | −123.58 | −139.07 | −89.9 | −112.8 |
| Valesamina | −96.14 | −94.62 | −90.52 | −96.13 |
| Jerantinine b | −91.65 | −104.49 | −112.84 | −96.27 |
| Prunifoline f | −103.05 | −87.92 | −92.2 | −21.34 |
| Kopsiloscine e | −117.29 | −79.36 | −118.67 | −65.31 |
| 12-Methoxykopsinaline | −92.33 | −76.26 | −95.59 | −29.34 |
| Jerantinine d | −99.78 | −108.72 | −120.78 | −71.75 |
| Methyl-3-oxo-12-methoxy-n(1)-decarbomethoxy- 14,15-didehydrochanofruticosinate | −109.89 | −112.23 | −110.54 | −89.49 |
| Secohomoaromoline | −116.04 | −129.7 | −154.8 | −97.94 |
| Kopsidarine | −73.72 | −96.13 | −101.26 | −46.91 |
| Alpneumine h | −61.29 | −93 | −84.58 | −103.19 |
| Aknadilactam | −88.94 | −101.18 | −83.19 | −38.56 |
| Lapidilectinol | −85.2 | −89.25 | −72.05 | −47.22 |
| Mersidasine c | −88.12 | −60.28 | −84.52 | −33.61 |
| Telikovinone | −62.13 | −64.49 | −91.73 | −39.23 |
| Kopsiloscine B | −100.81 | −105.86 | −87.98 | −87.67 |
| Disinomenine | −139.05 | −110.51 | −92.5 | −25.65 |
| Lahadinine B | −95.1 | −88.11 | −75.79 | −36.6 |
| Neotrilobine | −104.96 | −94.71 | −88.33 | −103.53 |
| Kopsofinone | −113.3 | −76.16 | −101.8 | −62.85 |
| N-oxide | −89.86 | −78.11 | −76.28 | −40.13 |
| Kopsiloscine j | −93.87 | −87.86 | −105.7 | −24.82 |
| 10-Demethoxykopsidasinine | −76.25 | −62.32 | −79.99 | −7.03 |
| Pauciflorine a | −106.69 | −103.1 | −103.19 | −76.25 |
| Paprazine | −81.84 | −70.23 | −87.35 | −78.93 |
| Kopsinol | −75.91 | −55.94 | −97.69 | −25.31 |
| Kopsinganol | −107.59 | −100.54 | −119.99 | −52.4 |
| Alpneumine g | −88.68 | −81.46 | −85.93 | −36.89 |
| N-Methylasimilobine-2-O-b-D-glucopyranoside | −103.54 | −106.85 | −120.85 | −103.47 |
| N-Oxide c | −89.91 | −97.29 | −122.26 | −53.85 |
| Pauciflorine b | −90.1 | −57.84 | −67.57 | −50.26 |
| Kopsifoline a | −96.18 | −104.64 | −114.36 | −61.7 |
| Alstonamic acid | −70.89 | −85.09 | −83.92 | −82.8 |
| Isoprostephabyssine | −75.29 | −93.55 | −83.42 | −30.14 |
| Dehatridine | −147.13 | −113.85 | −103.04 | 13.7 |
| Jerantinine e | −95.71 | −106.3 | −109.52 | −90.28 |
| Mersifoline a | −86.97 | −89.24 | −104.91 | −57.09 |
| 16(S)-10-Metoxi-epi-isositsirikina | −105.02 | −105.39 | −111.84 | −90.46 |

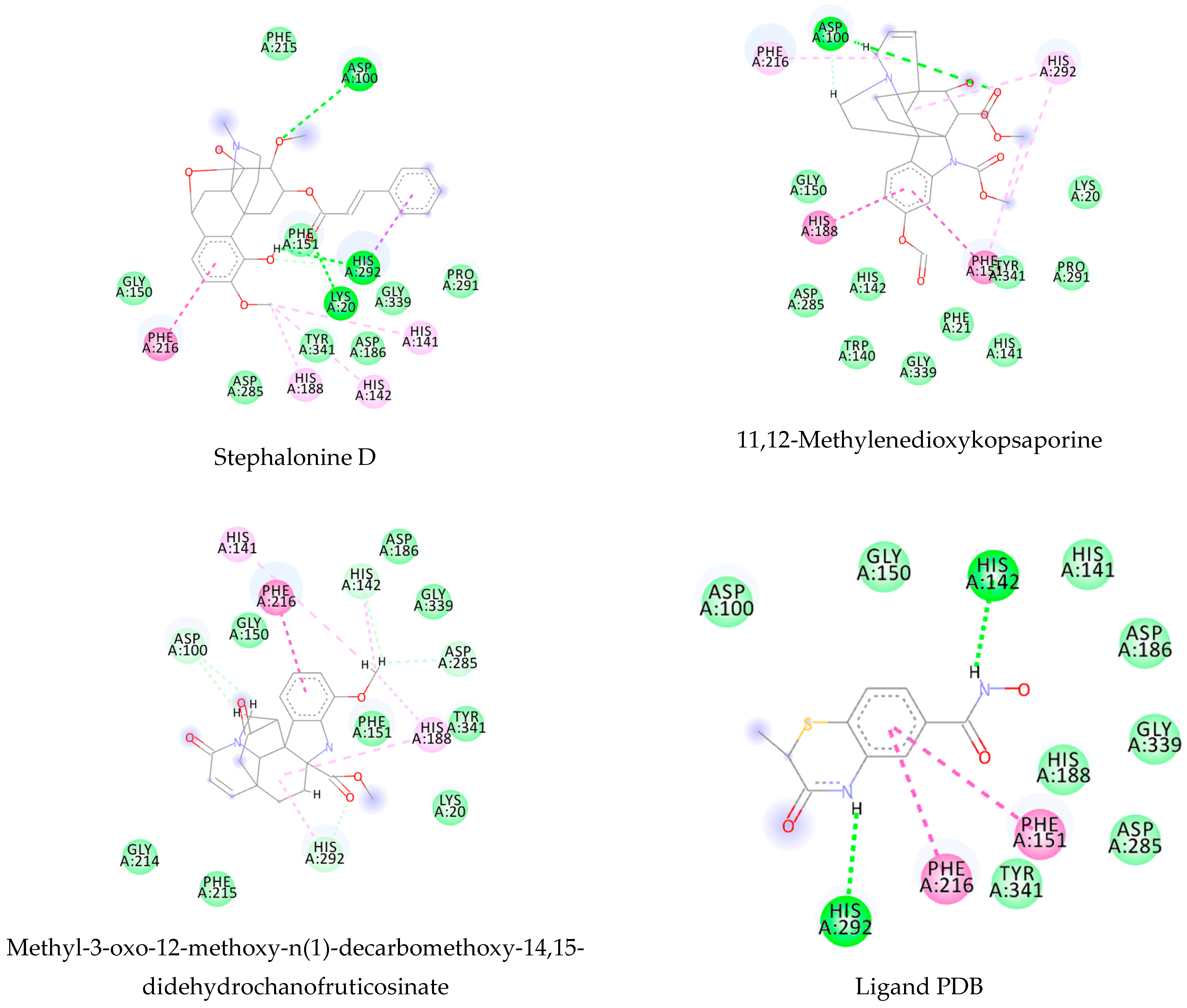
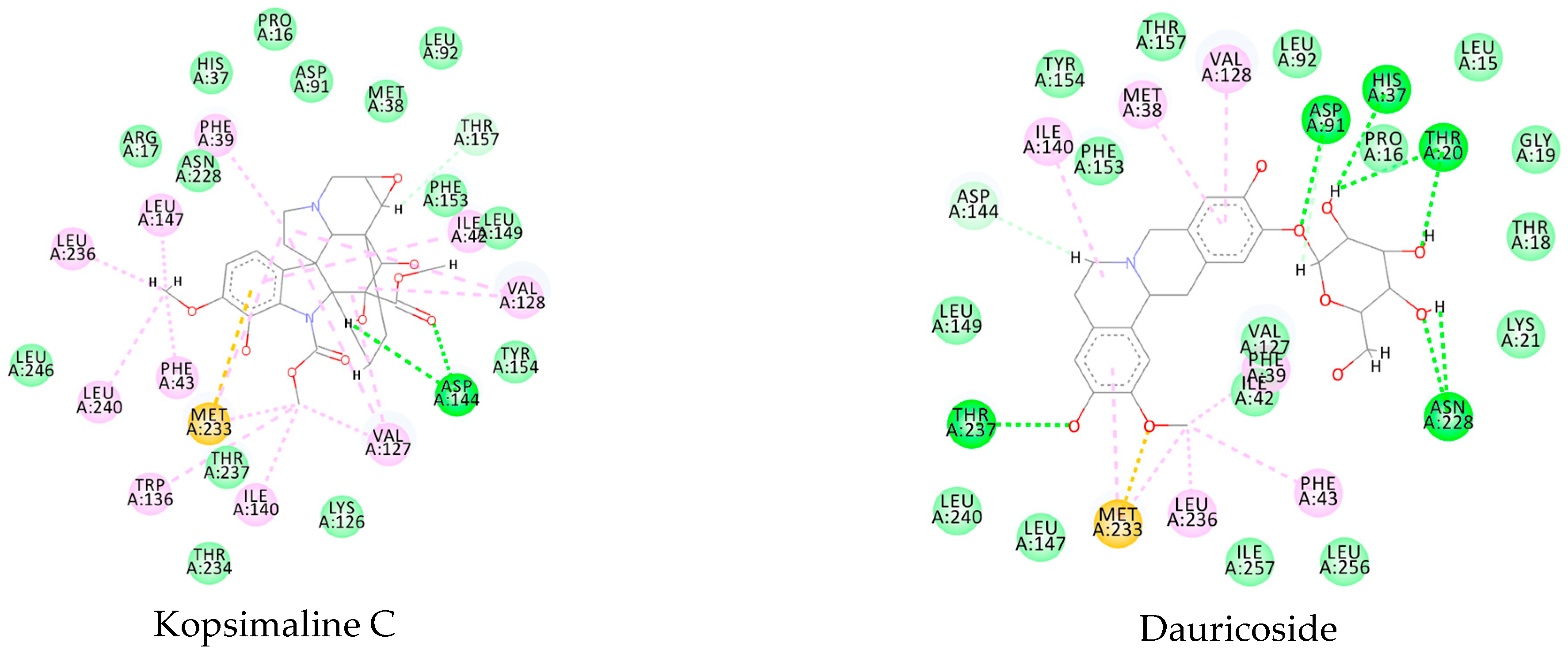
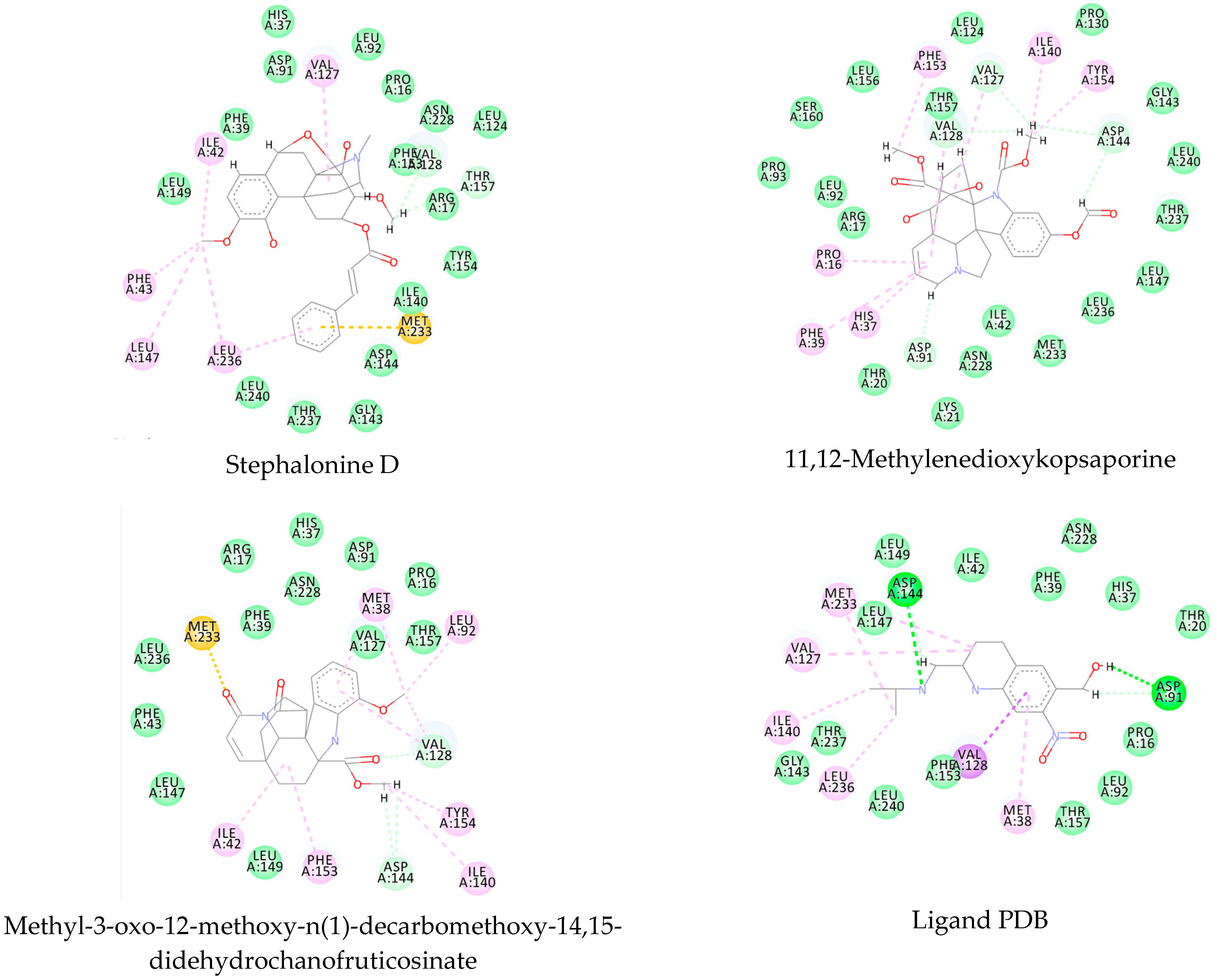


References
- Acharya, A.S.; Kaur, R.; Goel, A.D. Neglected tropical diseases—Challenges and opportunities in India. Indian J. Med. Spec. 2017, 8, 102–108. [Google Scholar] [CrossRef]
- Hotez, P.J.; Botazzi, M.E.; Franco-Paredes, C.; Ault, S.K.; Periago, M.R. The neglected tropical diseases of Latin America and the Caribbean: A review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl. Trop. Dis. 2008, 2, e300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meireles, C.B.; Maia, L.C.; Soares, G.C.; Teodoro, I.P.P.; Gadelha, M.D.S.V.; da Silva, C.G.L.; de Lima, M.A.P. Atypical presentations of cutaneous leishmaniasis: A systematic review. Acta Trop. 2017, 172, 240–254. [Google Scholar] [CrossRef] [PubMed]
- WHO. Schistosomiasis Fact Sheet. Available online: https://www.who.int/en/news-room/fact-sheets/detail/schistosomiasis (accessed on 19 March 2021).
- Adenowo, A.F.; Oyinloye, B.E.; Ogunyinka, B.I.; Kappo, A.P. Impact of human schistosomiasis in sub-Saharan Africa. Brazilian J. Infect. Dis. 2015, 19, 196–205. [Google Scholar] [CrossRef] [Green Version]
- Andrews, P.; Thomas, H.; Pohlke, R.; Seubert, J. Praziquantel. Med. Res. Rev. 1983, 3, 147–200. [Google Scholar] [CrossRef]
- King, C.H.; Mahmoud, A.A. Drugs five years later: Praziquantel. Ann. Intern. Med. 1989, 110, 290–296. [Google Scholar] [CrossRef]
- Chai, J.Y. Praziquantel treatment in trematode and cestode infections: An update. J. Infect. Chemother. 2013, 45, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Aboki, A.O.; Ibikoule, M.; Boko, P.M.; Savassi, B.S.; Tougoue, J.J.; Kaboré, A. Human schistosomiasis in Benin: Countrywide evidence of Schistosoma haematobium predominance. Acta Trop. 2019, 191, 185–197. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Barros, R.P.C.; da Cunha, E.V.L.; Catão, R.M.R.; Scotti, L.; Souza, M.S.R.; Brás, A.A.Q.; Scotti, M.T. Virtual screening of secondary metabolites of the genus Solanum with potential antimicrobial activity. Rev. Bras. Farmacogn. 2018, 28, 686–691. [Google Scholar] [CrossRef]
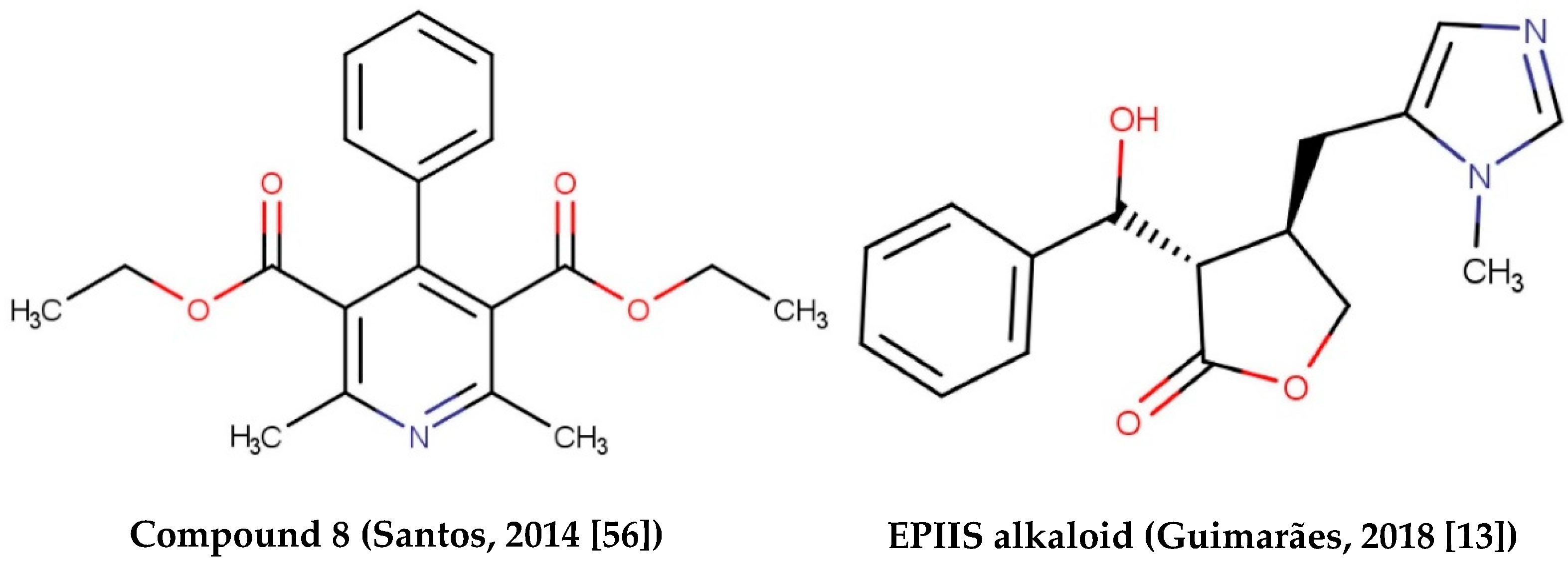
- Guimarães, M.A.; de Oliveira, R.N.; de Almeida, R.L.; Mafud, A.C.; Sarkis, A.L.V.; Ganassin, R.; da Silva, M.P.; Roquini, D.B.; Veras, L.M.; Sawada, T.C.H.; et al. Epiisopilosine alkaloid has activity against Schistosoma mansoni in mice without acute toxicity. PLoS ONE 2018, 13, e0196667. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.U.; Ali, H.S.; Jafari, B.; Zaib, S.; Hameed, A.; Al-Kahraman, Y.M.S.A.; Langer, P.; Iqbal, J. Structure-based virtual screening of dipeptidyl peptidase 4 inhibitors and their in vitro analysis. Comput. Biol. Chem. 2021, 91, 107326. [Google Scholar] [CrossRef]
- Pérez-Villanueva, J.; Yépez-Mulia, L.; Rodríguez-Villar, K.; Cortés-Benítez, F.; Palacios-Espinosa, J.F.; Soria-Arteche, O. The giardicidal activity of lobendazole, fabomotizole, tenatoprazole and ipriflavone: A ligand-based virtual screening and in vitro study. Eur. J. Med. Chem. 2021, 211, 113110. [Google Scholar] [CrossRef]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL web services: Streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015, 43, W612–W620. [Google Scholar] [CrossRef] [Green Version]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, 1100–1107. [Google Scholar] [CrossRef] [Green Version]
- Willighagen, E.L.; Waagmeester, A.; Spjuth, O.; Ansell, P.; Williams, A.J.; Tkachenko, V.; Hastings, J.; Chen, B.; Wild, D.J. The ChEMBL database as linked open data. J. Cheminform. 2013, 5, 23. [Google Scholar] [CrossRef]
- Costa, R.P.O.; Lucena, L.F.; Silva, L.M.A.; Zocolo, G.J.; Herrera-Acevedo, C.; Scotti, L.; Da-Costa, F.B.; Ionov, N.; Poroikov, V.; Muratov, E.N.; et al. The SistematX Web Portal of Natural Products: An Update. J. Chem. Inf. Model. 2021, 61, 2516–2522. [Google Scholar] [CrossRef]
- Scotti, M.T.; Herrera-Acevedo, C.; Oliveira, T.B.; Costa, R.P.O.; de Oliveira Santos, S.Y.K.; Rodrigues, R.P.; Scotti, L.; Da-Costa, F.B. SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites. Molecules 2018, 23, 103. [Google Scholar] [CrossRef] [Green Version]
- ChemAxon Marvin; ChemAxon Ltd.: Busdapest, Hungary, 2021.
- ChemAxon Standardizer Software; ChemAxon Ltd.: Busdapest, Hungary, 2021.
- Fourches, D.; Muratov, E.; Tropsha, A. Curation of chemogenomics data. Nat. Chem. Biol. 2015, 11, 535. [Google Scholar] [CrossRef] [PubMed]
- Fourches, D.; Muratov, E.; Tropsha, A.; Stork, C.; Chen, Y.; Šícho, M.; Kirchmair, J. Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model. 2016, 56, 1243–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourches, D.; Muratov, E.; Tropsha, A. Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010, 50, 1189–1204. [Google Scholar] [CrossRef] [PubMed]
- Crivori, P.; Cruciani, G.; Carrupt, P.-A.; Testa, B. Predicting blood− brain barrier permeation from three-dimensional molecular structure. J. Med. Chem. 2000, 43, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, G.; Pastor, M.; Guba, W. VolSurf: A new tool for the pharmacokinetic optimization of lead compounds. Eur. J. Pharm. Sci. 2000, 11, S29–S39. [Google Scholar] [CrossRef]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Thiel, K.; Wiswedel, B. KNIME-the Konstanz information miner: Version 2.0 and beyond. ACM SIGKDD Explor. Newsl. 2009, 11, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Fourches, D.; Pu, D.; Tassa, C.; Weissleder, R.; Shaw, S.Y.; Mumper, R.J.; Tropsha, A. Quantitative nanostructure—Activity relationship modeling. ACS Nano 2010, 4, 5703–5712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.C.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [Green Version]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. BBA-Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Scotti, M.T.; Scotti, L.; Ishiki, H.M.; Peron, L.M.; de Rezende, L.; do Amaral, A.T. Variable-selection approaches to generate QSAR models for a set of antichagasic semicarbazones and analogues. Chemom. Intell. Lab. Syst. 2016, 154, 137–149. [Google Scholar] [CrossRef]
- Aptula, A.O.; Roberts, D.W. Mechanistic applicability domains for nonanimal-based prediction of toxicological end points: General principles and application to reactive toxicity. Chem. Res. Toxicol. 2006, 19, 1097–1105. [Google Scholar] [CrossRef]
- Alves, V.M.; Golbraikh, A.; Capuzzi, S.J.; Liu, K.; Lam, W.I.; Korn, D.R.; Pozefsky, D.; Andrade, C.H.; Muratov, E.N.; Tropsha, A. Multi-Descriptor Read Across (MuDRA): A Simple and Transparent Approach for Developing Accurate Quantitative Structure–Activity Relationship Models. J. Chem. Inf. Model. 2018, 58, 1214–1223. [Google Scholar] [CrossRef]
- Low, Y.; Sedykh, A.; Fourches, D.; Golbraikh, A.; Whelan, M.; Rusyn, I.; Tropsha, A. Integrative Chemical–Biological Read-Across Approach for Chemical Hazard Classification. Chem. Res. Toxicol. 2013, 26, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.K.; Aha, D.W. Analyses of Instace-Based Learning Algorithms. AAAI-91 Proc. 1991, 553–558. Available online: https://www.aaai.org/Papers/AAAI/1991/AAAI91-086.pdf (accessed on 5 December 2021).
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Berman, H.M.; Bhat, T.N.; Bourne, P.E.; Feng, Z.; Gilliland, G.; Weissig, H.; Westbrook, J. The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 2000, 7, 957–959. [Google Scholar] [CrossRef]
- Angelucci, F.; Kenneth, A.J.; Baiocco, P.; Adriana, E.M.; Brunori, M.; Valle, C.; Vigorosi, F.; Troiani, A.R.; Liberti, P.; Cioli, D.; et al. Schistosoma mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure. Biochemistry 2004, 43, 13000–13011. [Google Scholar] [CrossRef] [PubMed]
- Marek, M.; Kannan, S.; Hauser, A.-T.; Mourão, M.M.; Caby, S.; Cura, V.; Stolfa, D.A.; Schmidtkunz, K.; Lancelot, J.; Andrade, L.; et al. Structural Basis for the Inhibition of Histone Deacetylase 8 (HDAC8), a Key Epigenetic Player in the Blood Fluke Schistosoma mansoni. PLoS Pathog. 2013, 9, e1003645. [Google Scholar] [CrossRef] [PubMed]
- Valentim, C.L.L.; Cioli, D.; Chevalier, F.D.; Cao, X.; Taylor, A.B.; Holloway, S.P.; Pica-Mattoccia, L.; Guidi, A.; Basso, A.; Tsai, I.J.; et al. Genetic and molecular basis of drug resistance and species-specific drug action in Schistosome parasites. Science 2013, 342, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, I.; Lyu, H.; Fata, F.; Boumis, G.; Miele, A.E.; Ardini, M.; Ippoliti, R.; Bellelli, A.; Jadhav, A.; Lea, W.A.; et al. Fragment-Based Discovery of a Regulatory Site in Thioredoxin Glutathione Reductase Acting as “Doorstop” for NADPH Entry. ACS Chem. Biol. 2018, 13, 2190–2202. [Google Scholar] [CrossRef]
- Onodera, K.; Satou, K.; Hirota, H. Evaluations of molecular docking programs for virtual screening. J. Chem. Inf. Model. 2007, 47, 1609–1618. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Discovery Studio Visualizer; Dassault Systèmes S.A.: Velizy-Villacoublay, France, 2020.
- Company, C. Bio Molegro Virtual Docker-User Manual; Copyright CLC Bio Company: Aarhus, Denmark, 2013. [Google Scholar]
- Cruciani, G.; Milani, N.; Benedetti, P.; Lepri, S.; Cesarini, L.; Baroni, M.; Spyrakis, F.; Tortorella, S.; Mosconi, E.; Goracci, L. From Experiments to a Fast Easy-to-Use Computational Methodology to Predict Human Aldehyde Oxidase Selectivity and Metabolic Reactions. J. Med. Chem. 2017, 61, 360–371. [Google Scholar] [CrossRef]
- Talete srl Dragon-Software for Molecular Descriptor Calculation; Version 7; Talete srl: Milano, Italy, 2012.
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Kovdienko, N.A.; Polishchuk, P.G.; Muratov, E.N.; Artemenko, A.G.; Kuz’min, V.E.; Gorb, L.; Hill, F.; Leszczynski, J. Application of Random Forest and Multiple Linear Regression Techniques to QSPR Prediction of an Aqueous Solubility for Military Compounds. Mol. Inform. 2010, 29, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Barros, R.P.C.; Scotti, L.; Scotti, M.T. Exploring secondary metabolites database of apocynaceae, menispermaceae, and annonaceae to select potential anti-HCV compounds. Curr. Top. Med. Chem. 2019, 19, 900–913. [Google Scholar] [CrossRef]
- Acevedo, C.H.; Scotti, L.; Scotti, M.T. In Silico Studies Designed to Select Sesquiterpene Lactones with Potential Antichagasic Activity from an In-House Asteraceae Database. ChemMedChem 2018, 13, 634–645. [Google Scholar] [CrossRef] [Green Version]
- Razzaghi-Asl, N.; Mirzayi, S.; Mahnam, K.; Sepehri, S. Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation. J. Mol. Graph. Model. 2018, 83, 138–152. [Google Scholar] [CrossRef]
- Plewczynski, D.; Łaźniewski, M.; Augustyniak, R.; Ginalski, K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J. Comput. Chem. 2011, 32, 742–755. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276. [Google Scholar] [CrossRef]
- Dos Santos, A.F.; Fonseca, S.A.; César, F.A.; De Azevedo Albuquerque, M.C.P.; Santana, J.V.; Santana, A.E.G. A penta-substituted pyridine alkaloid from the rhizome of Jatropha elliptica (Pohl) Muell. Arg. is active against Schistosoma mansoni and Biomphalaria glabrata. Parasitol. Res. 2014, 113, 1077–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, J.A.; Rego, N.C.S.; Carvalho, B.T.S.; Silva, F.I.; Sousa, J.A.; Ramos, R.M.; Passos, I.N.G.; de Moraes, J.; Leite, J.R.S.A.; Lima, F.C.A. Computational quantum chemistry, molecular docking, and ADMET predictions of imidazole alkaloids of Pilocarpus microphyllus with schistosomicidal properties. PLoS ONE 2018, 13, e0198476. [Google Scholar] [CrossRef]
- Devlin, F.J.; Stephens, P.J.; Österle, C.; Wiberg, K.B.; Cheeseman, J.R.; Frisch, M.J. Configurational and Conformational Analysis of Chiral Molecules Using IR and VCD Spectroscopies: Spiropentylcarboxylic Acid Methyl Ester and Spiropentyl Acetate. J. Org. Chem. 2002, 67, 8090–8096. [Google Scholar] [CrossRef]
- Scotti, M.; Speck-Planche, A.; Tavares, J.; da Silva, M.S.; Cordeiro, M.; Scotti, L. Virtual screening of alkaloids from Apocynaceae with potential antitrypanosomal Activity. Curr. Bioinform. 2015, 10, 509–519. [Google Scholar] [CrossRef]
- Oprea, T.I. Property distribution of drug-related chemical databases. J. Comput. Aided. Mol. Des. 2000, 14, 251–264. [Google Scholar] [CrossRef]
- Walters, W.P.; Murcko, M.A. Prediction of “drug-likeness”. Adv. Drug Deliv. Rev. 2002, 54, 255–271. [Google Scholar] [CrossRef]
- Zheng, S.; Luo, X.; Chen, G.; Zhu, W.; Shen, J.; Chen, K.; Jiang, H. A new rapid and effective chemistry space filter in recognizing a druglike database. J. Chem. Inf. Model. 2005, 45, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Rishton, G.M. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today 2003, 8, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2009; ISBN 9783527628766. [Google Scholar]
- Lorenzo, V.; Lúcio, A.; Scotti, L.; Tavares, J.; Filho, J.; Lima, T.; Rocha, J.; Scotti, M. Structure- and Ligand-Based Approaches to Evaluate Aporphynic Alkaloids from Annonaceae as Multi-Target Agent Against Leishmania donovani. Curr. Pharm. Des. 2016, 22, 5196–5203. [Google Scholar] [CrossRef] [PubMed]
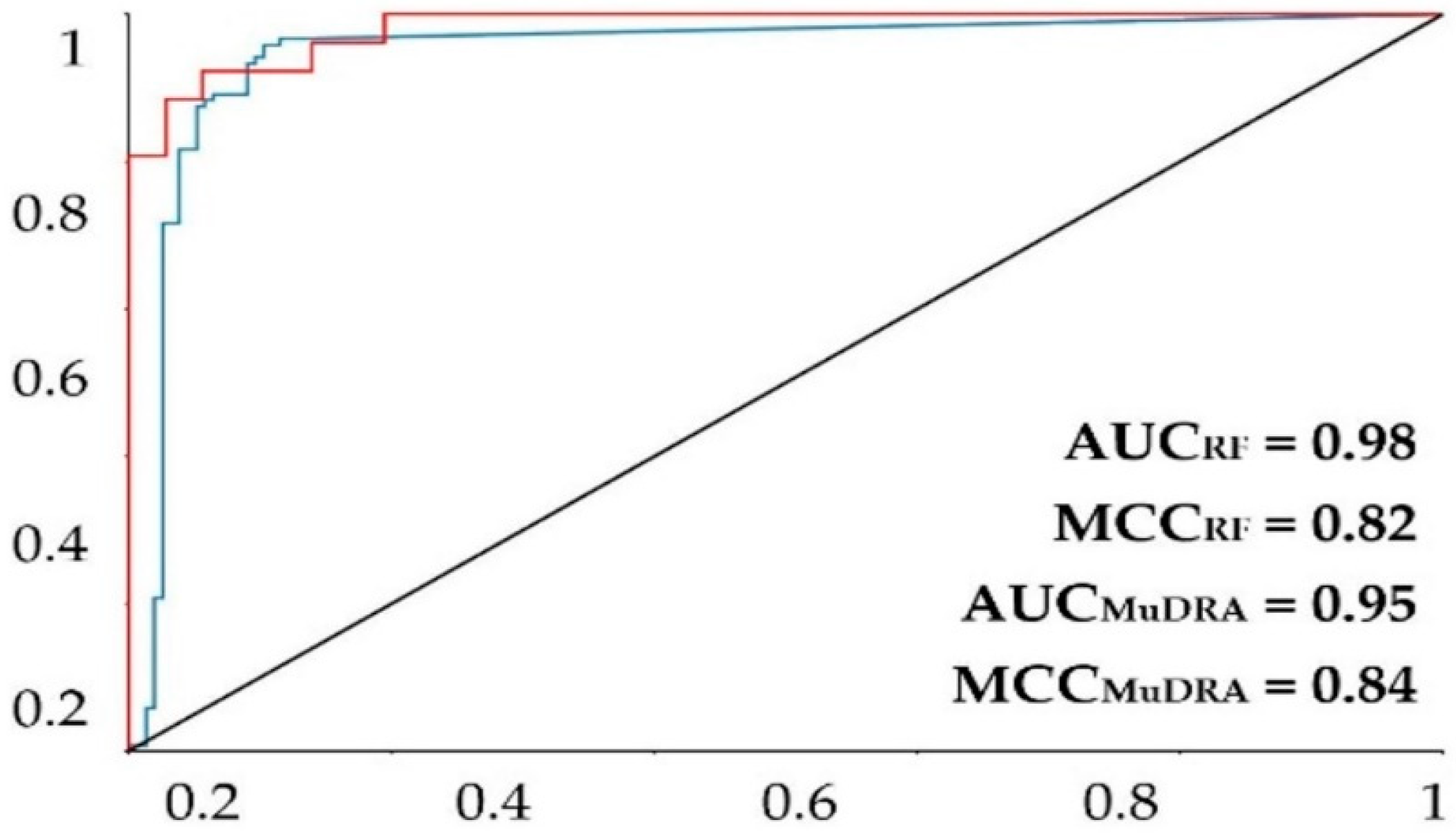



| Modeling Set | External Cross-Validation | |||||
| Fold | nº Compounds | Accuracy (%) | No. Compounds | Accuracy (%) | Sensitivity (%) | Specificity (%) |
| 1 | 247 | 89 | 62 | 90 | 96 | 86 |
| 2 | 247 | 90 | 62 | 89 | 89 | 89 |
| 3 | 247 | 89 | 62 | 90 | 92 | 89 |
| 4 | 247 | 91 | 62 | 90 | 88 | 91 |
| 5 | 248 | 92 | 61 | 91 | 85 | 97 |
| Confusion Matrices—External Cross-Validation | ||||||
| Fold | No. Compounds | True Positive | False Positive | True Negative | False Negative | |
| 1 | 62 | 25 | 5 | 31 | 1 | |
| 2 | 62 | 23 | 4 | 32 | 3 | |
| 3 | 62 | 24 | 4 | 32 | 2 | |
| 4 | 62 | 23 | 3 | 33 | 3 | |
| 5 | 61 | 22 | 1 | 35 | 3 | |
| Models | Specificity | Sensitivity | Accuracy | PPV | NPV |
|---|---|---|---|---|---|
| RF | 0.91 | 0.90 | 0.90 | 0.88 | 0.92 |
| Mudra | 0.90 | 0.93 | 0.91 | 0.88 | 0.94 |
| Protein Name | PDB ID | Best EALK | RMSD | EligPDB (Crystallized Ligand) | EligPDB (Redocking) |
|---|---|---|---|---|---|
| Schistosoma mansoni 14 kDa fatty-acid-binding protein (Sm14) | 1VYF | −147.1 1 | 0.51 | −88.53 | −86.63 |
| Histone deacetylase 8 | 4BZ8 | −167.70 2 | 0.22 | −85.97 | −81.84 |
| Sulfotransferase | 4MUB | −154.80 3 | 0.26 | −74.81 | −71.55 |
| Thioredoxin glutathione reductase | 6FTC | −125.82 4 | 0.48 | −76.24 | −72.78 |
| Molecule | Pcm | Index | 1VYF | 4BZ8 | 4MUB | 6FTC |
|---|---|---|---|---|---|---|
| Kopsimaline C | 0.73 | Ps Pc | 0.80 0.76 | 0.64 0.70 | 0.83 0.77 | 0.92 0.80 |
| Dauricoside | 0.72 | Ps Pc | 0.82 0.75 | 1 0.82 | 0.84 0.76 | 0.77 0.74 |
| Stephalonine D | 0.70 | Ps Pc | 0.81 0.74 | 0.87 0.76 | 0.84 0.75 | 0.89 0.77 |
| 11,12-Methylenedioxykopsaporine | 0.69 | Ps Pc | 0.66 0.68 | 0.83 0.74 | 0.68 0.69 | 0.87 0.75 |
| Methyl-3-oxo-12-methoxy-n(1)-decarbomethoxy-14,15-didehydrochanofruticosinate | 0.64 | Ps Pc | 0.74 0.68 | 0.66 0.65 | 0.71 0.67 | 0.70 0.66 |
| Substrate | Metabolite 1 | Metabolite 2 | Metabolite 3 | Metabolite 4 | Metabolite 5 |
|---|---|---|---|---|---|
 a |  O-Dealkylation |  N-Dealkylation |  N-Dealkylation |  Iminium Formation | 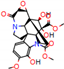 Aliphatic Carbonylation |
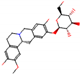 b |  O-Dealkylation |  O-Dealkylation | 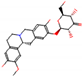 Alcoholic Oxidation |  Alcoholic Oxidation | 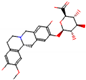 Alcoholic Oxidation |
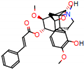 c | 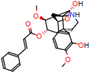 N-Dealkylation | 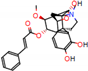 O-Dealkylation | 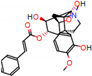 O-Dealkylation |  N-Dealkylation | 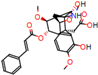 N-Dealkylation |
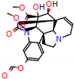 d |  N-Dealkylation | 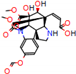 N-Dealkylation | 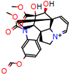 Iminium Formation | 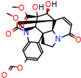 Aliphatic Carbonilation |  N-Dealkylation |
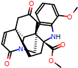 e | 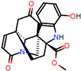 O-Dealkylation | 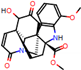 Aliphatic Hydroxylation | 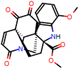 Aliphatic Hydroxylation | 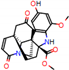 Aromatic Hydroxylation | 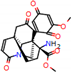 N-Dealkylation |
| Molecule | Mutagenic | Tumorigenic | Reproductive Toxicity | Dermal Toxicity |
|---|---|---|---|---|
| Kopsimaline C | None | None | None | None |
| Dauricoside | None | None | None | None |
| Stephalonine D | None | None | None | High risk |
| 11,12-Methylenedioxykopsaporine | None | None | None | None |
| Methyl-3-oxo-12-methoxy-n(1)-decarbomethoxy-14,15-didehydrochanofruticosinate | None | None | None | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menezes, R.P.B.d.; Viana, J.d.O.; Muratov, E.; Scotti, L.; Scotti, M.T. Computer-Assisted Discovery of Alkaloids with Schistosomicidal Activity. Curr. Issues Mol. Biol. 2022, 44, 383-408. https://doi.org/10.3390/cimb44010028
Menezes RPBd, Viana JdO, Muratov E, Scotti L, Scotti MT. Computer-Assisted Discovery of Alkaloids with Schistosomicidal Activity. Current Issues in Molecular Biology. 2022; 44(1):383-408. https://doi.org/10.3390/cimb44010028
Chicago/Turabian StyleMenezes, Renata Priscila Barros de, Jéssika de Oliveira Viana, Eugene Muratov, Luciana Scotti, and Marcus Tullius Scotti. 2022. "Computer-Assisted Discovery of Alkaloids with Schistosomicidal Activity" Current Issues in Molecular Biology 44, no. 1: 383-408. https://doi.org/10.3390/cimb44010028
APA StyleMenezes, R. P. B. d., Viana, J. d. O., Muratov, E., Scotti, L., & Scotti, M. T. (2022). Computer-Assisted Discovery of Alkaloids with Schistosomicidal Activity. Current Issues in Molecular Biology, 44(1), 383-408. https://doi.org/10.3390/cimb44010028










