Diversification of Ferredoxins across Living Organisms
Abstract
1. Introduction
2. Methodology
2.1. Species and Database
2.2. Genome Data Mining and Annotation of Ferredoxins
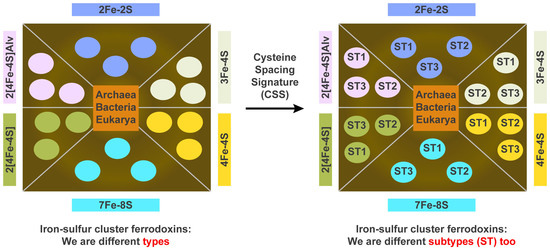
2.3. Ferredoxins Subtype Classification and Nomenclature
2.4. Assigning Fe-S Cluster Subtypes to the Ferredoxins Retrieved from the Literature
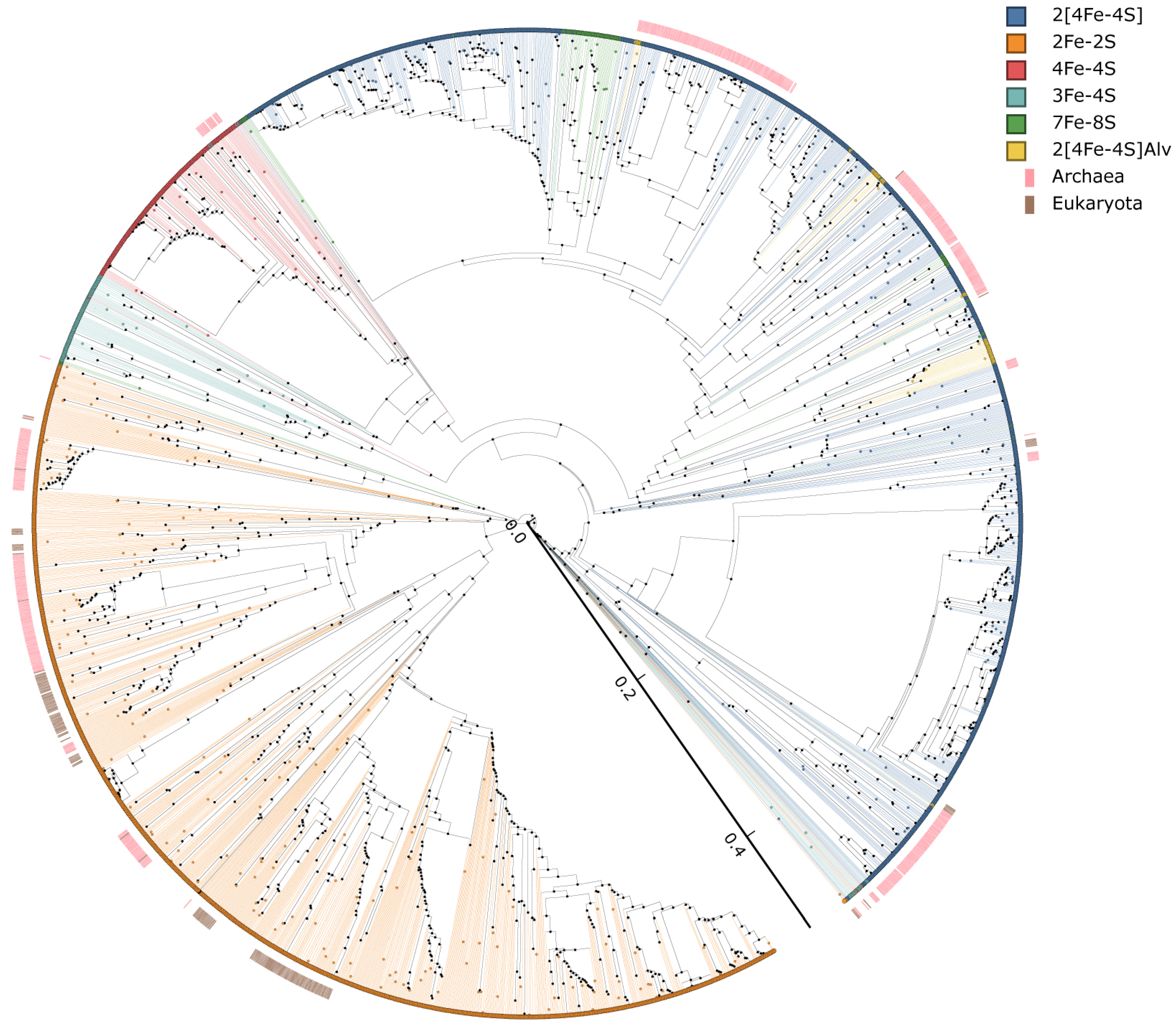
2.5. Phylogenetic Analysis of Ferredoxins
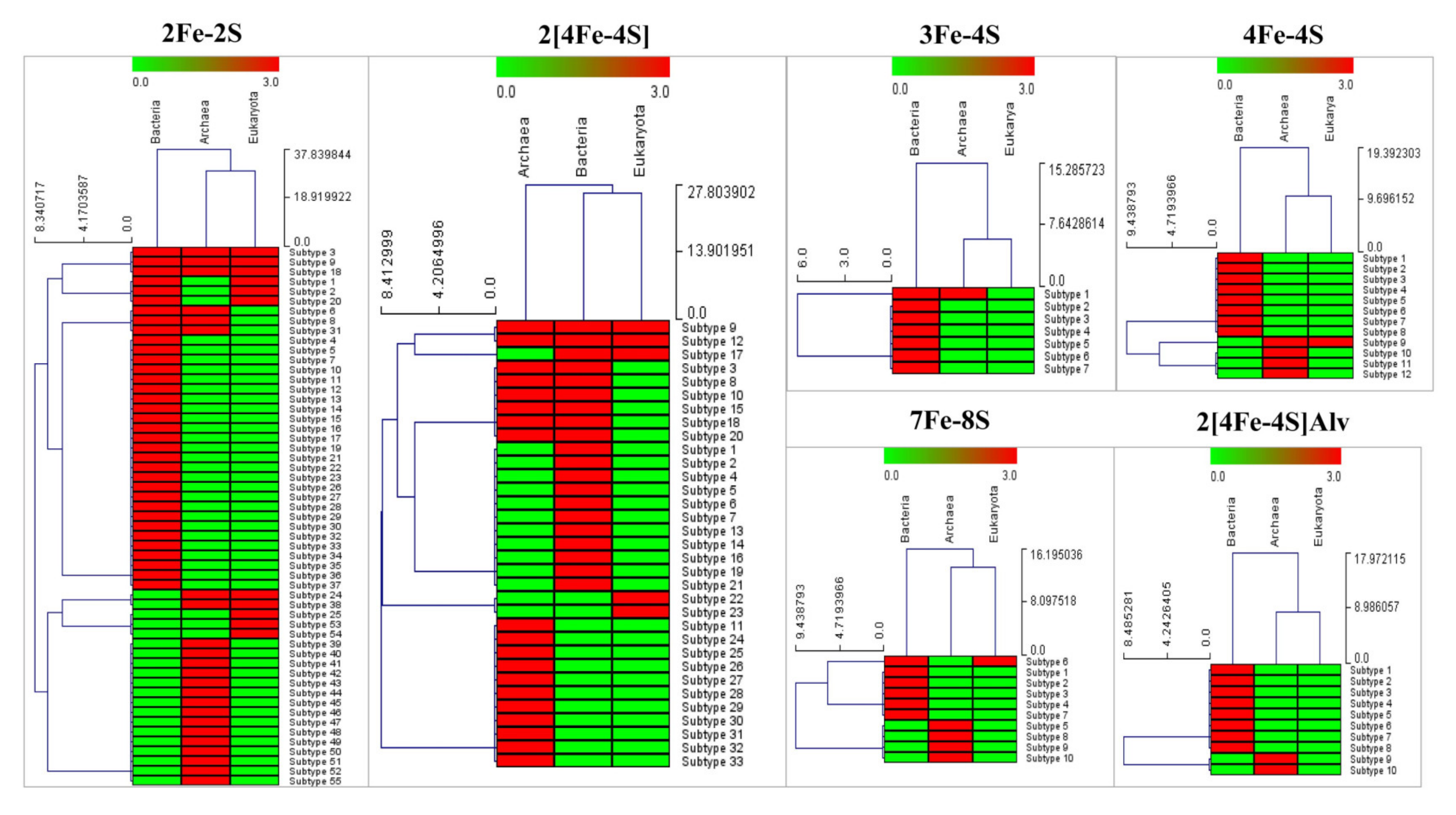
2.6. Generation of Ferredoxin Subtype Profile Heat Maps
3. Results and Discussion
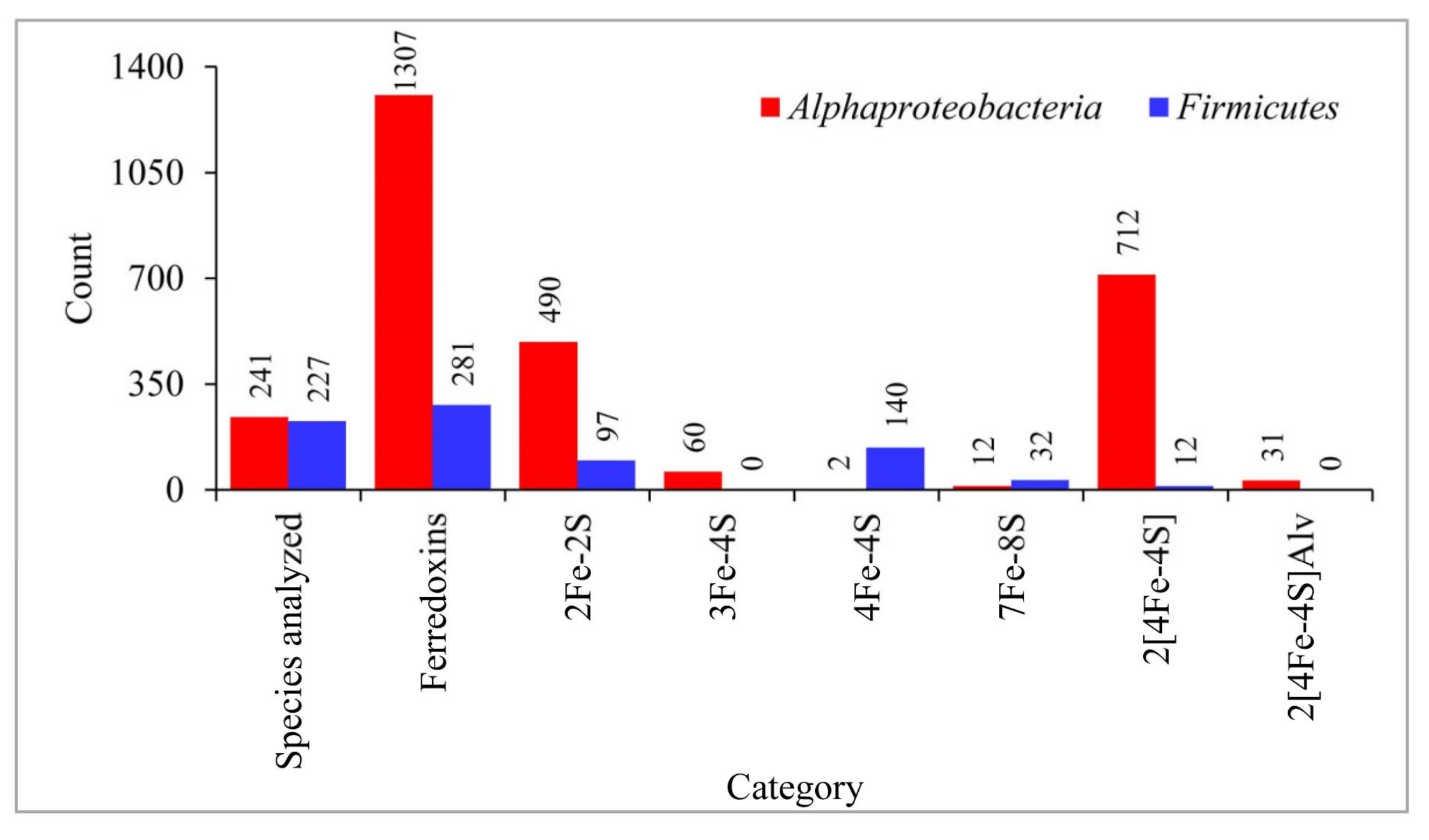
3.1. Alphaproteobacterial and Firmicutes Species Have Different Fe-S Cluster Type Ferredoxins in Their Genomes
3.2. Highly Diverse and Common Ancestral Origin of Ferredoxins between Alphaproteobacteria and Firmicutes
3.2.1. 2Fe-2S
3.2.2. 3Fe-4S
3.2.3. 4Fe-4S
3.2.4. 7Fe-8S
3.2.5. 2[4Fe-4S]
3.2.6. 2[4Fe-4S]Alv
3.3. Ferredoxin Fe-S Cluster Types Canonical Motifs
3.4. Evolutionary Linkage of Ferredoxins Subtype Classification
3.5. LGT of Ferredoxins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, D.; Cammack, R.; Rao, K. Role for ferredoxins in the origin of life and biological evolution. Nature 1971, 233, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Venkateswara Rao, P.; Holm, R. Synthetic analogues of the active sites of iron—Sulfur proteins. Chem. Rev. 2004, 104, 527–560. [Google Scholar] [CrossRef] [PubMed]
- Cammack, R. Evolution and diversity in the iron-sulphur proteins. Chem. Scr. 1983, 21, 87–95. [Google Scholar]
- Romero Romero, M.L.; Rabin, A.; Tawfik, D.S. Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew. Chem. Int. Ed. 2016, 55, 15966–15971. [Google Scholar] [CrossRef]
- Eck, R.V.; Dayhoff, M.O. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 1966, 152, 363–366. [Google Scholar] [CrossRef]
- Meyer, J. The evolution of ferredoxins. Trends Ecol. Evol. 1988, 3, 222–226. [Google Scholar] [CrossRef]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems—Biological variations of electron transport chains. Biochim. Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Chiliza, Z.E.; Martínez-Oyanedel, J.; Syed, K. An overview of the factors playing a role in cytochrome P450 monooxygenase and ferredoxin interactions. Biophys. Rev. 2020, 12, 1217–1222. [Google Scholar] [CrossRef]
- Li, S.; Du, L.; Bernhardt, R. Redox Partners: Function Modulators of Bacterial P450 Enzymes. Trends Microbiol. 2020, 28, 10. [Google Scholar] [CrossRef]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: Biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Burkhart, B.W.; Febvre, H.P.; Santangelo, T.J. Distinct physiological roles of the three ferredoxins encoded in the hyperthermophilic archaeon Thermococcus kodakarensis. mBio 2019, 10, e02807–e02818. [Google Scholar] [CrossRef]
- Cassier-Chauvat, C.; Chauvat, F. Function and regulation of ferredoxins in the cyanobacterium, Synechocystis PCC6803: Recent advances. Life 2014, 4, 666–680. [Google Scholar] [CrossRef]
- Onda, Y.; Matsumura, T.; Kimata-Ariga, Y.; Sakakibara, H.; Sugiyama, T.; Hase, T. Differential interaction of maize root ferredoxin: NADP+ oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 2000, 123, 1037–1046. [Google Scholar] [CrossRef]
- Terauchi, A.M.; Lu, S.-F.; Zaffagnini, M.; Tappa, S.; Hirasawa, M.; Tripathy, J.N.; Knaff, D.B.; Farmer, P.J.; Lemaire, S.D.; Hase, T. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 2009, 284, 25867–25878. [Google Scholar] [CrossRef]
- Ewen, K.M.; Hannemann, F.; Khatri, Y.; Perlova, O.; Kappl, R.; Krug, D.; Hüttermann, J.; Müller, R.; Bernhardt, R. Genome mining in Sorangium cellulosum So ce56: Identification and characterization of the homologous electron transfer proteins of a myxobacterial cytochrome P450. J. Biol. Chem. 2009, 284, 28590–28598. [Google Scholar] [CrossRef]
- Ortega Ugalde, S.; de Koning, C.P.; Wallraven, K.; Bruyneel, B.; Vermeulen, N.P.E.; Grossmann, T.N.; Bitter, W.; Commandeur, J.N.M.; Vos, J.C. Linking cytochrome P450 enzymes from Mycobacterium tuberculosis to their cognate ferredoxin partners. Appl. Microbiol. Biotechnol. 2018, 102, 9231–9242. [Google Scholar] [CrossRef]
- Petitjean, C.; Moreira, D.; López-García, P.; Brochier-Armanet, C. Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol. Biol. 2012, 12, 1–14. [Google Scholar] [CrossRef]
- Mustila, H.; Allahverdiyeva, Y.; Isojärvi, J.; Aro, E.; Eisenhut, M. The bacterial-type [4Fe–4S] ferredoxin 7 has a regulatory function under photooxidative stress conditions in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta BBA-Bioenerg. 2014, 1837, 1293–1304. [Google Scholar] [CrossRef]
- Mortenson, L.; Valentine, R.; Carnahan, J. An electron transport factor from Clostridiumpasteurianum. Biochem. Biophys. Res. Commun. 1962, 7, 448–452. [Google Scholar] [CrossRef]
- Campbell, I.J.; Bennett, G.N.; Silberg, J.J. Evolutionary relationships between low potential ferredoxin and flavodoxin electron carriers. Front. Energy Res. 2019, 7, 79. [Google Scholar] [CrossRef]
- Cai, K.; Tonelli, M.; Frederick, R.O.; Markley, J.L. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron–sulfur cluster biosynthesis. Biochemistry 2017, 56, 487–499. [Google Scholar] [CrossRef]
- Ewen, K.M.; Kleser, M.; Bernhardt, R. Adrenodoxin: The archetype of vertebrate-type [2Fe–2S] cluster ferredoxins. Biochim. Biophys. Acta BBA-Proteins Proteom. 2011, 1814, 111–125. [Google Scholar] [CrossRef]
- Pochapsky, T.C.; Jain, N.U.; Kuti, M.; Lyons, T.A.; Heymont, J. A refined model for the solution structure of oxidized putidaredoxin. Biochemistry 1999, 38, 4681–4690. [Google Scholar] [CrossRef]
- Sawyer, A.; Winkler, M. Evolution of Chlamydomonas reinhardtii ferredoxins and their interactions with [FeFe]-hydrogenases. Photosynth. Res. 2017, 134, 307–316. [Google Scholar] [CrossRef]
- Zhang, W.; Du, L.; Li, F.; Zhang, X.; Qu, Z.; Han, L.; Li, Z.; Sun, J.; Qi, F.; Yao, Q. Mechanistic Insights into Interactions between Bacterial Class I P450 Enzymes and Redox Partners. ACS Catal. 2018, 8, 9992–10003. [Google Scholar] [CrossRef]
- Saridakis, E.; Giastas, P.; Efthymiou, G.; Thoma, V.; Moulis, J.-M.; Kyritsis, P.; Mavridis, I.M. Insight into the protein and solvent contributions to the reduction potentials of [4Fe–4S] 2+/+ clusters: Crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin. JBIC J. Biol. Inorg. Chem. 2009, 14, 783–799. [Google Scholar] [CrossRef]
- Zanetti, G.; Pandini, V. Ferredoxin. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 296–298. [Google Scholar]
- Bertini, I.; Luchinat, C.; Provenzani, A.; Rosato, A.; Vasos, P.R. Browsing gene banks for Fe2S2 ferredoxins and structural modeling of 88 plant-type sequences: An analysis of fold and function. Proteins Struct. Funct. Bioinform. 2002, 46, 110–127. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef]
- Meyer, J. Iron–sulfur protein folds, iron–sulfur chemistry, and evolution. JBIC J. Biol. Inorg. Chem. 2008, 13, 157–170. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, B.; Golbeck, J.H. Understanding of the binding interface between PsaC and the PsaA/PsaB heterodimer in photosystem I. Biochemistry 2009, 48, 5405–5416. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, K.-L.; Yu, L.-K.; Chen, Y.-H.; Cheng, Y.-H.; Hsieh, Y.-C.; Ke, S.-C.; Hung, K.-W.; Chen, C.-J.; Huang, T.-H. FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J. Bacteriol. 2013, 195, 4726–4734. [Google Scholar] [CrossRef] [PubMed]
- Mutter, A.C.; Tyryshkin, A.M.; Campbell, I.J.; Poudel, S.; Bennett, G.N.; Silberg, J.J.; Nanda, V.; Falkowski, P.G. De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. USA 2019, 116, 14557–14562. [Google Scholar] [CrossRef]
- Yasunobu, K.T.; Tanaka, M. The evolution of iron-sulfur protein containing organisms. Syst. Zool. 1973, 22, 570–589. [Google Scholar] [CrossRef]
- Harayama, S.; Polissi, A.; Rekik, M. Divergent evolution of chloroplast-type ferredoxins. FEBS Lett. 1991, 285, 85–88. [Google Scholar] [CrossRef]
- Bruschi, M.; Guerlesquin, F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol. Lett. 1988, 54, 155–175. [Google Scholar] [CrossRef]
- Nixon, J.E.; Wang, A.; Field, J.; Morrison, H.G.; McArthur, A.G.; Sogin, M.L.; Loftus, B.J.; Samuelson, J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell 2002, 1, 181–190. [Google Scholar] [CrossRef]
- Pfeifer, F.; Griffig, J.; Oesterhelt, D. The fdx gene encoding the [2Fe-2S] ferredoxin of Halobacterium salinarium (H. halobium). Mol. Gen. Genet. MGG 1993, 239, 66–71. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Nzuza, N.; Padayachee, T.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species. Int. J. Mol. Sci. 2021, 22, 5542. [Google Scholar] [CrossRef]
- Padayachee, T.; Nzuza, N.; Chen, W.; Nelson, D.R.; Syed, K. Impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10. [Google Scholar] [CrossRef]
- wwPDB Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Klint, J.; Rasmussen, U. The first protein map of Synechococcus sp. strain PCC 7942. Microbiology 2006, 75, 664–672. [Google Scholar] [CrossRef]
- Lau, I.C.; Feyereisen, R.; Nelson, D.R.; Bell, S.G. Analysis and preliminary characterisation of the cytochrome P450 monooxygenases from Frankia sp. EuI1c (Frankia inefficax sp.). Arch. Biochem. Biophys. 2019, 669, 11–21. [Google Scholar] [CrossRef]
- Child, S.A.; Bradley, J.M.; Pukala, T.L.; Svistunenko, D.A.; Le Brun, N.E.; Bell, S.G. Electron transfer ferredoxins with unusual cluster binding motifs support secondary metabolism in many bacteria. Chem. Sci. 2018, 9, 7948–7957. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 2002, 417, 141. [Google Scholar] [CrossRef]
- McLean, K.J.; Warman, A.J.; Seward, H.E.; Marshall, K.R.; Girvan, H.M.; Cheesman, M.R.; Waterman, M.R.; Munro, A.W. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry 2006, 45, 8427–8443. [Google Scholar] [CrossRef]
- Frazão, C.; Aragão, D.; Coelho, R.; Leal, S.S.; Gomes, C.M.; Teixeira, M.; Carrondo, M.A. Crystallographic analysis of the intact metal centres [3Fe–4S] 1+/0 and [4Fe–4S] 2+/1+ in a Zn2+-containing ferredoxin. FEBS Lett. 2008, 582, 763–767. [Google Scholar] [CrossRef]
- Macedo-Ribeiro, S.; Martins, B.M.; Pereira, P.B.; Buse, G.; Huber, R.; Soulimane, T. New insights into the thermostability of bacterial ferredoxins: High-resolution crystal structure of the seven-iron ferredoxin from Thermus thermophilus. JBIC J. Biol. Inorg. Chem. 2001, 6, 663–674. [Google Scholar] [CrossRef]
- Dauter, Z.; Wilson, K.S.; Sieker, L.C.; Meyer, J.; Moulis, J.-M. Atomic resolution (0.94 Å) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein. Biochemistry 1997, 36, 16065–16073. [Google Scholar] [CrossRef]
- Saeki, K.; Suetsugu, Y.; Tokuda, K.-I.; Miyatake, Y.; Young, D.; Marrs, B.; Matsubara, H. Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J. Biol. Chem. 1991, 266, 12889–12895. [Google Scholar] [CrossRef]
- Moulis, J.M.; Sieker, L.C.; Wilson, K.S.; Dauter, Z. Crystal structure of the 2 [4Fe-4S] ferredoxin from Chromatium vinosum: Evolutionary and mechanistic inferences for [3/4Fe-4S] ferredoxins. Protein Sci. 1996, 5, 1765–1775. [Google Scholar] [CrossRef][Green Version]
- Unciuleac, M.; Boll, M.; Warkentin, E.; Ermler, U. Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 388–391. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Khumalo, M.J.; Nzuza, N.; Padayachee, T.; Chen, W.; Yu, J.H.; Nelson, D.R.; Syed, K. Comprehensive Analyses of Cytochrome P450 Monooxygenases and Secondary Metabolite Biosynthetic Gene Clusters in Cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Kryś, J.D.; Gront, D. VisuaLife: Library for interactive visualization in rich web applications. Bioinformatics 2021. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef]
- Hosseinzadeh, P.; Lu, Y. Design and fine-tuning redox potentials of metalloproteins involved in electron transfer in bioenergetics. Biochim Biophys. Acta BBA-Bioenerg. 2016, 1857, 557–581. [Google Scholar] [CrossRef]
- Kim, J.Y.; Nakayama, M.; Toyota, H.; Kurisu, G.; Hase, T. Structural and mutational studies of an electron transfer complex of maize sulfite reductase and ferredoxin. J. Biochem. 2016, 160, 101–109. [Google Scholar] [CrossRef]

| 2Fe-2S. | ||
|---|---|---|
| GenBank Accession Number or PDB Code | Species Name | Reference |
| NP_004100.1 (Adrenodoxin) | Homo sapiens | [22] |
| 1PDX (Putidaredoxin) | Pseudomonas putida | [24] |
| ABB56370.1 | Synechococcus elongatus PCC 7942 = FACHB-805 | [50] |
| ABB56928.1 | Synechococcus elongatus PCC 7942 = FACHB-805 | [50] |
| WP_013424358.1 (FraEuI1c_3227) | Frankia sp. EuI1c (Frankia inefficax sp.) | [51] |
| WP_012394830.1 (mmi:MMAR_3155) | Mycobacterium marinum | [52] |
| 3Fe-4S/4Fe-4S | ||
| CAB59502.1 | Streptomyces coelicolor A3(2) | [53] |
| WP_013425251.1 (FraEuI1c_4132) | Frankia sp. EuI1c (Frankia inefficax sp.) | [51] |
| WP_013426476.1 (FraEuI1c_5370) | Frankia sp. EuI1c (Frankia inefficax sp.) | [51] |
| WP_011740769.1 (Mmar_2879) | Mycobacterium marinum | [52] |
| WP_012395565.1 (Mmar_3973) | Mycobacterium marinum | [52] |
| WP_012396301.1 (Mmar_4763) | Mycobacterium marinum | [52] |
| NP_215277.1 (FdX-Rv0763c) | Mycobacterium tuberculosis H37Rv | [17,54] |
| NP_216302.1 (FdxE-Rv1786) | Mycobacterium tuberculosis H37Rv | [17,54] |
| ABB57779.1 | Synechococcus elongatus PCC 7942 = FACHB-805 | [50] |
| 7Fe-8S | ||
| NP_215693 (FdxC-Rv1177), | Mycobacterium tuberculosis H37Rv | [17] |
| NP_216523.1 (FdxA-Rv2007c) | Mycobacterium tuberculosis H37Rv | [17] |
| 2VKR | Acidianus ambivalens | [55] |
| 1H98 | Thermus thermophilus | [56] |
| ABB56846.1 | Synechococcus elongatus PCC 7942 = FACHB-805 | [50] |
| 2[4Fe-4S] | ||
| 2ZVS | Escherichia coli K-12 | [27] |
| 2FDN | Clostridium acidurici | [57] |
| WP_013068980.1 (FDI) | Rhodobacter capsulatus | [58] |
| 2[4Fe-4S]Alv | ||
| 1BLU | Allochromatium vinosum | [59] |
| 2FGO | Pseudomonas aeruginosa | [57] |
| WP_023923722.1 (FDIII) | Rhodobacter capsulatus | [58] |
| 1RGV | Thauera aromatica K172 | [60] |
| Subtypes | Cysteine Spacing Signature | No. AA | No of Ferredoxins | ||||
|---|---|---|---|---|---|---|---|
| Alphaproteobacteria | Firmicutes | From Literature | |||||
| Archaea | Bacteria | Eukarya | |||||
| 2Fe-2S | |||||||
| Subtype 1 | CX5CX2CX36C | 47 | 296 | 2 | 64 | ||
| Subtype 2 | CX5CX2CX37C | 48 | 81 | 1 | 4 | 22 | |
| Subtype 3 | CX4CX2CX29C | 39 | 11 | 1 | 7 | 11 | 55 |
| Subtype 4 | CX5CX2CX35C | 46 | 12 | 3 | 2 | ||
| Subtype 5 | CX5CX2CX38C | 49 | 12 | 1 | |||
| Subtype 6 | CX4CX2CX34C | 44 | 11 | 11 | 7 | 1 | |
| Subtype 7 | CX4CX2CX51C | 61 | 8 | 1 | |||
| Subtype 8 | CX4CX2CX31C | 41 | 7 | 6 | 1 | 1 | |
| Subtype 9 | CX4CX2CX33C | 43 | 2 | 3 | 19 | 2 | |
| Subtype 10 | CX5CX2CX39C | 50 | 2 | 1 | |||
| Subtype 11 | CX4CX2CX35C | 46 | 1 | ||||
| Subtype 12 | CX4CX2CX25C | 35 | 1 | ||||
| Subtype 13 | CX7CX34CX3C | 48 | 1 | ||||
| Subtype 14 | CX4CX29C | 36 | 1 | ||||
| Subtype 15 | CX7CX29C | 39 | 1 | ||||
| Subtype 16 | CX7CX35C | 45 | 1 | ||||
| Subtype 17 | CX5CX2CX33C | 44 | 1 | ||||
| Subtype 18 | CX5CX2CX34C | 45 | 2 | 20 | 2 | 1 | 1 |
| Subtype 19 | CX5CX2CX35C | 46 | 1 | ||||
| Subtype 20 | CX5CX2CX32C | 43 | 44 | 1 | 5 | ||
| Subtype 21 | CX4CX31CX3C | 42 | 5 | ||||
| Subtype 22 | CX2CX41CX3C | 50 | 2 | ||||
| Subtype 23 | CX7CX38CX3C | 52 | 1 | ||||
| Subtype 24 | CX4CX2CX30C | 40 | 70 | 1 | |||
| Subtype 25 | CX5CX2CX30C | 41 | 1 | ||||
| Subtype 26 | CX12CX30CX3C | 49 | 21 | ||||
| Subtype 27 | CX12CX31CX3C | 50 | 4 | ||||
| Subtype 28 | CX8CX44CX3C | 59 | 3 | ||||
| Subtype 29 | CX8CX33CX3C | 48 | 3 | ||||
| Subtype 30 | CX8CX32CX3C | 47 | 2 | ||||
| Subtype 31 | CX4CX36CX3C | 47 | 1 | 1 | |||
| Subtype 32 | CX8CX38CX3C | 53 | 1 | ||||
| Subtype 33 | CX8CX39CX3C | 54 | 1 | ||||
| Subtype 34 | CX9CX33CX3C | 49 | 1 | ||||
| Subtype 35 | CX12CX33CX3C | 52 | 1 | ||||
| Subtype 36 | CX3CX1CX38C | 46 | 1 | ||||
| Subtype 37 | CX12CX32CX3C | 51 | 1 | ||||
| Subtype 38 | CX4CX2CX28C | 38 | 40 | 3 | |||
| Subtype 39 | CX4CX2CX46C | 56 | 2 | ||||
| Subtype 40 | CX4CX2CX49C | 59 | 3 | ||||
| Subtype 41 | CX4CX2CX45C | 55 | 2 | ||||
| Subtype 42 | CX4CX2CX65C | 75 | 1 | ||||
| Subtype 43 | CX4CX2CX50C | 60 | 2 | ||||
| Subtype 44 | CX4CX2CX47C | 57 | 2 | ||||
| Subtype 45 | CX4CX2CX48C | 58 | 1 | ||||
| Subtype 46 | CX5CX2CX52C | 63 | 2 | ||||
| Subtype 47 | CX5CX2CX31C | 42 | 1 | ||||
| Subtype 48 | CX5CX2CX28C | 39 | 2 | ||||
| Subtype 49 | CX5CX2CX27C | 38 | 10 | ||||
| Subtype 50 | CX5CX2CX82C | 93 | 2 | ||||
| Subtype 51 | CX5CX2CX29C | 40 | 1 | ||||
| Subtype 52 | CX4CX2CX32C | 42 | 1 | ||||
| Subtype 53 | CX5CX2CX42C | 53 | 1 | ||||
| Subtype 54 | CX4CX2CX22C | 32 | 2 | ||||
| Subtype 55 | CX4CX2CX29C | 39 | 2 | ||||
| 3Fe-4S | |||||||
| Subtype 1 | CX5CX38CP | 47 | 26 | 2 | 7 | ||
| Subtype 2 | CX5CX37CP | 46 | 16 | 13 | |||
| Subtype 3 | CX5CX36CP | 45 | 14 | 2 | |||
| Subtype 4 | CX5CX40CP | 49 | 3 | ||||
| Subtype 5 | CX5CX36CP | 45 | 1 | ||||
| Subtype 6 | CX5CX35CP | 44 | 5 | ||||
| Subtype 7 | CX5CX49CP | 58 | 2 | ||||
| 4Fe-4S | |||||||
| Subtype 1 | CX5CX3CX33CP | 46 | 2 | ||||
| Subtype 2 | CX2CX2CX43CP | 52 | 107 | ||||
| Subtype 3 | CX2CX2CX45CP | 54 | 24 | 1 | |||
| Subtype 4 | CX2CX2CX37CP | 46 | 6 | ||||
| Subtype 5 | CX2CX2CX44CP | 53 | 2 | ||||
| Subtype 6 | CX2CX2CX39CP | 48 | 1 | ||||
| Subtype 7 | CX2CX2CX36CP | 45 | 2 | ||||
| Subtype 8 | CX2CX2CX34CP | 43 | 1 | ||||
| Subtype 9 | CX2CX2CX38CP | 47 | 1 | 1 | |||
| Subtype 10 | CX5CX3CX32CP | 45 | 1 | ||||
| Subtype 11 | CX5CX3CX30CP | 43 | 12 | ||||
| Subtype 12 | CX5CX3CX31CP | 44 | 2 | ||||
| 7Fe-8S | |||||||
| Subtype 1 | CX7CX3CPX17CX2CX2CX3CP * | 43 | 6 | 32 | 13 | ||
| Subtype 2 | CX5CX3CPX40CX2CX2CX3CP | 64 | 4 | ||||
| Subtype 3 | CX5CX3CPX20CX2CX2CX3CP | 44 | 1 | ||||
| Subtype 4 | CX10CX3CPX22CX2CX2CX3CP | 51 | 1 | ||||
| Subtype 5 | CX5CX3CPX26CX2CX2CX3CP | 50 | 1 | ||||
| Subtype 6 | CX5CX3CPX24CX2CX2CX3CP | 48 | 1 | 1 | |||
| Subtype 7 | CX10CX3CPX17CX2CX2CX3CP | 46 | 1 | ||||
| Subtype 8 | CX5CX3CPX22CX2CX2CX3CP | 46 | 9 | ||||
| Subtype 9 | CX5CX3CPX18CX2CX2CX3C | 42 | 1 | ||||
| Subtype 10 | CX3CX3CPX22CX2CX2CX3CP | 44 | 2 | ||||
| 2[4Fe-4S] | |||||||
| Subtype 1 | CX2CX4CX3CX18CX2CX2CX3C | 42 | 267 | ||||
| Subtype 2 | CX2CX2CX3CX18CX2CX8CX3C | 46 | 90 | 4 | |||
| Subtype 3 | CX2CX2CX3CX20CX2CX2CX3C | 42 | 33 | 2 | 52 | 1 | |
| Subtype 4 | CX7CX2CX3CX23CX2CX2CX3C | 50 | 5 | ||||
| Subtype 5 | CX2CX2CX3CX42CX2CX2CX3C | 64 | 3 | ||||
| Subtype 6 | CX2CX2CX3CX18CX2CX7CX3C | 45 | 2 | ||||
| Subtype 7 | CX2CX2CX3CX18CX2CX6CX3C | 44 | 2 | ||||
| Subtype 8 | CX2CX2CX3CX24CX2CX2CX3C | 46 | 2 | 2 | |||
| Subtype 9 | CX2CX2CX3CX18CX2CX2CX3C | 40 | 1 | 6 | 78 | 3 | 1 |
| Subtype 10 | CX2CX2CX3CX21CX2CX2CX3C | 43 | 6 | 2 | |||
| Subtype 11 | CX2CX2CX3CX18CX3CX2CX3C | 42 | 2 | ||||
| Subtype 12 | CX2CX2CX3CX28CX2CX2CX3C | 50 | 132 | 1 | 2 | ||
| Subtype 13 | CX2CX2CX3CX27CX2CX2CX3C | 49 | 2 | ||||
| Subtype 14 | CX5CX2CX3CX20CX2CX2CX3C | 45 | 21 | ||||
| Subtype 15 | CX2CX2CX3CX19CX2CX2CX3C | 41 | 1 | 4 | 58 | ||
| Subtype 16 | CX2CX2CX3CX40CX2CX2CX3C | 50 | 1 | ||||
| Subtype 17 | CX2CX2CX3CX29CX2CX2CX3C | 51 | 144 | 2 | |||
| Subtype18 | CX4CX2CX3CX18CX2CX2CX3C | 42 | 1 | 1 | |||
| Subtype 19 | CX3CX2CX3CX20CX2CX2CX3C | 43 | 2 | ||||
| Subtype 20 | CX2CX2CX3CX17CX2CX2CX3C | 39 | 24 | 1 | |||
| Subtype 21 | CX3CX3CX3CX37CX1CX3CX3C | 61 | 1 | ||||
| Subtype 22 | CX2CX2CX3CX26CX2CX2CX3C | 48 | 6 | ||||
| Subtype 23 | CX2CX2CX3CX30CX2CX2CX3C | 52 | 1 | ||||
| Subtype 24 | CX2CX2CX3CX33CX2CX2CX3C | 55 | 22 | ||||
| Subtype 25 | CX2CX2CX3CX32CX2CX2CX3C | 54 | 23 | ||||
| Subtype 26 | CX2CX2CX3CX23CX2CX2CX3C | 45 | 2 | ||||
| Subtype 27 | CX2CX2CX3CX34CX2CX2CX3C | 56 | 1 | ||||
| Subtype 28 | CX2CX2CX3CX14CX2CX2CX3C | 36 | 2 | ||||
| Subtype 29 | CX2CX2CX3CX22CX2CX2CX3C | 44 | 2 | ||||
| Subtype 30 | CX2CX2CX2CX38CX2CX2CX3C | 59 | 1 | ||||
| Subtype 31 | CX4CX2CX3CX19CX2CX2CX3C | 43 | 24 | ||||
| Subtype 32 | CX5CX2CX3CX19CX2CX2CX3C | 44 | 3 | ||||
| Subtype 33 | CX2CX2CX3CX16CX2CX2CX3C | 38 | 16 | ||||
| 2[4Fe-4S]Alv | |||||||
| Subtype 1 | CX2CX2CX3CX18CX2CX8CX3CX3C | 50 | 10 | 5 | |||
| Subtype 2 | CX2CX2CX3CX39CX2CX2CX3CX3C | 65 | 9 | ||||
| Subtype 3 | CX2CX2CX3CX43CX2CX2CX3CX3C | 69 | 5 | 1 | |||
| Subtype 4 | CX2CX2CX3CX42CX2CX2CX3CX3C | 68 | 1 | ||||
| Subtype 5 | CX2CX2CX3CX40CX2CX2CX3CX3C | 66 | 2 | ||||
| Subtype 6 | CX2CX2CX3CX38CX2CX2CX3CX3C | 64 | 1 | ||||
| Subtype 7 | CX2CX2CX3CX46CX2CX2CX3CX3C | 72 | 2 | ||||
| Subtype 8 | CX2CX2CX3CX44CX2CX2CX3CX3C | 70 | 1 | ||||
| Subtype 9 | CX2CX2CX3CX30CX2CX2CX3CX3C | 56 | 2 | ||||
| Subtype 10 | CX2CX2CX3CX19CX2CX2CX3CX3C | 45 | 8 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nzuza, N.; Padayachee, T.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Diversification of Ferredoxins across Living Organisms. Curr. Issues Mol. Biol. 2021, 43, 1374-1390. https://doi.org/10.3390/cimb43030098
Nzuza N, Padayachee T, Chen W, Gront D, Nelson DR, Syed K. Diversification of Ferredoxins across Living Organisms. Current Issues in Molecular Biology. 2021; 43(3):1374-1390. https://doi.org/10.3390/cimb43030098
Chicago/Turabian StyleNzuza, Nomfundo, Tiara Padayachee, Wanping Chen, Dominik Gront, David R. Nelson, and Khajamohiddin Syed. 2021. "Diversification of Ferredoxins across Living Organisms" Current Issues in Molecular Biology 43, no. 3: 1374-1390. https://doi.org/10.3390/cimb43030098
APA StyleNzuza, N., Padayachee, T., Chen, W., Gront, D., Nelson, D. R., & Syed, K. (2021). Diversification of Ferredoxins across Living Organisms. Current Issues in Molecular Biology, 43(3), 1374-1390. https://doi.org/10.3390/cimb43030098