Growth Performance, Biochemical Composition and Nutrient Recovery Ability of Twelve Microalgae Consortia Isolated from Various Local Organic Wastes Grown on Nano-Filtered Pig Slurry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Nano-Filtered Permeate (NFP)
2.2. Organic Waste Sources of Microalgae Consortia
2.3. Microalgae Consortia Growth
2.4. Nutrient Mass Balance
2.5. Biochemical Composition of AC Biomasses
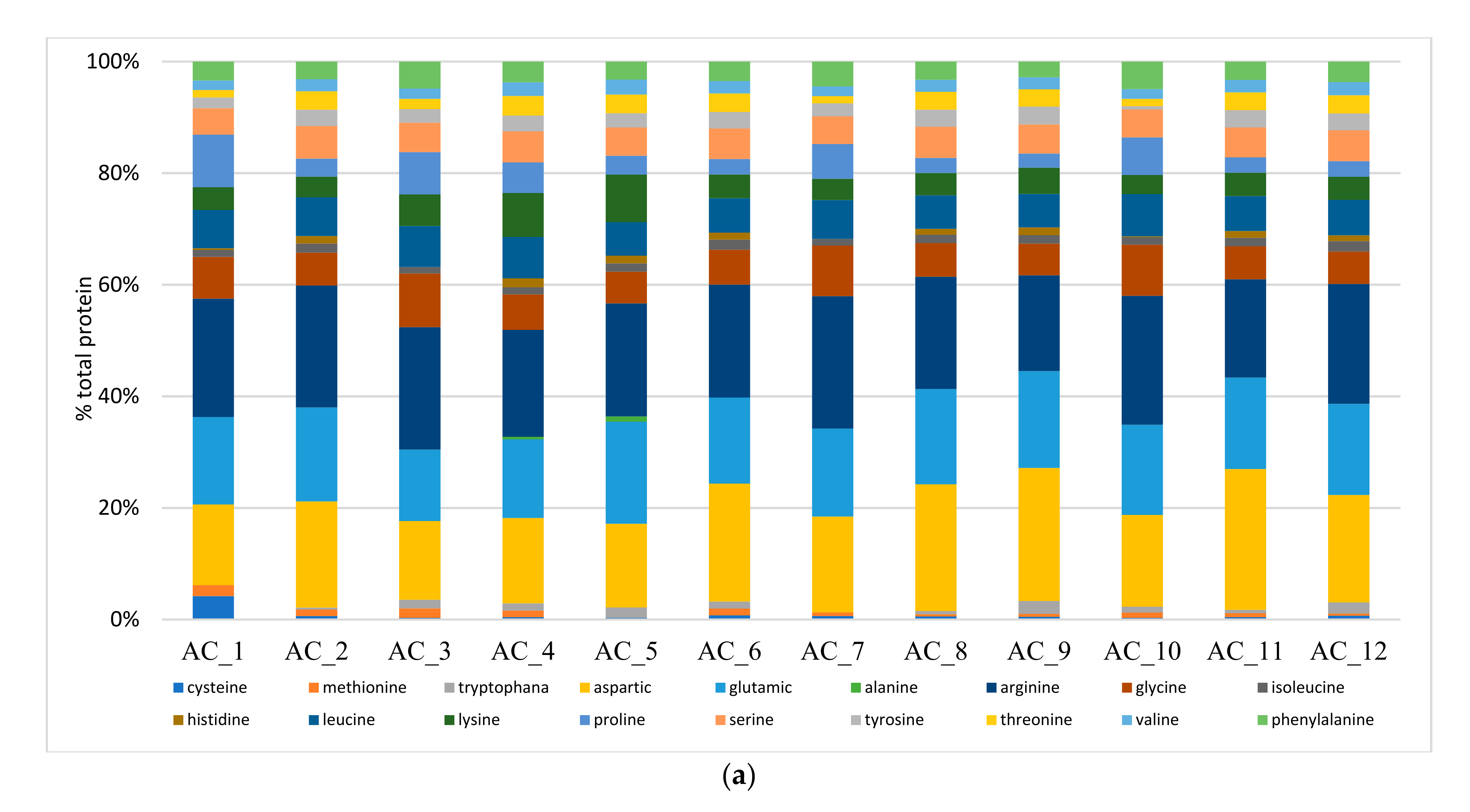
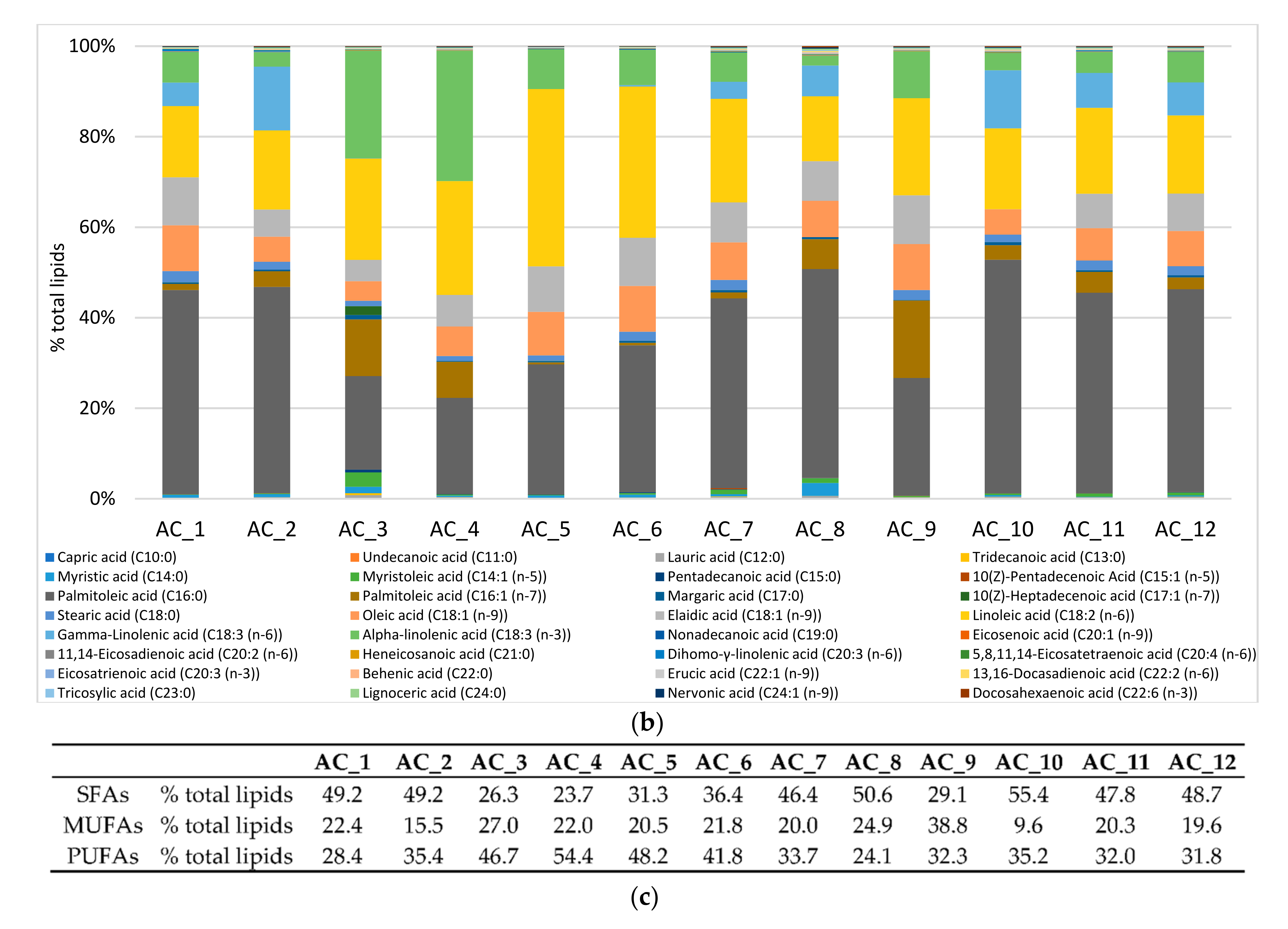
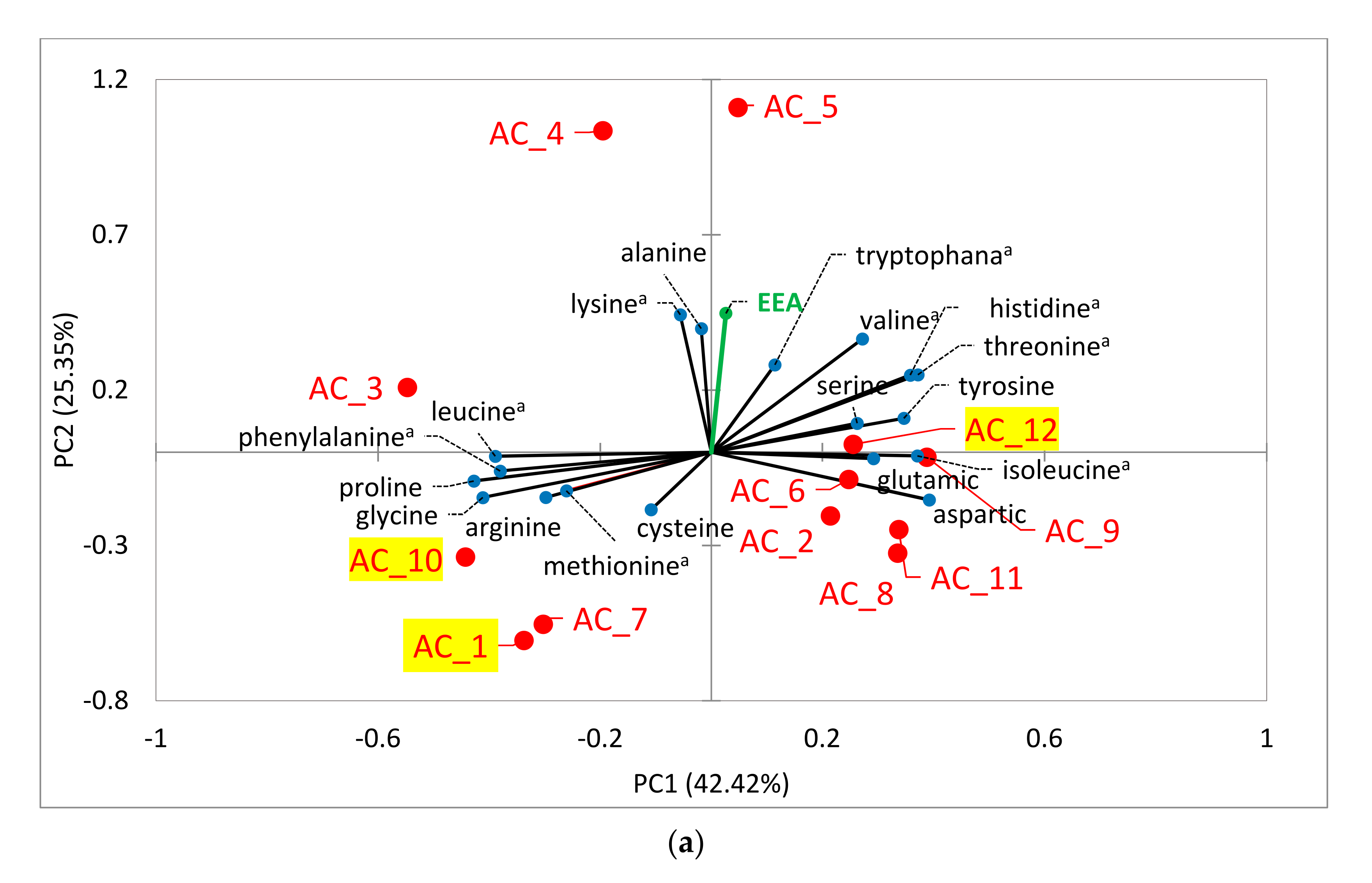
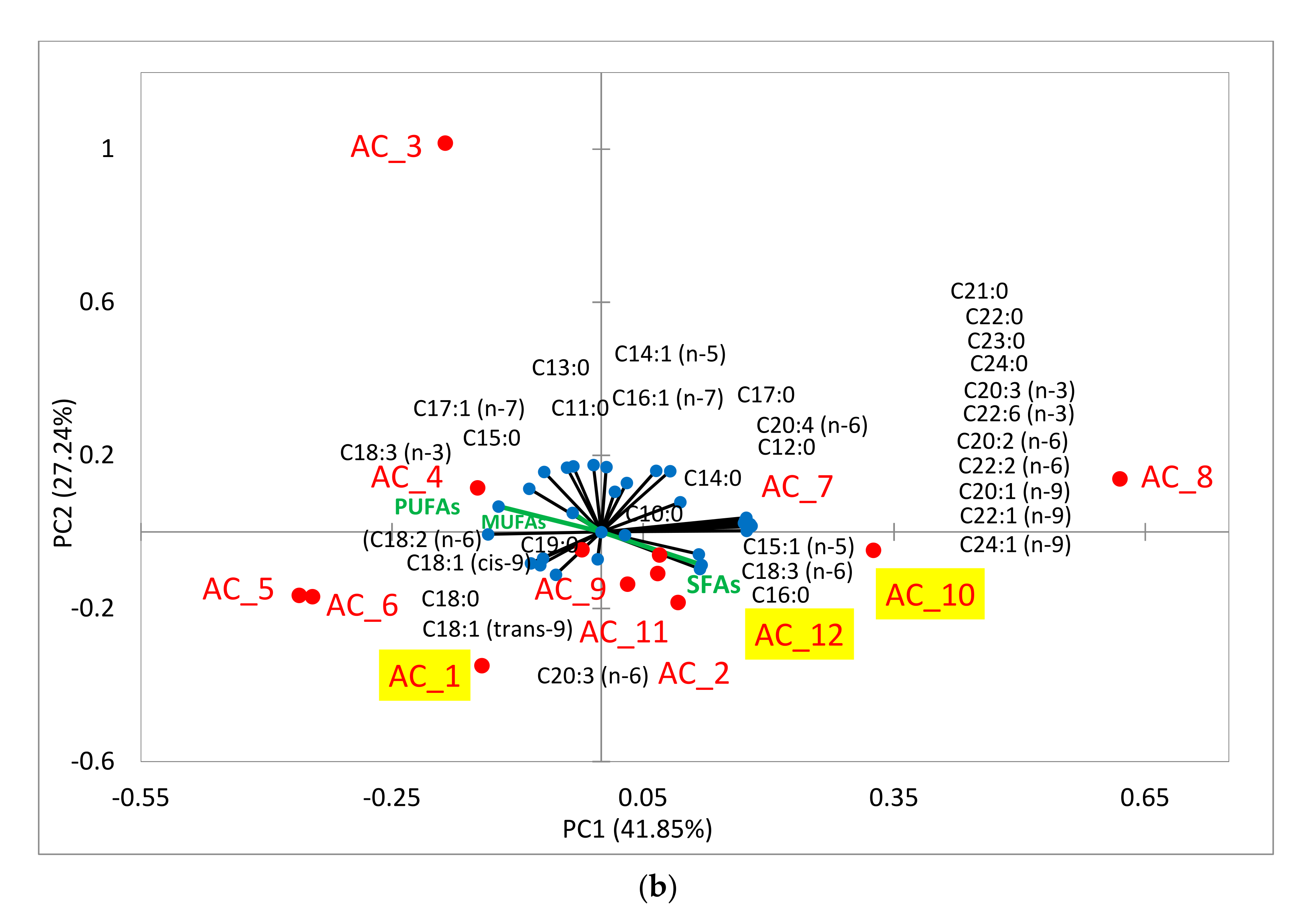
2.6. Amino Acids (AA) and Fatty Acids (FA) Speciation
2.6.1. Amino Acids (AA)
2.6.2. Fatty Acids (FA)
2.7. Economic Benefits
3. Materials and Methods
3.1. Nanofiltered Permeate Sampling and Characterization
3.2. Microalgae Consortia and Preparation of Inoculum
3.3. Microalgae Consortia Molecular Characterization
3.4. Experimental Set-Up and Cultivation
3.5. Microalgae Growth Determination
3.6. Biochemical Analysis
3.7. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Stiles, W.A.V.; Styles, D.; Chapman, S.P.; Esteves, S.; Bywater, A.; Melville, L.; Silkina, A.; Lupatsch, I.; Fuentes Grünewald, C.; Lovitt, R.; et al. Using microalgae in the circular economy to valorise anaerobic digestate: Challenges and opportunities. Bioresour. Technol. 2018, 267, 732–742. [Google Scholar] [CrossRef] [Green Version]
- Acién Fernández, F.G.; Gómez-Serrano, C.; Fernández-Sevilla, J.M. Recovery of nutrients from wastewaters using microalgae. Front. Sustain. Food Syst. 2018, 2, 59. [Google Scholar] [CrossRef] [Green Version]
- Olguín, E.J. Dual purpose microalgae-bacteria-based systems that treat wastewater and produce biodiesel and chemical products within a Biorefinery. Biotechnol. Adv. 2012, 30, 1031–1046. [Google Scholar] [CrossRef]
- Batista, A.P.; Gouveia, L.; Bandarra, N.M.; Franco, J.M.; Raymundo, A. Comparison of microalgal biomass profiles as novel functional ingredients for food products. Algal Res. 2013, 2, 164–173. [Google Scholar] [CrossRef] [Green Version]
- Kerckhof, F.M.; Courtens, E.N.P.; Geirnaert, A.; Hoefman, S.; Ho, A.; Vilchez-Vargas, R.; Pieper, D.H.; Jauregui, R.; Vlaeminck, S.E.; Van De Wiele, T.; et al. Optimized cryopreservation of mixed microbial communities for conserved functionality and diversity. PLoS ONE 2014, 9, e99517. [Google Scholar] [CrossRef]
- Kumar, A.; Bera, S. Revisiting nitrogen utilization in algae: A review on the process of regulation and assimilation. Bioresour. Technol. Rep. 2020, 12, 100584. [Google Scholar] [CrossRef]
- Collos, Y.; Harrison, P.J. Acclimation and toxicity of high ammonium concentrations to unicellular algae. Mar. Pollut. Bull. 2014, 80, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Dell’Orto, M.; D’Imporzano, G.; Bani, A.; Dumbrell, A.J.; Adani, F. The structure and diversity of microalgae-microbial consortia isolated from various local organic wastes. Bioresour. Technol. 2021, 126416. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.; Kwan, T.A.; Zimmerman, J.B.; Peccia, J. Ammonia inhibition in oleaginous microalgae. Algal Res. 2016, 19, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Rossi, S.; Díez-Montero, R.; Rueda, E.; Castillo Cascino, F.; Parati, K.; García, J.; Ficara, E. Free ammonia inhibition in microalgae and cyanobacteria grown in wastewaters: Photo-respirometric evaluation and modelling. Bioresour. Technol. 2020, 305, 123046. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Zhai, J.; Wei, H.; Wang, Q. Effect of ammonium nitrogen on microalgal growth, biochemical composition and photosynthetic performance in mixotrophic cultivation. Bioresour. Technol. 2019, 273, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, W.; Chen, H.; Zhan, J.; He, C.; Wang, Q. Ammonium nitrogen tolerant Chlorella strain screening and its damaging effects on photosynthesis. Front. Microbiol. 2019, 9, 3250. [Google Scholar] [CrossRef]
- Markou, G.; Muylaert, K. Effect of light intensity on the degree of ammonia toxicity on PSII activity of Arthrospira platensis and Chlorella vulgaris. Bioresour. Technol. 2016, 216, 453–461. [Google Scholar] [CrossRef]
- Chuka-ogwude, D.; Ogbonna, J.; Borowitzka, M.A.; Moheimani, N.R. Screening, acclimation and ammonia tolerance of microalgae grown in food waste digestate. J. Appl. Phycol. 2020, 32, 3775–3785. [Google Scholar] [CrossRef]
- Arbib, Z.; Ruiz, J.; Álvarez-Díaz, P.; Garrido-Pérez, C.; Perales, J.A. Capability of different microalgae species for phytoremediation processes: Wastewater tertiary treatment, CO2 bio-fixation and low cost biofuels production. Water Res. 2014, 49, 465–474. [Google Scholar] [CrossRef]
- Ledda, C.; Idà, A.; Allemand, D.; Mariani, P.; Adani, F. Production of wild Chlorella sp. cultivated in digested and membrane-pretreated swine manure derived from a full-scale operation plant. Algal Res. 2015, 12, 68–73. [Google Scholar] [CrossRef]
- Liu, J.; Huang, J.; Sun, Z.; Zhong, Y.; Jiang, Y.; Chen, F. Differential lipid and fatty acid profiles of photoautotrophic and heterotrophic Chlorella zofingiensis: Assessment of algal oils for biodiesel production. Bioresour. Technol. 2011, 102, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Martinez, A.; Martin Garcia, N.; Romero, I.; Seco, A.; Ferrer, J. Microalgae cultivation in wastewater: Nutrient removal from anaerobic membrane bioreactor effluent. Bioresour. Technol. 2012, 126, 247–253. [Google Scholar] [CrossRef]
- Tam, N.F.Y.; Wong, Y.S. Effect of ammonia concentrations on growth of Chlorella vulgaris and nitrogen removal from media. Bioresour. Technol. 1996, 57, 45–50. [Google Scholar] [CrossRef]
- Tejido-Nuñez, Y.; Aymerich, E.; Sancho, L.; Refardt, D. Treatment of aquaculture effluent with Chlorella vulgaris and Tetradesmus obliquus: The effect of pretreatment on microalgae growth and nutrient removal efficiency. Ecol. Eng. 2019, 136, 1–9. [Google Scholar] [CrossRef]
- Gonçalves, V.D.; Fagundes-Klen, M.R.; Trigueros, D.E.G.; Schuelter, A.R.; Kroumov, A.D.; Módenes, A.N. Combination of Light Emitting Diodes (LEDs) for photostimulation of carotenoids and chlorophylls synthesis in Tetradesmus sp. Algal Res. 2019, 43, 101649. [Google Scholar] [CrossRef]
- Massa, M.; Buono, S.; Langellotti, A.L.; Castaldo, L.; Martello, A.; Paduano, A.; Sacchi, R.; Fogliano, V. Evaluation of anaerobic digestates from different feedstocks as growth media for Tetradesmus obliquus, Botryococcus braunii, Phaeodactylum tricornutum and Arthrospira maxima. New Biotechnol. 2017, 36, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Nadzir, S.M.; Yusof, N.; Nordin, N.; Abdullah, H.; Kamari, A. Combination effect of temperature and light intensity on lipid productivity of Tetradesmus obliquus. J. Phys. Conf. Ser. 2018, 1097, 012038. [Google Scholar] [CrossRef]
- Rajabi Islami, H.; Assareh, R. Effect of different iron concentrations on growth, lipid accumulation, and fatty acid profile for biodiesel production from Tetradesmus obliquus. J. Appl. Phycol. 2019, 31, 3421–3432. [Google Scholar] [CrossRef]
- Ray, A.; Nayak, M.; Ghosh, A. A review on co-culturing of microalgae: A greener strategy towards sustainable biofuels production. Sci. Total Environ. 2022, 802, 149765. [Google Scholar] [CrossRef] [PubMed]
- Pachacama, L.; Tirado, J.O.; Duchicela, J.; Manjunatha, B.; Kundapur, R.R.; Rajeswari, B. Evaluation of microalgae’s (Chlorella sp. and Synechocystis sp.) pollutant removal property: Pig effluent as a live stock discharge. J. Appl. Pharm. Sci. 2016, 6, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Lam, T.P.; Lee, T.M.; Chen, C.Y.; Chang, J.S. Strategies to control biological contaminants during microalgal cultivation in open ponds. Bioresour. Technol. 2018, 252, 180–187. [Google Scholar] [CrossRef]
- Delgadillo-Mirquez, L.; Lopes, F.; Taidi, B.; Pareau, D. Nitrogen and phosphate removal from wastewater with a mixed microalgae and bacteria culture. Biotechnol. Rep. 2016, 11, 18–26. [Google Scholar] [CrossRef]
- Prathima Devi, M.; Venkata Subhash, G.; Venkata Mohan, S. Heterotrophic cultivation of mixed microalgae for lipid accumulation and wastewater treatment during sequential growth and starvation phases: Effect of nutrient supplementation. Renew. Energy 2012, 43, 276–283. [Google Scholar] [CrossRef]
- Su, Y.; Mennerich, A.; Urban, B. Municipal wastewater treatment and biomass accumulation with a wastewater-born and settleable algal-bacterial culture. Water Res. 2011, 45, 3351–3358. [Google Scholar] [CrossRef]
- Markou, G. Fed-batch cultivation of Arthrospira and Chlorella in ammonia-rich wastewater: Optimization of nutrient removal and biomass production. Bioresour. Technol. 2015, 193, 35–41. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, Q.; Wang, L. Comparative metabolomic analysis of the green microalga Chlorella sorokiniana cultivated in the single culture and a consortium with bacteria for wastewater remediation. Appl. Biochem. Biotechnol. 2017, 183, 1062–1075. [Google Scholar] [CrossRef]
- del Mar Morales-Amaral, M.; Gómez-Serrano, C.; Acién, F.G.; Fernández-Sevilla, J.M.; Molina-Grima, E. Production of microalgae using centrate from anaerobic digestion as the nutrient source. Algal Res. 2015, 9, 297–305. [Google Scholar] [CrossRef]
- Vonshak, A. Spirulina Platensis Arthrospira: Physiology, Cell-Biology and Biotechnology; CRC Press: London, UK, 1997; pp. 43–65. ISBN 0-7484-0674-3. [Google Scholar]
- Silva-Benavides, A.M.; Torzillo, G. Nitrogen and phosphorus removal through laboratory batch cultures of microalga Chlorella vulgaris and cyanobacterium Planktothrix isothrix grown as monoalgal and as co-cultures. J. Appl. Phycol. 2012, 24, 267–276. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Chen, P.; Min, M.; Chen, Y.; Zhu, J.; Ruan, R.R. Anaerobic digested dairy manure as a nutrient supplement for cultivation of oil-rich green microalgae Chlorella sp. Bioresour. Technol. 2010, 101, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Safi, C.; Charton, M.; Pignolet, O.; Silvestre, F.; Vaca-Garcia, C.; Pontalier, P.Y. Influence of microalgae cell wall characteristics on protein extractability and determination of nitrogen-to-protein conversion factors. J. Appl. Phycol. 2013, 25, 523–529. [Google Scholar] [CrossRef] [Green Version]
- Ansari, F.A.; Singh, P.; Guldhe, A.; Bux, F. Microalgal cultivation using aquaculture wastewater: Integrated biomass generation and nutrient remediation. Algal Res. 2017, 21, 169–177. [Google Scholar] [CrossRef]
- Ajala, S.O.; Alexander, M.L. Assessment of Chlorella vulgaris, Scenedesmus obliquus, and Oocystis minuta for removal of sulfate, nitrate, and phosphate in wastewater. Int. J. Energy Environ. Eng. 2020, 11, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Michelon, W.; Da Silva, M.L.B.; Mezzari, M.P.; Pirolli, M.; Prandini, J.M.; Soares, H.M. Effects of nitrogen and phosphorus on biochemical composition of microalgae polyculture harvested from phycoremediation of piggery wastewater digestate. Appl. Biochem. Biotechnol. 2016, 178, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Nordin, N.; Samsudin, S.; Yusof, N. Isolation and identification of microalgae from high nitrate landfill leachate. J. Teknol. 2019, 81, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Ji, Y.; Hu, W.; Pei, H.; Nie, C.; Ma, G.; Song, M. Adjusting irradiance to enhance growth and lipid production of Chlorella vulgaris cultivated with monosodium glutamate wastewater. J. Photochem. Photobiol. B Biol. 2016, 162, 619–624. [Google Scholar] [CrossRef]
- Buono, S.; Colucci, A.; Angelini, A.; Langellotti, A.L.; Massa, M.; Martello, A.; Fogliano, V.; Dibenedetto, A. Productivity and biochemical composition of Tetradesmus obliquus and Phaeodactylum tricornutum: Effects of different cultivation approaches. J. Appl. Phycol. 2016, 28, 3179–3192. [Google Scholar] [CrossRef]
- Gu, Z.; Liu, Y.; Zou, G.; Zhang, Q.; Lu, R.; Yan, H.; Cao, L.; Liu, T.; Ruan, R. Enhancement of nutrients removal and biomass accumulation of Chlorella vulgaris in pig manure anaerobic digestate effluent by the pretreatment of indigenous bacteria. Bioresour. Technol. 2021, 328, 124846. [Google Scholar] [CrossRef]
- Zhu, S.; Feng, S.; Xu, Z.; Qin, L.; Shang, C.; Feng, P.; Wang, Z.; Yuan, Z. Cultivation of Chlorella vulgaris on unsterilized dairy-derived liquid digestate for simultaneous biofuels feedstock production and pollutant removal. Bioresour. Technol. 2019, 285, 121353. [Google Scholar] [CrossRef]
- Breuer, G.; Lamers, P.P.; Martens, D.E.; Draaisma, R.B.; Wijffels, R.H. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 2012, 124, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Li, Y.; El-Dalatony, M.M.; Zhang, C.; Li, X.; Salama, E.S. A complete characterization of microalgal biomass through FTIR/TGA/CHNS analysis: An approach for biofuel generation and nutrients removal. Renew. Energy. 2021, 163, 1973–1982. [Google Scholar] [CrossRef]
- Becker, E.W. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Lage, S.; Gojkovic, Z.; Funk, C.; Gentili, F.G. Algal biomass from wastewater and flue gases as a source of bioenergy. Energies 2018, 11, 664. [Google Scholar] [CrossRef] [Green Version]
- Fleurence, J.; Morançais, M.; Dumay, J. 9—Seaweed proteins. Proteins Food Process. In Woodhead Publishing Series in Food Science, Technology and Nutrition, 2nd ed.; Yoda, R.Y., Ed.; Woodhead Publishing: Cambridge, UK, 2018; Volume 10, pp. 245–262. ISBN 978-0-0810-0722-8. [Google Scholar]
- Dong, T.; Van Wychen, S.; Nagle, N.; Pienkos, P.T.; Laurens, L.M.L. Impact of biochemical composition on susceptibility of algal biomass to acid-catalyzed pretreatment for sugar and lipid recovery. Algal Res. 2016, 18, 69–77. [Google Scholar] [CrossRef] [Green Version]
- González-González, L.M.; De-Bashan, L.E. Toward the enhancement of microalgal metabolite production through microalgae–bacteria consortia. Biology 2021, 10, 282. [Google Scholar] [CrossRef]
- Choix, F.J.; de-Bashan, L.E.; Bashan, Y. Enhanced accumulation of starch and total carbohydrates in alginate-immobilized Chlorella spp. induced by Azospirillum brasilense: I. Autotrophic conditions. Enzyme Microb. Technol. 2012, 51, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kuo-Dahab, W.C.; Dolan, S.; Park, C. Kinetics of nutrient removal and expression of extracellular polymeric substances of the microalgae, Chlorella sp. and Micractinium sp. in wastewater treatment. Bioresour. Technol. 2014, 154, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Gorgônio, C.; Aranda, D.; Couri, S. Morphological and chemical aspects of Chlorella pyrenoidosa, Dunaliella tertiolecta, Isochrysis galbana and Tetraselmis gracilis microalgae. Nat. Sci. 2013, 5, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, I.; Nakao, T.; Shigeno, I.; Ando, Y.; Hirayama, K. Application of unicellular algae Chlorella vulgaris for the mass-culture of marine rotifer Brachionus. In Live Food in Aquaculture. Developments in Hydrobiology; Hagiwara, A., Snell, T.W., Lubzens, E., Tamaru, C.S., Eds.; Springer: Dordrecht, The Netherlands, 1997; Volume 124, pp. 133–138. ISBN 978-94-017-2097-7. [Google Scholar]
- Shalaby, S. Quality of novel healthy processed cheese analogue enhanced with marine microalgae Chlorella vulgaris biomass. World Appl. Sci. J. 2017, 23, 914–925. [Google Scholar] [CrossRef]
- Lu, Z.; Zheng, L.; Liu, J.; Dai, J.; Song, L. A novel fed-batch strategy to boost cyst cells production based on the understanding of intracellular carbon and nitrogen metabolism in Haematococcus pluvialis. Bioresour. Technol. 2019, 289, 121744. [Google Scholar] [CrossRef] [PubMed]
- Grosse, J.; Brussaard, C.P.D.; Boschker, H.T.S. Nutrient limitation driven dynamics of amino acids and fatty acids in coastal phytoplankton. Limnol. Oceanogr. 2019, 64, 302–316. [Google Scholar] [CrossRef] [Green Version]
- El-Sheekh, M.; Eladel, H.; Battah, M.; Abd-Elal, S.M. Effect of different nitrogen sources on growth and biochemical composition of the green microalgae Scenedesmus obliquus and Chlorella kessleri. Int. Confer. Biol. Sci. 2004, 3, 419–432. [Google Scholar]
- Mišurcová, L.; Buňka, F.; Ambrožová, J.V.; Machů, L.; Samek, D.; Kráčmar, S. Amino acid composition of algal products and its contribution to RDI. Food Chem. 2014, 151, 120–125. [Google Scholar] [CrossRef]
- FAO/WHO. Energy and Protein Requirement; Report of a Joint FAO/WHO ad hoc Expert Committee, 52; FAO: Geneva, Switzerland, 1973. [Google Scholar]
- Rohit, M.V.; Venkata, M.S. Quantum Yield and Fatty Acid Profile Variations with Nutritional Mode During Microalgae Cultivation. Front. Bioeng. Biotechnol. 2018, 6, 111. [Google Scholar] [CrossRef]
- Melo, M.; Fernandes, S.; Caetano, N.; Borges, M.T. Chlorella vulgaris ( SAG 211-12 ) biofilm formation capacity and proposal of a rotating flat plate photobioreactor for more sustainable biomass production. J. Appl. Phycol. 2018, 30, 887–899. [Google Scholar] [CrossRef]
- Han, S.F.; Jin, W.; Abomohra, A.E.F.; Tu, R.; Zhou, X.; He, Z.; Chen, C.; Xie, G. Municipal wastewater enriched with trace metals for enhanced lipid production of the biodiesel-promising microalga Scenedesmus obliquus. Bioenergy Res. 2019, 12, 1127–1133. [Google Scholar] [CrossRef]
- Sharma, J.; Kumar, V.; Kumar, S.S.; Malyan, S.K.; Mathimani, T.; Bishnoi, N.R.; Pugazhendhi, A. Microalgal consortia for municipal wastewater treatment—Lipid augmentation and fatty acid profiling for biodiesel production. J. Photochem. Photobiol. B Biol. 2020, 202, 111638. [Google Scholar] [CrossRef] [PubMed]
- Anto, S.; Pugazhendhi, A.; Mathimani, T. Lipid enhancement through nutrient starvation in Chlorella sp. and its fatty acid profiling for appropriate bioenergy feedstock. Biocatal. Agric. Biotechnol. 2019, 20, 101179. [Google Scholar] [CrossRef]
- Mahapatra, D.M.; Chanakya, H.N.; Ramachandra, T.V. Bioremediation and lipid synthesis through mixotrophic algal consortia in municipal wastewater. Bioresour. Technol. 2014, 168, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Mathimani, T.; Uma, L.; Prabaharan, D. Formulation of low-cost seawater medium for high cell density and high lipid content of Chlorella vulgaris BDUG 91771 using central composite design in biodiesel perspective. J. Clean. Prod. 2018, 198, 575–586. [Google Scholar] [CrossRef]
- Deshmukh, S.; Kumar, R.; Bala, K. Microalgae biodiesel: A review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Process. Technol. 2019, 191, 232–247. [Google Scholar] [CrossRef]
- Martins, D.A.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K.M. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P.; DiNicolantonio, J.J. The importance of a balanced ω-6 to ω-3 ratio in the prevention and management of obesity. Open Heart 2016, 20, e000385. [Google Scholar] [CrossRef] [Green Version]
- Maltsev, Y.; Maltseva, K. Fatty acids of microalgae: Diversity and applications. Rev. Environ. Sci. Biotechnol. 2021, 20, 515–547. [Google Scholar] [CrossRef]
- Barsanti, L.; Gualtieri, P. Algae: Anatomy, Biochemistry and Biotechnology, 1st ed.; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar] [CrossRef]
- Europe Microalgae Products Market. Global Industry Report, TMRGL82608. 2021. Available online: http://dsnyulesbcj.com/europe-microalgae-products-market.html (accessed on 7 January 2022).
- Travieso, L.; Benítez, F.; Sánchez, E.; Borja, R.; Martín, A.; Colmenarejo, M.F. Batch mixed culture of Chlorella vulgaris using settled and diluted piggery waste. Ecol. Eng. 2006, 28, 158–165. [Google Scholar] [CrossRef]
- Salati, S.; D’Imporzano, G.; Menin, B.; Veronesi, D.; Scaglia, B.; Abbruscato, P.; Mariani, P.; Adani, F. Mixotrophic cultivation of Chlorella for local protein production using agro-food by-products. Bioresour. Technol. 2017, 230, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Sforza, E.; Cipriani, R.; Morosinotto, T.; Bertucco, A.; Giacometti, G.M. Excess CO2 supply inhibits mixotrophic growth of Chlorella protothecoides and Nannochloropsis salina. Bioresour. Technol. 2012, 104, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, A.C. Recovery of Dairy Manure Nutrients by Benthic Freshwater Algae Recovery of dairy manure nutrients by benthic freshwater algae. Bioresour. Technol. 2002, 84, 81–91. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
| NFP | BG-11 | ||
|---|---|---|---|
| pH | 8.5 ± 0 | 7.4 | |
| TN | mg L−1 | 136 ± 0 | 247 |
| NH4-N | mg L−1 | 132 ± 2 | 19 |
| COD | mg L−1 O2 | 77 ± 4 | - |
| P | mg L−1 | 7.61 a | 7.11 |
| Na | mg L−1 | 249 ± 3 | 414 |
| Mg | mg L−1 | 5.4 ± 0.1 | 7.39 |
| K | mg L−1 | 188 ± 8 | 17.95 |
| Ca | mg L−1 | 9.7 ± 0.2 | 9.81 |
| Fe | mg L−1 | 1.42 b | 1.42 |
| B | mg L−1 | 0.5 ± 0.1 | 0.50 |
| Al | mg L−1 | 0.6 ± 0.0 | n.p. e |
| Cr | μg L−1 | 4.7 ± 0.6 | n.p. |
| Co | μg L−1 | 4.8 ± 1.2 | 10 |
| Cu | μg L−1 | 30.7 ± 0.4 | 30 |
| Zn | μg L−1 | 57 ± 16 | 50 |
| Se | μg L−1 | 5.2 ± 0.4 | n.p. |
| Mo | μg L−1 | 19 ± 5 | 150 |
| Cd | μg L−1 | 7 c | n.p. |
| Pb | μg L−1 | 6.5 ± 2 | n.p. |
| As | μg L−1 | u.d.l d | n.p. |
| Mn | μg L−1 | u.d.l | 500 |
| Ni | μg L−1 | u.d.l | n.p. |
| Eukaryotic Genus | Prokaryotic Genus | µ | |||
|---|---|---|---|---|---|
| Algae % a,f | Other Eukaryotes % a,f | Algae % b,g | Other Prokaryotes % b,g | d−1 | |
| AC_1 | Chlorella 99.1% | n.f. c | n.f. | Paludisphaera (Planctomycetota) 36% | 0.55 ± 0.04a e |
| AC_2 | Chlorella 8.4% | Nuclearia 40.6%; Vahlkampfia 30.7%; Colpoda 15.6% | Synechocystis 35.9% | Truepera (Deinococcata) 21% | 0.22 ± 0.03b |
| AC_3 | Chlorella 85% | - | Synechocystis 19.6% | SM1A02 (Planctomycetota) 36.8% | 0.25 ± 0.04b |
| AC_4 | Chlorella 76.4% | Colpoda 10.3% | Synechocystis 27.9% | SM1A02 (Planctomycetota) 34.5% | 0.31 ± 0.12b |
| AC_5 | Chlorella 30.6% | Colpoda 36.1%; Nuclearia 17.7% | Synechocystis 84.8% | n.f. | 0.29 ± 0.04b |
| AC_6 | Tetradesmus 85.4% | Colpoda 9% | n.f. | Others d 61% | 0.31 ± 0.02b |
| AC_7 | Tetradesmus 42.6% | Colpoda 34.8% | Synechocystis 21.4% | Chloronema (Chloroflexi) 22.9% | 0.24 ± 0.02b |
| AC_8 | Scenedesmus 8.1%; Chlorella 6.3% | Colpoda 69.3% | n.f. | SM1A02 (Planctomycetota) 42.5% | 0.28 ± 0.08b |
| AC_9 | Chlorella 82.3% | Vermamoeba 11.9% | Synechocystis 35.4% | SM1A02 (Planctomycetota) 34.3% | 0.31 ± 0.02b |
| AC_10 | Tetradesmus 98.4% | n.f. | Synechocystis 54.2% | n.f. | 0.52 ± 0.06a |
| AC_11 | Chlorella 34.5% | Cyclidium 34.1% | Synechocystis 9.2% | SM1A02 (Planctomycetota) 57.7% | 0.18 ± 0.01b |
| AC_12 | Chlorella 39.6%; Tetradesmus 32.6% | Vermamoeba 9.4% | Synechocystis 3.6% | Sandaracinus (Proteobacteria) 29.8%; Others c 52.5% | 0.58 ± 0.06a |
| AC | TNinitial a | TNfinal b | Nbiomass c | N Taken Up by Biomass | Pinitial d | Pfinal e | P Taken Up by Biomass | Proteins | Lipids | Carbohydrates |
|---|---|---|---|---|---|---|---|---|---|---|
| mg L−1 | mg L−1 | g kg−1 DM f | % TNinitial a | mg L−1 | mg L−1 | % Pinitial d | g kg−1 DM | g kg−1 DM | g kg−1 DM | |
| AC_1 | 136 ± 0 | 35 ± 6 | 41 ± 0 | 54 ± 0 | 7.61 ± 0 | 0.49 ± 0.01 | 94 ± 0 | 257 ± 0g g | 119 ± 1fg | 596 ± 4 |
| AC_2 | 136 ± 0 | 23 ± 1 | 74 ± 0.6 | 65 ± 1 | 7.61 ± 0 | 1.13 ± 0.16 | 85 ± 2 | 460 ± 4b | 105 ± 7g | 405 ± 9 |
| AC_3 | 136 ± 0 | 12 ± 3 | 90 ± 0.2 | 59 ± 0 | 7.61 ± 0 | 0.72 ± 0.37 | 91 ±5 | 561 ± 1a | 152 ± 10cde | 254 ± 11 |
| AC_4 | 136 ± 0 | 20 ± 4 | 68 ± 0.4 | 78 ± 0 | 7.61 ± 0 | 0.40 ± 0.14 | 95 ± 2 | 422 ± 3c | 177 ± 3b | 359 ± 6 |
| AC_5 | 136 ± 0 | 45 ± 13 | 43 ± 0.1 | 76 ± 0 | 7.61 ± 0 | 0.47 ± 0.08 | 94 ± 1 | 266 ± 0g | 153 ± 9bcde | 565 ± 9 |
| AC_6 | 136 ± 0 | 29 ± 4 | 49 ± 0.7 | 65 ± 1 | 7.61 ± 0 | 0.45 ± 0.17 | 94 ± 2 | 305 ± 5f | 173 ± 11bc | 486 ± 12 |
| AC_7 | 136 ± 0 | 3.4 ± 0.5 | 53 ± 0.7 | 46 ± 0 | 7.61 ± 0 | 0.54 ± 0.03 | 93 ± 0 | 334 ± 4e | 128 ± 8efg | 512 ± 9 |
| AC_8 | 136 ± 0 | 38 ± 2 | 67 ± 0.4 | 53 ± 0 | 7.61 ± 0 | 0.51 ± 0.11 | 93 ± 1 | 420 ± 2c | 177 ± 9b | 369 ± 10 |
| AC_9 | 136 ± 0 | 35 ± 7 | 64 ± 0.3 | 87 ± 0 | 7.61 ± 0 | 0.36 ± 0.08 | 95 ± 1 | 398 ± 2cd | 178 ± 8b | 406 ± 8 |
| AC_10 | 136 ± 0 | 31 ± 1 | 61 ± 0.1 | 62 ± 0 | 7.61 ± 0 | 0.43 ± 0.08 | 94 ± 1 | 382 ± 7d | 156 ± 9bcd | 425 ± 9 |
| AC_11 | 136 ± 0 | 27 ± 5 | 55 ± 0.3 | 29 ± 0 | 7.61 ± 0 | 1.37 ± 0.37 | 82 ± 5 | 341 ± 2e | 135 ± 4def | 494 ± 5 |
| AC_12 | 136 ± 0 | 21 ± 1 | 44 ± 0.6 | 53 ± 11 | 7.61 ± 0 | 0.46 ± 0.01 | 94 ± 0 | 273 ± 22g | 230 ± 15a | 472 ± 27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.; Dell’Orto, M.; Scaglia, B.; D’Imporzano, G.; Bani, A.; Adani, F. Growth Performance, Biochemical Composition and Nutrient Recovery Ability of Twelve Microalgae Consortia Isolated from Various Local Organic Wastes Grown on Nano-Filtered Pig Slurry. Molecules 2022, 27, 422. https://doi.org/10.3390/molecules27020422
Su M, Dell’Orto M, Scaglia B, D’Imporzano G, Bani A, Adani F. Growth Performance, Biochemical Composition and Nutrient Recovery Ability of Twelve Microalgae Consortia Isolated from Various Local Organic Wastes Grown on Nano-Filtered Pig Slurry. Molecules. 2022; 27(2):422. https://doi.org/10.3390/molecules27020422
Chicago/Turabian StyleSu, Min, Marta Dell’Orto, Barbara Scaglia, Giuliana D’Imporzano, Alessia Bani, and Fabrizio Adani. 2022. "Growth Performance, Biochemical Composition and Nutrient Recovery Ability of Twelve Microalgae Consortia Isolated from Various Local Organic Wastes Grown on Nano-Filtered Pig Slurry" Molecules 27, no. 2: 422. https://doi.org/10.3390/molecules27020422
APA StyleSu, M., Dell’Orto, M., Scaglia, B., D’Imporzano, G., Bani, A., & Adani, F. (2022). Growth Performance, Biochemical Composition and Nutrient Recovery Ability of Twelve Microalgae Consortia Isolated from Various Local Organic Wastes Grown on Nano-Filtered Pig Slurry. Molecules, 27(2), 422. https://doi.org/10.3390/molecules27020422







