Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses
Abstract
:1. Introduction
2. Aptamers: A Potential Diagnostic and Therapeutic Alternative
3. Aptamers for Virus Diagnosis and Treatment
3.1. Aptamers against Human Immunodeficiency Virus (HIV)
3.1.1. Aptamers HIV in Diagnostics
3.1.2. Aptamers to HIV as Antiviral Agents
Aptamers to HIV Genome
Aptamer to HIV Proteins
Aptamers to Protease (PR)
Aptamers to Integrase (IN)
Aptamers to Reverse Transcriptase (RT)
Aptamers to Nucleocapsid Protein
Aptamers to Surface Glycoprotein (gp 120)
Aptamers to Gag Protein
Aptamers to Cell Proteins
Aptamers to CCR5
Aptamers to Nucleolin (NCL)
Use of Aptamers for Delivery of Therapeutic Molecules
3.2. Aptamers against HBV
3.2.1. Aptamers HBV in Diagnostics
3.2.2. Aptamers to HBV as Antiviral Agents
3.3. Aptamers against HCV
3.3.1. Aptamers HCV in Diagnostics
3.3.2. Aptamers to HCV as Antiviral Agents
Aptamers to 5′ and 3′ Untranslated Regions (5′ and 3′UTR)
Aptamers to HCV Proteins
Aptamers to Nonstructural Protein 2 (NS2)
Aptamers to Nonstructural Protein 3 (NS3)
Aptamers to Nonstructural Protein 5A (NS5A)
Aptamers to Nonstructural Protein 5B (NS5B)
Aptamers to Structural Proteins E1, E2 and Core
3.4. Aptamers against Human Papilloma Virus (HPV)
3.4.1. Aptamers to HPV in Diagnostics
3.4.2. Aptamers to HPV as Antiviral Agents
3.5. Aptamers to Herpes Simplex Virus (HSV)
3.6. Aptamers to Influenza
3.6.1. Aptamers to Influenza Virus in Diagnostics
3.6.2. Aptamers to Influenza Virus as Antiviral Agents
3.7. Aptamers against Other Emerging Viruses
3.7.1. Aptamers to Rift Valley Fever Virus (RVFV)
3.7.2. Aptamers to Tick-Borne Encephalitis Virus (TBEV)
3.7.3. Aptamers to Dengue Virus (DENV)
3.7.4. Aptamers to Ebola Virus (EV)
3.7.5. Aptamers to Severe Acute Respiratory Syndrome (SARS)
4. Perspectives
Acknowledgments
Conflicts of Interest
References
- De Clercq, E.; Field, H.J. Antiviral prodrugs—The development of successful prodrug strategies for antiviral chemotherapy. Br. J. Pharmacol. 2006, 147, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Witthoft, T.; Moller, B.; Wiedmann, K.H.; Mauss, S.; Link, R.; Lohmeyer, J.; Lafrenz, M.; Gelbmann, C.M.; Huppe, D.; Niederau, C.; et al. Safety, tolerability and efficacy of peginterferon α-2A and ribavirin in chronic hepatitis c in clinical practice: The german open safety trial. J. Viral Hepat. 2007, 14, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Reesink, H.W.; Fanning, G.C.; Farha, K.A.; Weegink, C.; Van Vliet, A.; Van ‘t Klooster, G.; Lenz, O.; Aharchi, F.; Marien, K.; Van Remoortere, P.; et al. Rapid HCV-RNA decline with once daily TMC435: A phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 2010, 138, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Rudin, D.; Shah, S.M.; Kiss, A.; Wetz, R.V.; Sottile, V.M. Interferon and lamivudine vs. Interferon for hepatitis B E antigen-positive hepatitis B treatment: Meta-analysis of randomized controlled trials. Liver Int. 2007, 27, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Shum, K.T.; Zhou, J.; Rossi, J.J. Aptamer-based therapeutics: New approaches to combat human viral diseases. Pharmaceuticals 2013, 6, 1507–1542. [Google Scholar] [CrossRef] [PubMed]
- Wandtke, T.; Wozniak, J.; Kopinski, P. Aptamers in diagnostics and treatment of viral infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Blank, M.; Buhmann, R.; Resch, A. Aptamers—Basic research, drug development, and clinical applications. Appl. Microbiol. Biotechnol. 2005, 69, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T. Aptamers and SELEX: The technology. World Pat. Inf. 2003, 25, 123–129. [Google Scholar] [CrossRef]
- Marton, S.; Reyes-Darias, J.A.; Sanchez-Luque, F.J.; Romero-Lopez, C.; Berzal-Herranz, A. In vitro and ex vivo selection procedures for identifying potentially therapeutic DNA and RNA molecules. Molecules 2010, 15, 4610–4638. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, N.; Gorenstein, D.G. Strategies for the discovery of therapeutic aptamers. Expert Opin. Drug Discov. 2011, 6, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Sullenger, B.A.; Rusconi, C.P. Developing aptamers into therapeutics. J. Clin. Investig. 2000, 106, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Trujillo, C.A.; Nery, A.A.; Alves, J.M.; Majumder, P.; Resende, R.R.; Martins, A.H. DNA and RNA aptamers: From tools for basic research towards therapeutic applications. Comb. Chem. High Throughput Screen 2006, 9, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Jenison, R.D.; Jennings, S.D.; Walker, D.W.; Bargatze, R.F.; Parma, D. Oligonucleotide inhibitors of p-selectin-dependent neutrophil-platelet adhesion. Antisense Nucleic Acid Drug Dev. 1998, 8, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Kubik, M.F.; Stephens, A.W.; Schneider, D.; Marlar, R.A.; Tasset, D. High-affinity RNA ligands to human α-thrombin. Nucleic Acids Res. 1994, 22, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Padmapriya, A.; Morden, K.M.; Jayasena, S.D. Peptide conjugation to an in vitro-selected DNA ligand improves enzyme inhibition. Proc. Natl. Acad. Sci. USA 1995, 92, 11044–11048. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.A. How does HIV cause aids? Science 1993, 260, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Gokengin, D.; Geretti, A.M.; Begovac, J.; Palfreeman, A.; Stevanovic, M.; Tarasenko, O.; Radcliffe, K. 2014 european guideline on HIV testing. Int. J. STD AIDS 2014, 25, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Luzi, E.; Mascini, M. Aptamer-based biosensors for the detection of HIV-1 tat protein. Bioelectrochemistry 2005, 67, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Rahim Ruslinda, A.; Tanabe, K.; Ibori, S.; Wang, X.; Kawarada, H. Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 tat protein. Biosens. Bioelectron. 2013, 40, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Satheesan, S.; Li, H.; Weinberg, M.S.; Morris, K.V.; Burnett, J.C.; Rossi, J.J. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015, 22, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Butovskaya, E.; Lago, S.; Garzino-Demo, A.; Pannecouque, C.; Palu, G.; Richter, S.N. The G-quadruplex-forming aptamer as1411 potently inhibits HIV-1 attachment to the host cell. Int. J. Antimicrob. Agents 2016, 47, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Shibata, T.; Kabashima, T.; Kai, M. Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem. 2012, 56, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Srisawat, C.; Engelke, D.R. Selection of RNA aptamers that bind HIV-1 LTR DNA duplexes: Strand invaders. Nucleic Acids Res. 2010, 38, 8306–8315. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Luque, F.J.; Stich, M.; Manrubia, S.; Briones, C.; Berzal-Herranz, A. Efficient HIV-1 inhibition by a 16 NT-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014, 4, 6242. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Manchester, M.; Swanstrom, R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 1994, 68, 6782–6786. [Google Scholar] [PubMed]
- Wiegers, K.; Rutter, G.; Kottler, H.; Tessmer, U.; Hohenberg, H.; Krausslich, H.G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 1998, 72, 2846–2854. [Google Scholar] [PubMed]
- Duclair, S.; Gautam, A.; Ellington, A.; Prasad, V.R. High-affinity RNA aptamers against the HIV-1 protease inhibit both in vitro protease activity and late events of viral replication. Mol. Ther. Nucleic Acids 2015, 4, e228. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Riccardi, C.; Montesarchio, D. G-quadruplex forming oligonucleotides as anti-HIV agents. Molecules 2015, 20, 17511–17532. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.K.; Davies, D.R. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 2004, 4, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Ojwang, J.O.; Buckheit, R.W.; Pommier, Y.; Mazumder, A.; De Vreese, K.; Este, J.A.; Reymen, D.; Pallansch, L.A.; Lackman-Smith, C.; Wallace, T.L.; et al. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 1995, 39, 2426–2435. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Maddali, K.; Metifiot, M.; Pommier, Y. HIV-1 in inhibitors: 2010 update and perspectives. Curr. Top. Med. Chem. 2009, 9, 1016–1037. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Hogan, M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998, 273, 34992–34999. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Rando, R.F.; Pommier, Y.; Hogan, M.E. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry 1997, 36, 12498–12505. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; De Clercq, E.; Rando, R.F.; Pallansch, L.; Lackman-Smith, C.; Lee, S.; Hogan, M.E. Stability-activity relationships of a family of G-tetrad forming oligonucleotides as potent HIV inhibitors. A basis for anti-HIV drug design. J. Biol. Chem. 2000, 275, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and function of HIV-1 reverse transcriptase: Molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [PubMed]
- DeStefano, J.J.; Nair, G.R. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides 2008, 18, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Michalowski, D.; Chitima-Matsiga, R.; Held, D.M.; Burke, D.H. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008, 36, 7124–7135. [Google Scholar] [CrossRef] [PubMed]
- Andreola, M.L.; Pileur, F.; Calmels, C.; Ventura, M.; Tarrago-Litvak, L.; Toulme, J.J.; Litvak, S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 2001, 40, 10087–10094. [Google Scholar] [CrossRef] [PubMed]
- Metifiot, M.; Leon, O.; Tarrago-Litvak, L.; Litvak, S.; Andreola, M.L. Targeting HIV-1 integrase with aptamers selected against the purified RNase H domain of HIV-1 RT. Biochimie 2005, 87, 911–919. [Google Scholar] [CrossRef] [PubMed]
- De Soultrait, V.R.; Lozach, P.Y.; Altmeyer, R.; Tarrago-Litvak, L.; Litvak, S.; Andreola, M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002, 324, 195–203. [Google Scholar] [CrossRef]
- Levin, J.G.; Mitra, M.; Mascarenhas, A.; Musier-Forsyth, K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010, 7, 754–774. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, M.Y.; Lee, J.H.; You, J.C.; Jeong, S. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2002, 291, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.R.; Vickers, T.A.; Roberson, J.L.; Buckheit, R.W., Jr.; Klimkait, T.; DeBaets, E.; Davis, P.W.; Rayner, B.; Imbach, J.L.; Ecker, D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA 1994, 91, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Hotoda, H.; Koizumi, M.; Koga, R.; Kaneko, M.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Sone, J.; et al. Biologically active oligodeoxyribonucleotides. 5. 5′-end-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998, 41, 3655–3663. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Koga, R.; Hotoda, H.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Abe, K.; Kosaka, T.; et al. Biologically active oligodeoxyribonucleotides—IX. Synthesis and anti-HIV-1 activity of hexadeoxyribonucleotides, TGGGAG, bearing 3′- and 5′-end-modification. Bioorg. Med. Chem. 1997, 5, 2235–2243. [Google Scholar] [CrossRef]
- Koizumi, M.; Koga, R.; Hotoda, H.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Abe, K.; Kosaka, T.; Tsutsumi, S.; et al. Biologically active oligodeoxyribonucleotides. Part 11: The least phosphate-modification of quadruplex-forming hexadeoxyribonucleotide TGGGAG, bearing 3-and 5-end-modification, with anti-HIV-1 activity. Bioorg. Med. Chem. 1998, 6, 2469–2475. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Erra, E.; Martino, L.; Fabio, G.D.; Napoli, L.D.; Giancola, C.; Montesarchio, D. 5′-modified g-quadruplex forming oligonucleotides endowed with anti-HIV activity: Synthesis and biophysical properties. Bioconj. Chem. 2007, 18, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Di Fabio, G.; D’Onofrio, J.; Chiapparelli, M.; Hoorelbeke, B.; Montesarchio, D.; Balzarini, J.; de Napoli, L. Discovery of novel anti-HIV active G-quadruplex-forming oligonucleotides. Chem. Commun. 2011, 47, 2363–2365. [Google Scholar] [CrossRef] [PubMed]
- Romanucci, V.; Milardi, D.; Campagna, T.; Gaglione, M.; Messere, A.; D’Urso, A.; Crisafi, E.; La Rosa, C.; Zarrelli, A.; Balzarini, J.; et al. Synthesis, biophysical characterization and anti-HIV activity of d(TG3AG) quadruplexes bearing hydrophobic tails at the 5′-end. Bioorg. Med. Chem. 2014, 22, 960–966. [Google Scholar] [CrossRef] [PubMed]
- D’Atri, V.; Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Mayol, L.; Piccialli, V.; Haider, S.; Hoorelbeke, B.; Balzarini, J.; et al. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: Effect of the base sequence on anti-HIV activity. Chem. Commun. 2012, 48, 9516–9518. [Google Scholar] [CrossRef] [PubMed]
- Romanucci, V.; Gaglione, M.; Messere, A.; Potenza, N.; Zarrelli, A.; Noppen, S.; Liekens, S.; Balzarini, J.; di Fabio, G. Hairpin oligonucleotides forming G-quadruplexes: New aptamers with anti-HIV activity. Eur. J. Med. Chem. 2015, 89, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Khati, M.; Schuman, M.; Ibrahim, J.; Sattentau, Q.; Gordon, S.; James, W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′f-RNA aptamers. J. Virol. 2003, 77, 12692–12698. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Khati, M.; Tang, M.; Wyatt, R.; Lea, S.M.; James, W. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J. Virol. 2005, 79, 13806–13810. [Google Scholar] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-GP120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.; Forzan, M.; Sproat, B.; Pantophlet, R.; McGowan, I.; Burton, D.; James, W. An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of GP120 in the CCR5 binding site. Virology 2008, 381, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.A.; Freed, E.O. HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retrovir. 2012, 28, 54–75. [Google Scholar] [CrossRef] [PubMed]
- Lochrie, M.A.; Waugh, S.; Pratt, D.G., Jr.; Clever, J.; Parslow, T.G.; Polisky, B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res. 1997, 25, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, D.; Duclair, S.; Datta, S.A.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Hahn, U.; Rentmeister, A. Cell-specific aptamers as emerging therapeutics. J. Nucleic Acids 2011, 2011, 904750. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J.J. Therapeutic potential of aptamer-siRNA conjugates for treatment of HIV-1. BioDrugs 2012, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Cell-type-specific aptamer and aptamer-small interfering RNA conjugates for targeted human immunodeficiency virus type 1 therapy. J. Investig. Med. 2014, 62, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Swiderski, P.; Li, H.; Zhang, J.; Neff, C.P.; Akkina, R.; Rossi, J.J. Selection, characterization and application of new RNA HIV GP 120 aptamers for facile delivery of dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009, 37, 3094–3109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, H.; Li, S.; Zaia, J.; Rossi, J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008, 16, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci. Transl. Med. 2011, 3, 66ra66. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Neff, C.P.; Swiderski, P.; Li, H.; Smith, D.D.; Aboellail, T.; Remling-Mulder, L.; Akkina, R.; Rossi, J.J. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013, 21, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, L.A.; Trifonova, R.; Vrbanac, V.; Basar, E.; McKernan, S.; Xu, Z.; Seung, E.; Deruaz, M.; Dudek, T.; Einarsson, J.I.; et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Investig. 2011, 121, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Dienstag, J.L. Hepatitis B virus infection. N. Engl. J. Med. 2008, 359, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.K.; Song, S.; Oh, H.B.; Hwang, S.H.; Hah, S.S. Aptamer-based competitive binding assay for one-step quantitation of hepatitis B surface antigen. Analyst 2014, 139, 4310–4314. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, Y.; Hu, B.; Ma, Z.Y.; Huang, H.P.; Yu, Y.; Liu, S.P.; Lu, M.J.; Yang, D.L. Development of HBSAG-binding aptamers that bind HEPG2.2.15 cells via HBV surface antigen. Virologica Sinica 2010, 25, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Huang, R.; Li, Z.; He, N.; Wang, T.; Su, E.; Deng, Y. Selection of HBSAG-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis B virus infection. ACS Appl. Mater. Interfaces 2015, 7, 11215–11223. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.H. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS ONE 2011, 6, e27862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Pei, X.; Zhang, Q.; Lu, B.; Zhang, X.; Liu, J. An aptamer targets HBV core protein and suppresses HBV replication in HEPG2.2.15 cells. Int. J. Mol. Med. 2014, 34, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Orabi, A.; Bieringer, M.; Geerlof, A.; Bruss, V. An aptamer against the matrix binding domain on the hepatitis B virus capsid impairs virion formation. J. Virol. 2015, 89, 9281–9287. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, U.A.; Javed, T.; Rehman, S.; Nawaz, Z.; Riazuddin, S. An overview of HCV molecular biology, replication and immune responses. Virol. J. 2011, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, Y.S.; Jo, M.; Jin, M.; Lee, D.K.; Kim, S. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 2007, 358, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Li, D.; Chen, H.; Zhang, X.L. CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2. PLoS ONE 2009, 4, e8142. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Yu, X.; Gao, Y.; Xue, B.; Wu, X.; Wang, X.; Yang, D.; Zhu, H. Inhibition of hepatitis C virus production by aptamers against the core protein. J. Virol. 2014, 88, 1990–1999. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, L.; Shen, X. Development of a nucleic acid lateral flow strip for detection of hepatitis C virus (HCV) core antigen. Nucleotides Nucleic Acids 2013, 32, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jee, M.H.; Kwon, O.S.; Keum, S.J.; Jang, S.K. Infectivity of hepatitis c virus correlates with the amount of envelope protein E2: Development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology 2013, 439, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.S.; Lee, S.M.; Eom, K.; Lee, J.H.; Lee, Y.S.; Park, J.H.; Yoon, D.S.; Kim, T.S. Nanomechanical microcantilever operated in vibration modes with use of RNA aptamer as receptor molecules for label-free detection of HCV helicase. Biosens. Bioelectron. 2007, 23, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Roh, C.; Kim, S.E.; Jo, S.K. Label free inhibitor screening of hepatitis C virus (HCV) NS5B viral protein using RNA oligonucleotide. Sensors 2011, 11, 6685–6696. [Google Scholar] [CrossRef] [PubMed]
- Tamori, A.; Enomoto, M.; Kawada, N. Recent advances in antiviral therapy for chronic hepatitis C. Med. Inflamm. 2016, 2016, 6841628. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H.; Lee, S.W. Prospects for nucleic acid-based therapeutics against hepatitis C virus. World J. Gastroenterol. 2013, 19, 8949–8962. [Google Scholar] [CrossRef] [PubMed]
- Toulme, J.J.; Darfeuille, F.; Kolb, G.; Chabas, S.; Staedel, C. Modulating viral gene expression by aptamers to RNA structures. Biol. Cell 2003, 95, 229–238. [Google Scholar] [CrossRef]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Hwang, J.; Kuno, A.; Hasegawa, T.; Nishikawa, S. RNA aptamers targeted to domain II of hepatitis C virus ires that bind to its apical loop region. J. Biochem. 2003, 133, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Kuno, A.; Hasegawa, T.; Nishikawa, S. A hepatitis C virus (HCV) internal ribosome entry site (ires) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIID. Nucleic Acids Res. 2005, 33, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Umehara, T.; Nishikawa, F.; Fukuda, K.; Hasegawa, T.; Nishikawa, S. Increased inhibitory ability of conjugated RNA aptamers against the HCV ires. Biochem. Biophys. Res. Commun. 2009, 386, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Barroso-delJesus, A.; Puerta-Fernandez, E.; Berzal-Herranz, A. Interfering with hepatitis C virus ires activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005, 386, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Diaz-Gonzalez, R.; Barroso-delJesus, A.; Berzal-Herranz, A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J. Gen. Virol. 2009, 90, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Berzal-Herranz, B.; Gomez, J.; Berzal-Herranz, A. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antivir. Res. 2012, 94, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; Iizuka, M.; Fujita, S.; Nishikawa, S.; Hasegawa, T.; Fukuda, K. An RNA aptamer containing two binding sites against the HCV minus-IRES domain I. Nucleotides Nucleic Acids 2011, 30, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; Fujita, S.; Iizuka, M.; Nishikawa, S.; Hasegawa, T.; Fukuda, K. Isolation and characterization of RNA aptamers specific for the HCV minus-IRES domain I. Nucleic Acids Symp. Ser. 2008, 493–494. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Berzal-Herranz, B.; Garmendia, E.; Cueto, F.J.; Berzal-Herranz, A. Anti-HCV RNA aptamers targeting the genomic cis-acting replication element. Pharmaceuticals 2011, 5, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Romero-Lopez, C.; Berzal-Herranz, A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J. Viral Hepat. 2013, 20, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, X.; Xue, B.; Zhou, F.; Wang, X.; Yang, D.; Liu, N.; Xu, L.; Fang, X.; Zhu, H. Inhibition of hepatitis C virus infection by DNA aptamer against NS2 protein. PLoS ONE 2014, 9, e90333. [Google Scholar] [CrossRef] [PubMed]
- Urvil, P.T.; Kakiuchi, N.; Zhou, D.M.; Shimotohno, K.; Kumar, P.K.; Nishikawa, S. Selection of RNA aptamers that bind specifically to the NS3 protease of hepatitis C virus. Eur. J. Biochem. 1997, 248, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.K.; Machida, K.; Urvil, P.T.; Kakiuchi, N.; Vishnuvardhan, D.; Shimotohno, K.; Taira, K.; Nishikawa, S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology 1997, 237, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Vishnuvardhan, D.; Sekiya, S.; Hwang, J.; Kakiuchi, N.; Taira, K.; Shimotohno, K.; Kumar, P.K.; Nishikawa, S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000, 267, 3685–3694. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Fauzi, H.; Fukuda, K.; Sekiya, S.; Kakiuchi, N.; Taira, K.; Kusakabe, I.; Nishikawa, S. Analysis of aptamer binding site for HCV-NS3 protease by alanine scanning mutagenesis. Nucleic Acids Symp. Ser. 2000, 253–254. [Google Scholar] [CrossRef]
- Nishikawa, S.; Nishikawa, F.; Fukuda, K. In vitro selection of RNA aptamers against HCV-NS3 helicase and their structural similarity with 3′(+)UTR of HCV. Nucleic Acids Res. Suppl. 2003, 241–242. [Google Scholar] [CrossRef]
- Nishikawa, F.; Kakiuchi, N.; Funaji, K.; Fukuda, K.; Sekiya, S.; Nishikawa, S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003, 31, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Umehara, T.; Sekiya, S.; Kunio, K.; Hasegawa, T.; Nishikawa, S. An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun. 2004, 325, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, F.; Funaji, K.; Fukuda, K.; Nishikawa, S. In vitro selection of RNA aptamers against the HCV NS3 helicase domain. Oligonucleotides 2004, 14, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Umehara, T.; Fukuda, K.; Nishikawa, F.; Kohara, M.; Hasegawa, T.; Nishikawa, S. Rational design of dual-functional aptamers that inhibit the protease and helicase activities of HCV NS3. J. Biochem. 2005, 137, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gao, Y.; Xue, B.; Wang, X.; Yang, D.; Qin, Y.; Yu, R.; Liu, N.; Xu, L.; Fang, X.; et al. Inhibition of hepatitis C virus infection by NS5A-specific aptamer. Antiviral Res. 2014, 106, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Biroccio, A.; Hamm, J.; Incitti, I.; De Francesco, R.; Tomei, L. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002, 76, 3688–3696. [Google Scholar] [CrossRef] [PubMed]
- Bellecave, P.; Andreola, M.L.; Ventura, M.; Tarrago-Litvak, L.; Litvak, S.; Astier-Gin, T. Selection of DNA aptamers that bind the RNA-dependent RNA polymerase of hepatitis C virus and inhibit viral RNA synthesis in vitro. Oligonucleotides 2003, 13, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Bellecave, P.; Cazenave, C.; Rumi, J.; Staedel, C.; Cosnefroy, O.; Andreola, M.L.; Ventura, M.; Tarrago-Litvak, L.; Astier-Gin, T. Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: Mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother. 2008, 52, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Lee, Y.J.; Kim, J.H.; Lim, J.H.; Kim, J.H.; Han, W.; Lee, S.H.; Noh, G.J.; Lee, S.W. Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J. Virol. 2013, 87, 7064–7074. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Lee, S.H.; Kim, J.H.; Noh, Y.H.; Noh, G.J.; Lee, S.W. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol. Ther. Nucleic Acids 2015, 4, e254. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.A.; Clancy, L.E.; Rawlinson, W.D.; White, P.A. High-affinity aptamers to subtype 3A hepatitis C virus polymerase display genotypic specificity. Antimicrob. Agents Chemother. 2006, 50, 3019–3027. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Meng, X.; Yu, Q.; Xu, L.; Long, Y.; Liu, B.; Fang, X.; Zhu, H. Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein. Antimicrob. Agents Chemother. 2013, 57, 4937–4944. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H.; Kim, H.W.; Myung, H.; Lee, S.W. Hepatitis C virus replication-specific inhibition of microRNA activity with self-cleavable allosteric ribozyme. Nucleic Acid Therap. 2012, 22, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Ljubojevic, S.; Skerlev, M. HPV-associated diseases. Clin. Dermatol. 2014, 32, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Zur Hausen, H. Papillomaviruses in the causation of human cancers—A brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, T.A.; Nicol, C.; Cesur, O.; Trave, G.; Blair, G.E.; Stonehouse, N.J. An RNA aptamer targets the PDZ-binding motif of the HPV16 E6 oncoprotein. Cancers 2014, 6, 1553–1569. [Google Scholar] [CrossRef] [PubMed]
- Toscano-Garibay, J.D.; Benitez-Hess, M.L.; Alvarez-Salas, L.M. Isolation and characterization of an RNA aptamer for the HPV-16 E7 oncoprotein. Arch. Med. Res. 2011, 42, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Leija-Montoya, A.G.; Benitez-Hess, M.L.; Toscano-Garibay, J.D.; Alvarez-Salas, L.M. Characterization of an RNA aptamer against HPV-16 L1 virus-like particles. Nucleic Acid Therap. 2014, 24, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.C.; Zarbl, H. Use of cell-selex to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS ONE 2012, 7, e36103. [Google Scholar] [CrossRef] [PubMed]
- Nicol, C.; Cesur, O.; Forrest, S.; Belyaeva, T.A.; Bunka, D.H.; Blair, G.E.; Stonehouse, N.J. An RNA aptamer provides a novel approach for the induction of apoptosis by targeting the HPV16 E7 oncoprotein. PLoS ONE 2013, 8, e64781. [Google Scholar] [CrossRef] [PubMed]
- Cesur, O.; Nicol, C.; Groves, H.; Mankouri, J.; Blair, G.E.; Stonehouse, N.J. The subcellular localisation of the human papillomavirus (HPV) 16 E7 protein in cervical cancer cells and its perturbation by RNA aptamers. Viruses 2015, 7, 3443–3461. [Google Scholar] [CrossRef] [PubMed]
- Gourronc, F.A.; Rockey, W.M.; Thiel, W.H.; Giangrande, P.H.; Klingelhutz, A.J. Identification of RNA aptamers that internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Virology 2013, 446, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, F.; Deback, C.; Agut, H. Herpes simplex encephalitis: From virus to therapy. Infect. Disord. Drug Targets 2011, 11, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Corbin-Lickfett, K.A.; Chen, I.H.; Cocco, M.J.; Sandri-Goldin, R.M. The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures. Nucleic Acids Res. 2009, 37, 7290–7301. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Hayashi, K.; Kumar, P.K. Aptamer that binds to the GD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J. Virol. 2012, 86, 6732–6744. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Bunka, D.H.; Forzan, M.; Spear, P.G.; Stockley, P.G.; McGowan, I.; James, W. Generation of neutralizing aptamers against herpes simplex virus type 2: Potential components of multivalent microbicides. J. Gen. Virol. 2011, 92, 1493–1499. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Subbarao, K. Global epidemiology of influenza: Past and present. Annu. Rev. Med. 2000, 51, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Baltimore, D.; Doherty, P.C.; Markel, H.; Morens, D.M.; Webster, R.G.; Wilson, I.A. Reconstruction of the 1918 influenza virus: Unexpected rewards from the past. mBio 2012, 3, e00201-12. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Tharakaraman, K.; Sasisekharan, R. Influenza surveillance: 2014–2015 H1N1 “swine”-derived influenza viruses from india. Cell Host Microb. 2015, 17, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Watanabe, S.; Maher, E.A.; Neumann, G.; Kawaoka, Y. Pandemic potential of avian influenza a (H7N9) viruses. Trends Microbiol. 2014, 22, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Tang, T.H.; Chen, Y.; Citartan, M.; Tominaga, J.; Lakshmipriya, T. Sensing strategies for influenza surveillance. Biosens. Bioelectron. 2014, 61, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Kopylov, A. Aptamers to hemagglutinin: A novel tool for influenza virus recognition and neutralization. Curr. Pharm. Design 2016, 22, 4835–4853. [Google Scholar] [CrossRef]
- Misono, T.S.; Kumar, P.K. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005, 342, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Misono, T.S.; Kawasaki, K.; Mizuno, T.; Imai, M.; Odagiri, T.; Kumar, P.K. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006, 87, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Sakamaki, Y.; Kawasaki, K.; Kumar, P.K. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 2006, 139, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Kumar, P.K. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomaterialia 2013, 9, 8932–8941. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, J.; Jiang, T.; Kwon, Y.M.; Lu, H.; Jiao, P.; Liao, M.; Li, Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods 2013, 189, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.C.; Wang, C.H.; Liou, T.M.; Lee, G.B. Influenza a virus-specific aptamers screened by using an integrated microfluidic system. Lab Chip 2014, 14, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.T.; Wang, C.H.; Chang, C.P.; Lee, G.B. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens. Bioelectron. 2016, 82, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Chang, C.P.; Lee, G.B. Integrated microfluidic device using a single universal aptamer to detect multiple types of influenza viruses. Biosens. Bioelectron. 2016, 86, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Shojaei, T.R.; Tabatabaei, M.; Shawky, S.; Salleh, M.A.; Bald, D. A review on emerging diagnostic assay for viral detection: The case of avian influenza virus. Mol. Biol. Rep. 2015, 42, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.; Li, Y. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013, 42, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Callaway, Z.; Lum, J.; Wang, R.; Lin, J.; Li, Y. Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem. 2014, 86, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.; Li, Y. An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 2015, 15, 18565–18578. [Google Scholar] [CrossRef] [PubMed]
- Karash, S.; Wang, R.; Kelso, L.; Lu, H.; Huang, T.J.; Li, Y. Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J. Virol. Methods 2016, 236, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Seo, H.B.; Kim, B.C.; Kim, S.K.; Song, C.S.; Gu, M.B. Highly sensitive sandwich-type spr based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 2016, 86, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.H.; Kayhan, B.; Ben-Yedidia, T.; Arnon, R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 2004, 279, 48410–48419. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Dong, J.; Yao, L.; Chen, A.; Jia, R.; Huan, L.; Guo, J.; Shu, Y.; Zhang, Z. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008, 366, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.H.; Chiang, H.Y.; Yang, D.K.; Peck, K.; Chen, L.C. Selection of aptamers targeting the sialic acid receptor of hemagglutinin by epitope-specific SELEX. Chem. Commun. 2014, 50, 8719–8722. [Google Scholar] [CrossRef] [PubMed]
- Musafia, B.; Oren-Banaroya, R.; Noiman, S. Designing anti-influenza aptamers: Novel quantitative structure activity relationship approach gives insights into aptamer-virus interaction. PLoS ONE 2014, 9, e97696. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, X.; Yan, X.; Liu, K.; Deng, L. A DNA aptamer against influenza a virus: An effective inhibitor to the hemagglutinin-glycan interactions. Nucleic Acid Therap. 2016, 26, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Lee, C.; Lee, K.S.; Choe, S.Y.; Mo, I.P.; Seong, R.H.; Hong, S.; Jeon, S.H. DNA aptamers against the receptor binding region of hemagglutinin prevent avian influenza viral infection. Mol. Cells 2011, 32, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, E.; Kumar, P.K. An aptamer that binds efficiently to the hemagglutinins of highly pathogenic avian influenza viruses (H5N1 and H7N7) and inhibits hemagglutinin-glycan interactions. Acta Biomaterialia 2014, 10, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, Z.; Jiang, F.; Fu, P.; Shen, J.; Wu, W.; Li, J. Two DNA aptamers against avian influenza H9N2 virus prevent viral infection in cells. PLoS ONE 2015, 10, e0123060. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.M. Functions of the influenza a virus NS1 protein in antiviral defense. Curr. Opin. Virol. 2015, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.M.; Kim, K.S.; Lee, J.M.; Shim, H.S.; Cho, S.J.; Lee, W.K.; Ko, H.W.; Keum, Y.S.; Kim, S.Y.; Pathinayake, P.; et al. Single-stranded DNA aptamer that specifically binds to the influenza virus NS1 protein suppresses interferon antagonism. Antivir. Res. 2013, 100, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Boivin, S.; Cusack, S.; Ruigrok, R.W.; Hart, D.J. Influenza a virus polymerase: Structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 2010, 285, 28411–28417. [Google Scholar] [CrossRef] [PubMed]
- Elton, D.; Amorim, M.J.; Medcalf, L.; Digard, P. “Genome gating”; polarized intranuclear trafficking of influenza virus RNPS. Biol. Lett. 2005, 1, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Resa-Infante, P.; Jorba, N.; Coloma, R.; Ortin, J. The influenza virus RNA synthesis machine: Advances in its structure and function. RNA Biol. 2011, 8, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Reich, S.; Guilligay, D.; Pflug, A.; Malet, H.; Berger, I.; Crepin, T.; Hart, D.; Lunardi, T.; Nanao, M.; Ruigrok, R.W.; et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 2014, 516, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Zhang, N.; Singh, K.; Shuai, H.; Chu, H.; Zhou, J.; Chow, B.K.; Zheng, B.J. Cross-protection of influenza a virus infection by a DNA aptamer targeting the PA endonuclease domain. Antimicrob. Agents Chemother. 2015, 59, 4082–4093. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.Y.; Sun, H.Y.; Lee, K.H.; Oh, B.H.; Cha, Y.J.; Kim, B.H.; Yoo, J.Y. 5′-triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012, 40, 2724–2733. [Google Scholar] [CrossRef] [PubMed]
- Weber, F. The catcher in the RIG-I. Cytokine 2015, 76, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Aragon, T.; de la Luna, S.; Novoa, I.; Carrasco, L.; Ortin, J.; Nieto, A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 2000, 20, 6259–6268. [Google Scholar] [CrossRef] [PubMed]
- Burgui, I.; Aragon, T.; Ortin, J.; Nieto, A. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 2003, 84, 3263–3274. [Google Scholar] [CrossRef] [PubMed]
- Yanguez, E.; Rodriguez, P.; Goodfellow, I.; Nieto, A. Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 2012, 422, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.; Perez-Morgado, M.I.; Gonzalez, V.M.; Martin, M.E.; Nieto, A. Inhibition of influenza virus replication by DNA aptamers targeting a cellular component of translation initiation. Mol. Ther. Nucleic Acids 2016, 5, e308. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P.; Richarte, A.M.; Phillips, T.; Andrews, C.; Lee, J.S. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes 2012, 5, 633. [Google Scholar] [CrossRef] [PubMed]
- Balkhy, H.H.; Memish, Z.A. Rift valley fever: An uninvited zoonosis in the arabian peninsula. Int. J. Antimicrob. Agents 2003, 21, 153–157. [Google Scholar] [CrossRef]
- Mansfield, K.L.; Banyard, A.C.; McElhinney, L.; Johnson, N.; Horton, D.L.; Hernandez-Triana, L.M.; Fooks, A.R. Rift valley fever virus: A review of diagnosis and vaccination, and implications for emergence in europe. Vaccine 2015, 33, 5520–5531. [Google Scholar] [CrossRef] [PubMed]
- Ruigrok, R.W.; Crepin, T.; Kolakofsky, D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011, 14, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Ellenbecker, M.; Sears, L.; Li, P.; Lanchy, J.M.; Lodmell, J.S. Characterization of RNA aptamers directed against the nucleocapsid protein of rift valley fever virus. Antivir. Res. 2012, 93, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, L.; Vapalahti, O. Tick-borne encephalitis. Lancet 2008, 371, 1861–1871. [Google Scholar] [CrossRef]
- Kondratov, I.G.; Khasnatinov, M.A.; Potapova, U.V.; Potapov, V.V.; Levitskii, S.A.; Leonova, G.N.; Pavlenko, E.V.; Solovarov, I.S.; Denikina, N.N.; Kulakova, N.V.; et al. Obtaining aptamers to a fragment of surface protein E of tick-borne encephalitis virus. Dokl. Biochem. Biophys. 2013, 448, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Hsiao, W.H.; Lee, H.C.; Wu, S.C.; Cheng, J.W. Selection and characterization of DNA aptamers targeting all four serotypes of dengue viruses. PLoS ONE 2015, 10, e0131240. [Google Scholar] [CrossRef] [PubMed]
- Rodenhuis-Zybert, I.A.; Wilschut, J.; Smit, J.M. Dengue virus life cycle: Viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010, 67, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Balinsky, C.A.; Schmeisser, H.; Ganesan, S.; Singh, K.; Pierson, T.C.; Zoon, K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013, 87, 13094–13106. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang, J.; Ye, W.; Wang, Y.; Ye, C.; Weng, D.; Gao, H.; Zhang, F.; Xu, Z.; Lei, Y. Isolation of endogenously assembled RNA-protein complexes using affinity purification based on streptavidin aptamer S1. Int. J. Mol. Sci. 2015, 16, 22456–22472. [Google Scholar] [CrossRef] [PubMed]
- Binning, J.M.; Wang, T.; Luthra, P.; Shabman, R.S.; Borek, D.M.; Liu, G.; Xu, W.; Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Development of RNA aptamers targeting ebola virus VP35. Biochemistry 2013, 52, 8406–8419. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Moller, P.; Bick, M.J.; Wurr, S.; Becker, S.; Gunther, S.; Kummerer, B.M. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 2007, 81, 2391–2400. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, X.; Gao, G. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell 2010, 1, 752–759. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. Sars and mers: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.G.; Jeon, I.J.; Kim, J.D.; Song, M.S.; Han, S.R.; Lee, S.W.; Jung, H.; Oh, J.W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 2009, 134, 1896–1901. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Woo, H.M.; Kim, K.S.; Oh, J.W.; Jeong, Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011, 112, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Lee, N.R.; Yeo, W.S.; Jeong, Y.J.; Kim, D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus ntpase/helicase. Biochem. Biophys. Res. Commun. 2008, 366, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Shum, K.T.; Tanner, J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the sars coronavirus helicase. Chembiochem 2008, 9, 3037–3045. [Google Scholar] [CrossRef] [PubMed]
- Pecoul, B.; Chirac, P.; Trouiller, P.; Pinel, J. Access to essential drugs in poor countries: A lost battle? JAMA 1999, 281, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Wrenger, C. Disease-specific biomarker discovery by aptamers. Cytom. Part A 2009, 75A, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Scholte, F.E.; Chiang, C.; Albulescu, I.C.; Nichols, C.; He, Z.; Lin, R.; Snijder, E.J.; van Hemert, M.J.; Hiscott, J. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type i interferon-independent stimulation of the innate antiviral response. J. Virol. 2014, 88, 4180–4194. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Perez, N.; Ramos, E.; Garcia-Hernandez, M.; Pinto, C.; Soto, M.; Martin, M.E.; Gonzalez, V.M. Molecular and functional characterization of ssDNA aptamers that specifically bind leishmania infantum pabp. PLoS ONE 2015, 10, e0140048. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.; Gray, N.K. Poly(a)-binding protein (PABP): A common viral target. Biochem. J. 2010, 426, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Lakshmipriya, T.; Chen, Y.; Arshad, M.K.; Kerishnan, J.P.; Ruslinda, A.R.; Al-Douri, Y.; Voon, C.H.; Hashim, U. Cell-targeting aptamers act as intracellular delivery vehicles. Appl. Microbiol. Biotechnol. 2016, 100, 6955–6969. [Google Scholar] [CrossRef] [PubMed]
- Ogunwuyi, O.; Kumari, N.; Smith, K.A.; Bolshakov, O.; Adesina, S.; Gugssa, A.; Anderson, W.A.; Nekhai, S.; Akala, E.O. Antiretroviral drugs-loaded nanoparticles fabricated by dispersion polymerization with potential for HIV/AIDS treatment. Infect. Dis. 2016, 9, 21–32. [Google Scholar]
- Makita-Chingombe, F.; Kutscher, H.L.; DiTursi, S.L.; Morse, G.D.; Maponga, C.C. Poly(lactic-co-glycolic) acid-chitosan dual loaded nanoparticles for antiretroviral nanoformulations. J. Drug Deliv. 2016, 2016, 3810175. [Google Scholar] [CrossRef] [PubMed]
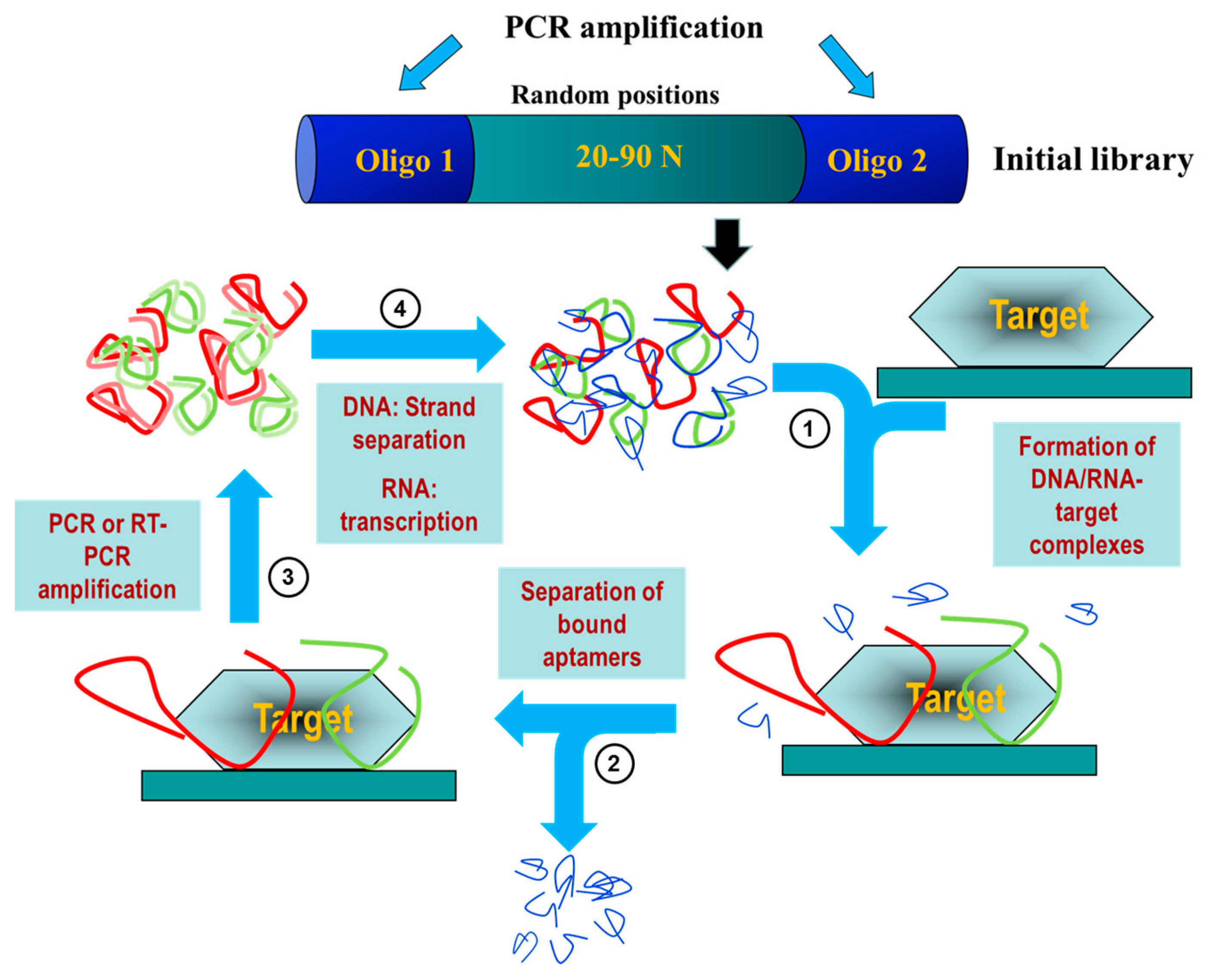
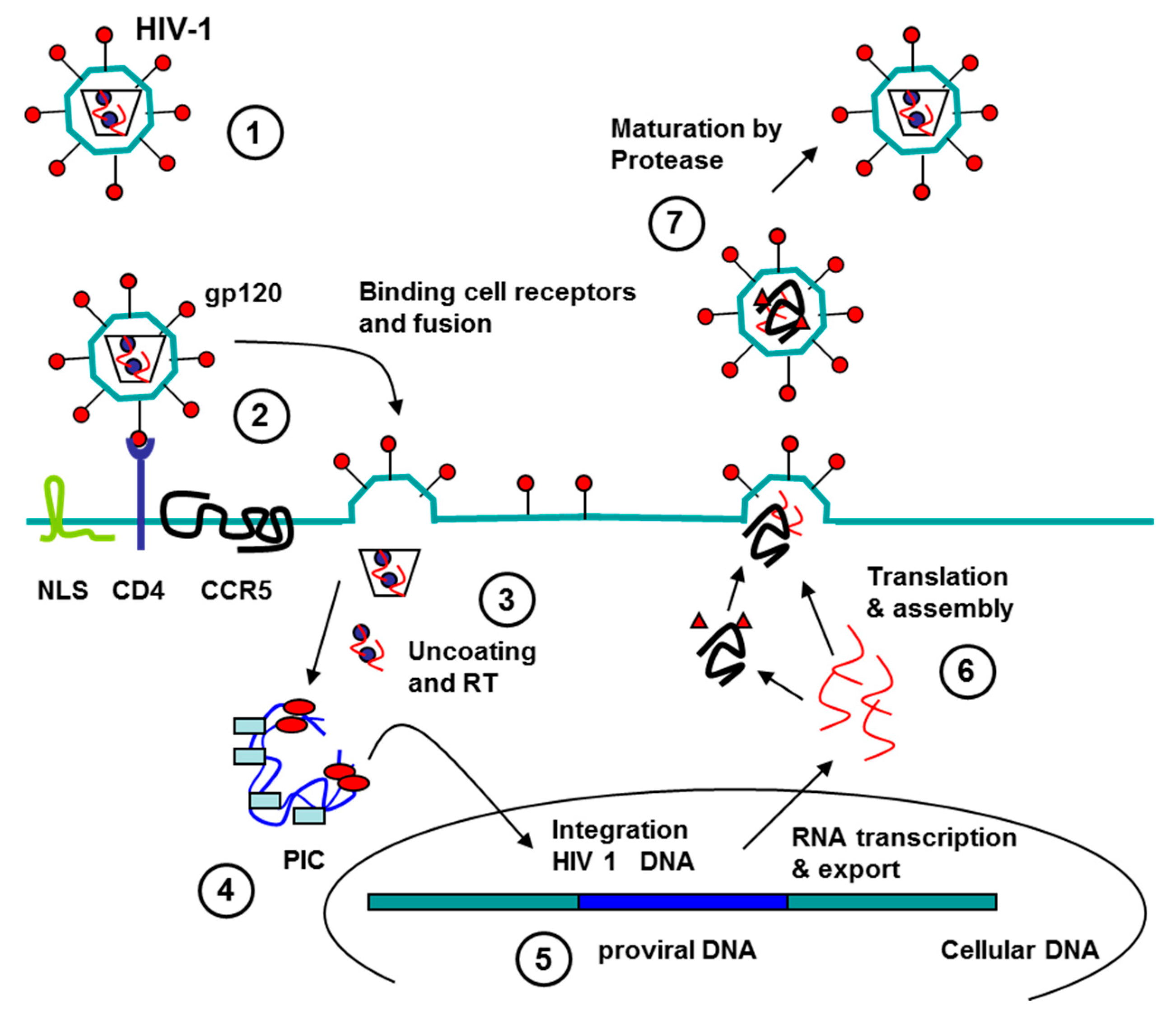
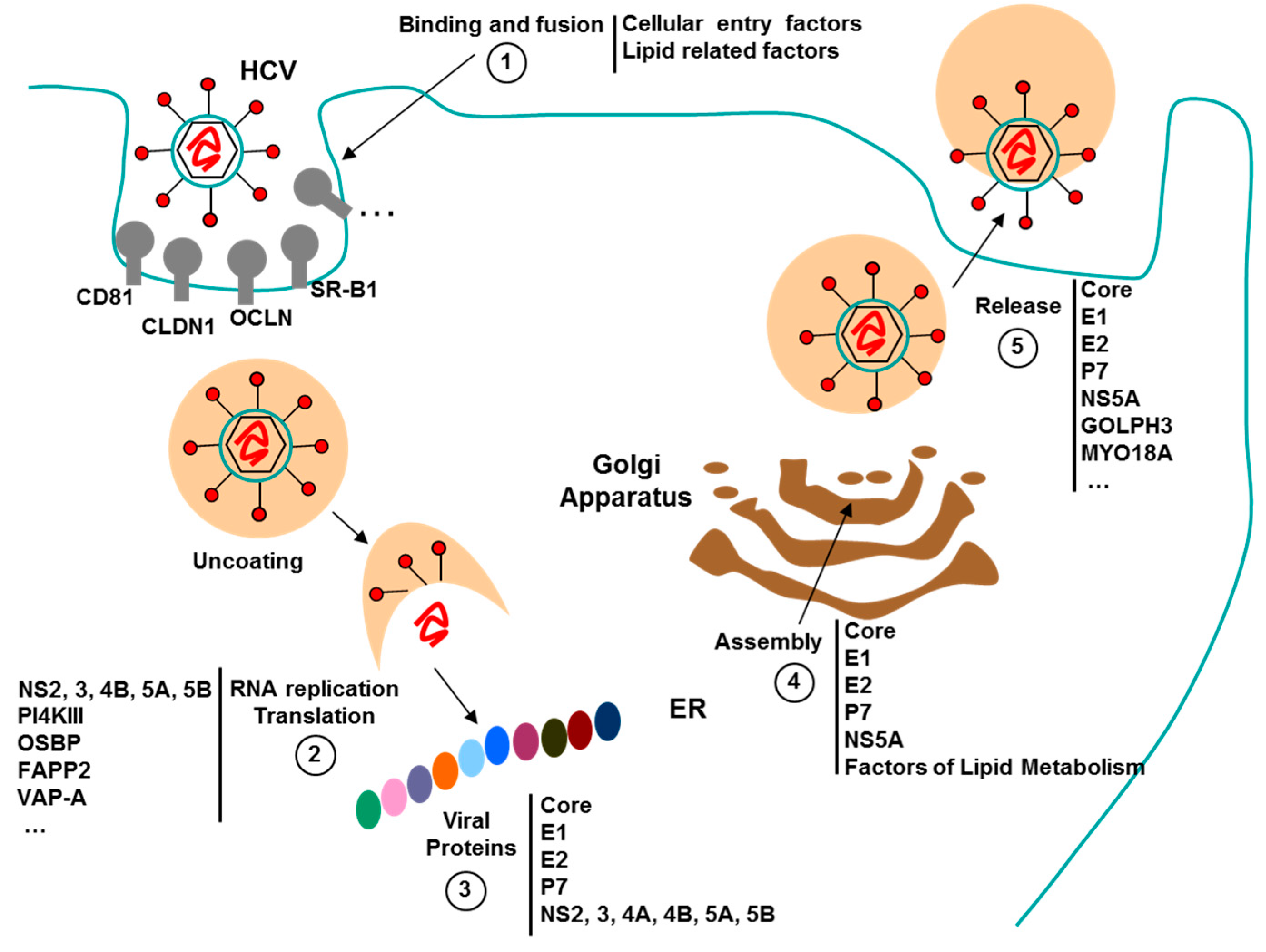
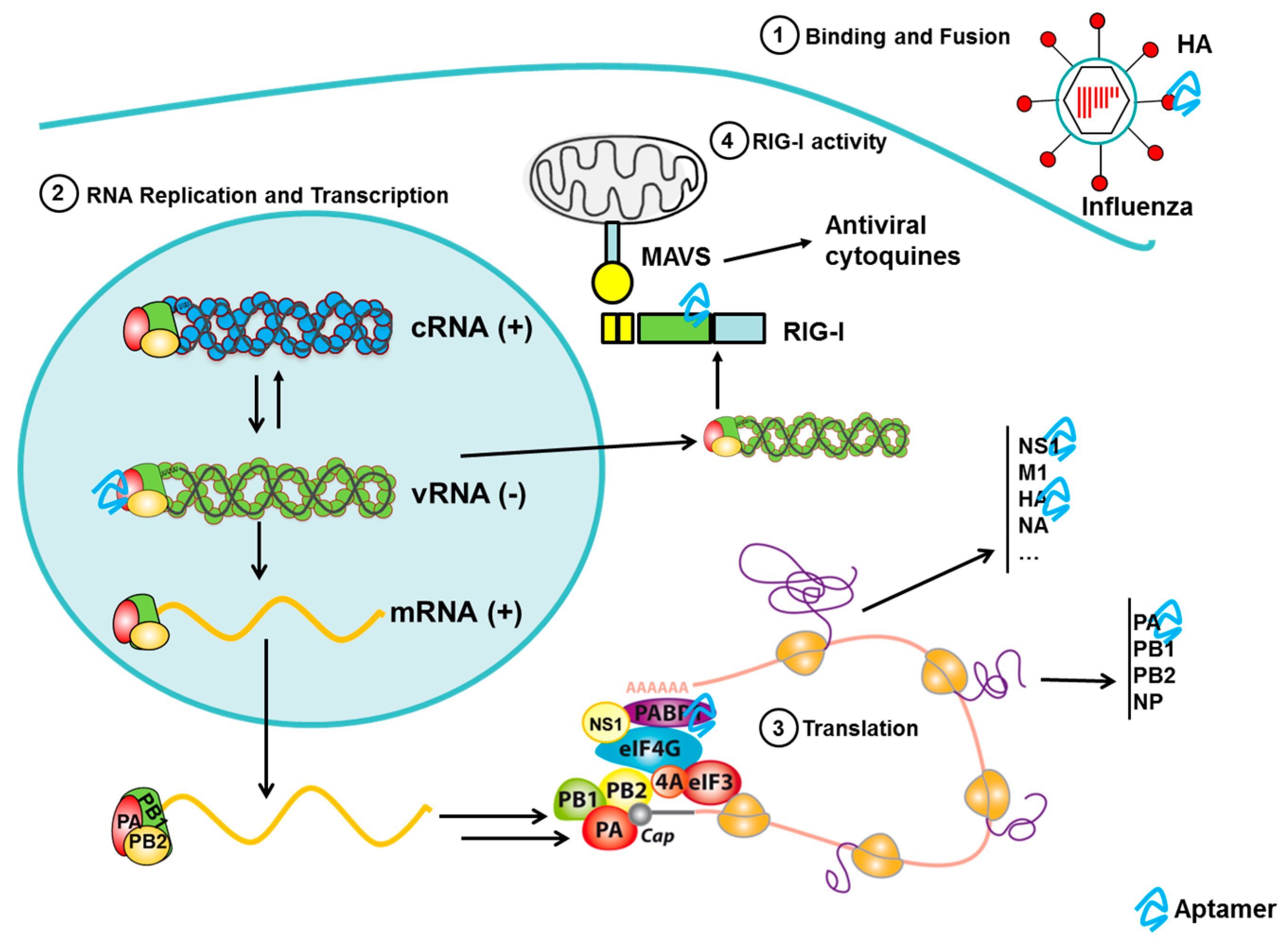
| Characteristic | Advantage |
|---|---|
| Aptamers are identified through an in vitro process (SELEX) | Selection conditions can be manipulated to obtain aptamers stable in a wide range of environmental conditions |
| Aptamers may be obtained against non-immunogenic proteins and toxins | |
| Aptamers are produced by chemical synthesis | Little or no batch to batch variation |
| Aptamers can be modified increasing their stability | |
| Reporter molecules can be attached to aptamers at precise locations not involved in binding | |
| Aptamers are oligonucleotides | They can be amplified to be easily detected |
| Denatured aptamers can be regenerated within minutes | |
| Aptamers are stable to long term storage and can be transported at ambient temperature | |
| They are not immunogenic | |
| Their small size allows for more efficient entry into the cell and its compartments |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| HIV | n.d. | DNA | Tat, Rev viral protein | [23,24] |
| ASn, ALn, BSn, BLn | RNA | LTR viral sequence | [28] | |
| RNApt16 | RNA | 5′-untranslated region of HIV-1 genome | [29] | |
| PR10.1, PR10.9, PR10.13, PR10.18 | RNA | Protease viral protein | [32] | |
| T30177, T30695 | DNA | Integrase viral protein | [35,37,38,39] | |
| 37NT | DNA | Reverse Transcriptase viral protein | [41] | |
| RT5, RT6 and RT47 | DNA | Reverse Transcriptase viral protein | [42] | |
| ODNs 93 and 112 93del, 112del | DNA | RNAse activity associated to RT viral protein | [43,44,45] | |
| aptamers 8–6, 8–10, 8–13 | RNA | Nucleocapsid viral protein | [47] | |
| Hotoda’s sequence and modifications | DNA | gp120 viral protein | [48,49,50,51,52,53,54,55,56] | |
| B4, B40, B40t77 | RNA | gp120 viral protein | [55,56,57,58,59] | |
| ML6.8t33-82, ML8.20t14-75. DP6-12 | RNA | Gag viral protein | [62,63] | |
| G3 | RNA | CCR5 cellular receptor | [25] | |
| AS1411 | DNA | NLC cellular protein | [14] | |
| n.d. | DNA | CD4 cellular receptor | [27] |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| HBV | anti-HBsAg RNA aptamer | RNA | HBsAg | [73] |
| HBs-A22 | RNA | HBsAg | [74] | |
| HO1, HO2, HO3 | DNA | HBsAg | [75] | |
| Class I, An & Class II, Sn | RNA | MiniP protein | [76] | |
| Apt No 28 | DNA | Core | [77] | |
| AO-01 | DNA | Capsid | [78] |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| HCV | 9-n | RNA | Core | [80] |
| ZEn | DNA | E2 Glycoprotein | [81] | |
| C4, 7, 42, 97, 103 & 104 | DNA | Core | [82] | |
| 9-15 | DNA | Core | [83] | |
| anti-HCVE2 aptamers | DNA (5-benzylaminocarbonyl-dUridine(Bz-dU) | E2 | [84] | |
| RNA aptamer | RNA | Helicase | [85] | |
| biotinylated RNA oligonucleotide | RNA | NS5B | [86] | |
| 2-02, 3-07, 0207 and 0702 aptamers | RNA | IRES element | [90,91,92] | |
| P6-n, HH363-n | RNA | IRES element | [93,94,95] | |
| Family I, II, III (AP30) | RNA | Minus-IRES domain I | [96,97] | |
| P58 and P78 aptamers | RNA | CRE element | [98,99] | |
| NS2-1, 2, & 3 | DNA | NS2 | [100] | |
| 10G-1, G6-16, G6-19 | RNA | NS3 | [101,102] | |
| G9-I, G9-II, G9-III | RNA | NS3 | [103,104] | |
| HDV-G9-II | RNA | NS3 | [105,106] | |
| NEO-III, DNEO-III. NEO-III-14U, 5′-14U-NEO-III | RNA | NS3 | [107] | |
| aptamer 5, 5 (1-30), 5ss, 5D, 5m, 5mS, 5mL1, 5mL2, ss5m-3′X, 3′(+) UTR, UC-3′X, 3′X | RNA | NS3 | [108] | |
| NEO-35-sX, G925-sX | RNA | NS3 | [109] | |
| NS5A-1,2,3,4 &5 | DNA | NS5A | [110] | |
| A,1, A.2, B.1, B,2, B,3, C.1 & C.2 | RNA | NS5B | [111] | |
| Class A, B, C & D (ODN n) | DNA | NS5B | [112,113] | |
| R-F, R-OH, Gal-PEG-R-F t2, Chol R-F t2, chol-aptamer | RNA | NS5B | [114,115] | |
| r10/N, r8a, r8b, r8c | DNA | NS5, genotipe 3a | [116] | |
| E1E2-1, 2, 3, 4, 5, 6 | DNA | E1 E2 | [117] |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| HPV | F2 and F4 | RNA | E6 | [121] |
| G5α3N.4 | RNA | E7 | [122] | |
| Sc5-c3 | RNA | HPV-16 L1 virus-like particles (VLPs) | [123] | |
| DNA | HPV-transformed cervical cancer cells | [124] | ||
| A2 | RNA | E7 | [125,126] | |
| RNA | HPV-16 E6/E7-human tonsillar epithelial cells (HTECs) | [127] | ||
| HSV | GC-rich RNA aptamer | RNA | ICP27 | [129] |
| aptamer-1 and aptamer-5 mini-1 aptamer (44-mer) | RNA | gD protein | [130] | |
| G7a | RNA | gD protein | [131] |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| Influenza H3N2 | Clone B | RNA | HA | [139] |
| Influenza H3N2 | PN30-10-16 PN30-10-1 | RNA | HA | [140,141] |
| Influenza H1N1 | D26, D12 | RNA | HA | [142] |
| Influenza H5N1 | (1), (2), (3) | DNA | HA (4 cycles) and virus | [143,148,149,150,151,152] |
| Influenza H1N1 | n.d. | DNA | virus | [144,145,146] |
| Influenza H5N1 | n.d. | DNA | HA | [148] |
| Influenza H5Nx | IF10, IF15, IF20, IF22, IF23 | DNA | Virus | [153] |
| Influenza H3N2 | A21, A22 | DNA | HA-(91–261) peptide | [154] |
| Influenza H5N1 | A05, A10 | DNA | HA from H5N1 | [155] |
| Influenza H1N1 | CP9P526, CP9P528, CP9P536, CP9P554, CP9P596 | DNA | Sialic Acid Receptor (SAR) epitope | [156] |
| BV42, BV35r | DNA | 2nd generation (A22) | [157] | |
| Influenza H1N1 | 1, 2 | DNA | HA | [158] |
| Influenza H9N2 | C7, C7-35M | DNA | H9 peptide (HA,101–257) | [159] |
| Influenza H5N1 | 8-1, 8-3, 8-10 | RNA | HA | [160] |
| Influenza H9N2 | A9, B4 | DNA | HA | [161] |
| Influenza A | n.d. | DNA | NS1 | [163] |
| Influenza H5N1 | PAN-1, PAN-2, PAN-3, PAN-4, PAN-5, PAN-6 | DNA | PA | [168] |
| Virus | Name | DNA/RNA | Target | References |
|---|---|---|---|---|
| Rift Valley fever | n.d. | RNA | nucleocapsid protein | [179] |
| Tick-borne encephalitis | Population | DNA | surface protein E | [181] |
| Dengue | S15 | DNA | DENV-2 envelop protein, domain III | [182] |
| Ebola | 1G8–14, 2F11-14 | RNA | protein 35 (VP35) | [186] |
| Ebola | 21E, 21K, 21I | RNA | zinc-finger antiviral protein | [188] |
| Severe acute respiratory syndrome | n.d. | DNA/RNA | nucleocapsid protein | [190,191] |
| Severe acute respiratory syndrome | n.d. | DNA/RNA | non-structural nsp13 protein | [192,193] |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, V.M.; Martín, M.E.; Fernández, G.; García-Sacristán, A. Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals 2016, 9, 78. https://doi.org/10.3390/ph9040078
González VM, Martín ME, Fernández G, García-Sacristán A. Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals. 2016; 9(4):78. https://doi.org/10.3390/ph9040078
Chicago/Turabian StyleGonzález, Víctor M., M. Elena Martín, Gerónimo Fernández, and Ana García-Sacristán. 2016. "Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses" Pharmaceuticals 9, no. 4: 78. https://doi.org/10.3390/ph9040078
APA StyleGonzález, V. M., Martín, M. E., Fernández, G., & García-Sacristán, A. (2016). Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals, 9(4), 78. https://doi.org/10.3390/ph9040078







