Non-Monotonic Survival of Staphylococcus aureus with Respect to Ciprofloxacin Concentration Arises from Prophage-Dependent Killing of Persisters
Abstract
:1. Introduction
2. Results and Discussion
2.1. Survival of S. aureus is Non-monotonic with Respect to Ciprofloxacin Concentration

2.2. Ciprofloxacin Induction of Cell Lysis and Bacteriophage Release is Quenched at High Concentrations
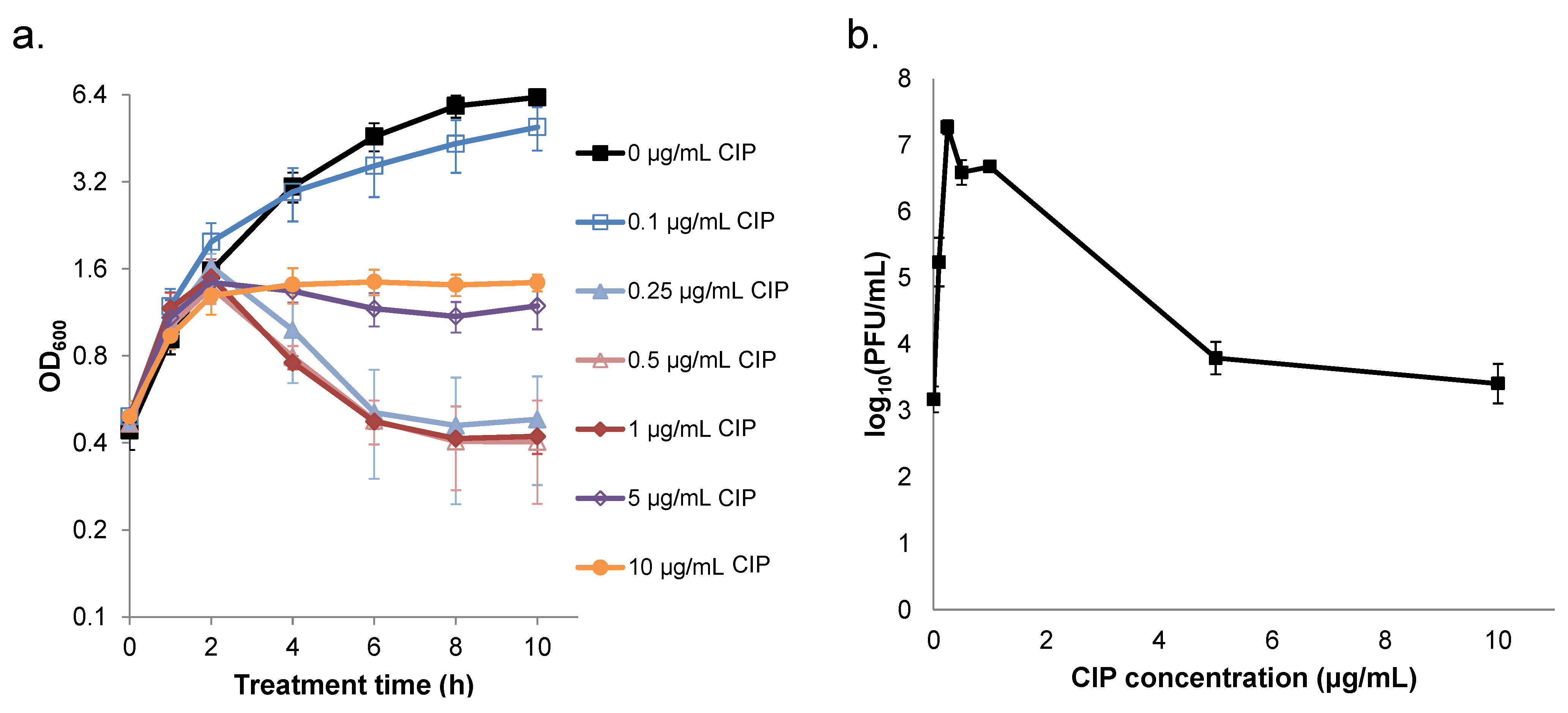
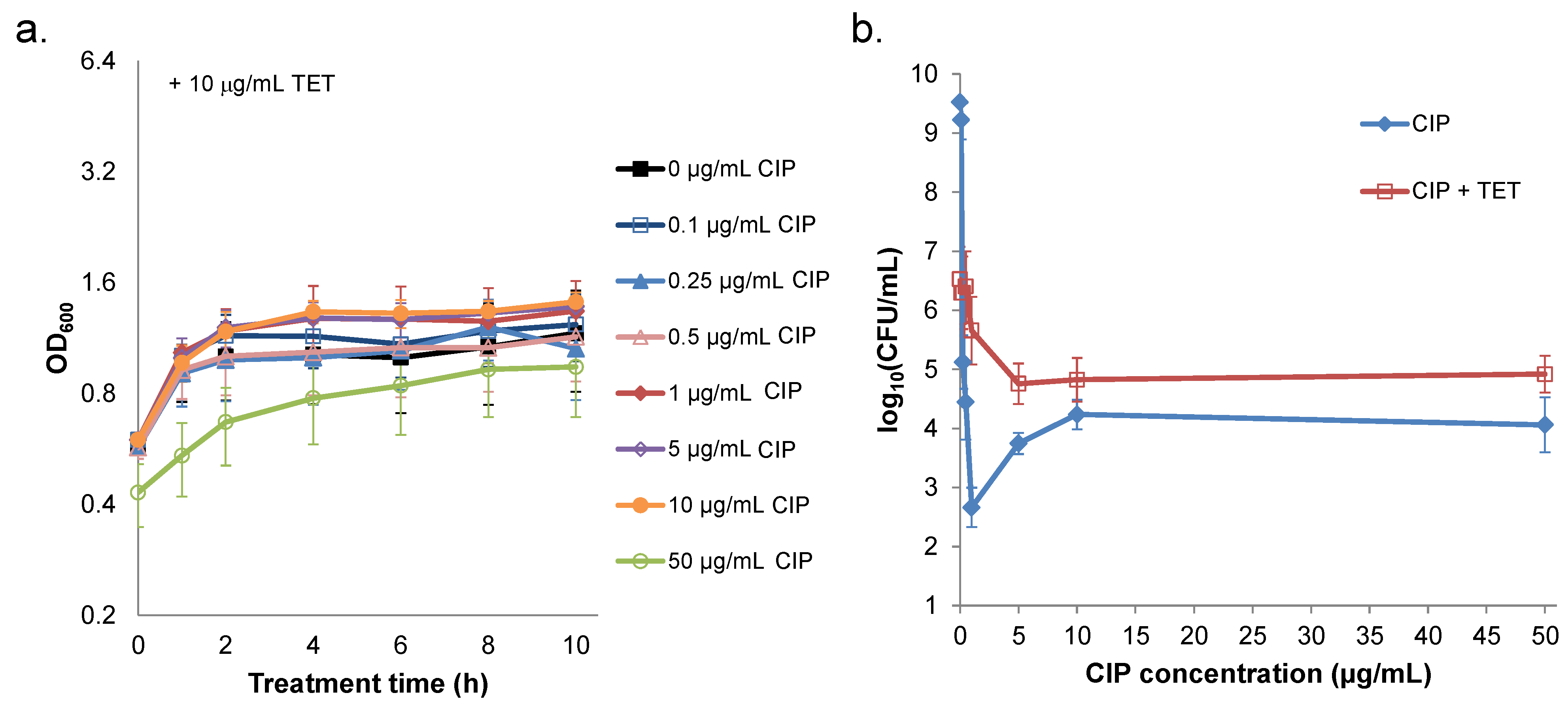
2.3. Prevention of Prophage Induction Protects Persisters and Leads to Monotonic Survival
2.4. Quenched Induction of DNA Damage-inducible GFP Reporter at High Ciprofloxacin Concentrations
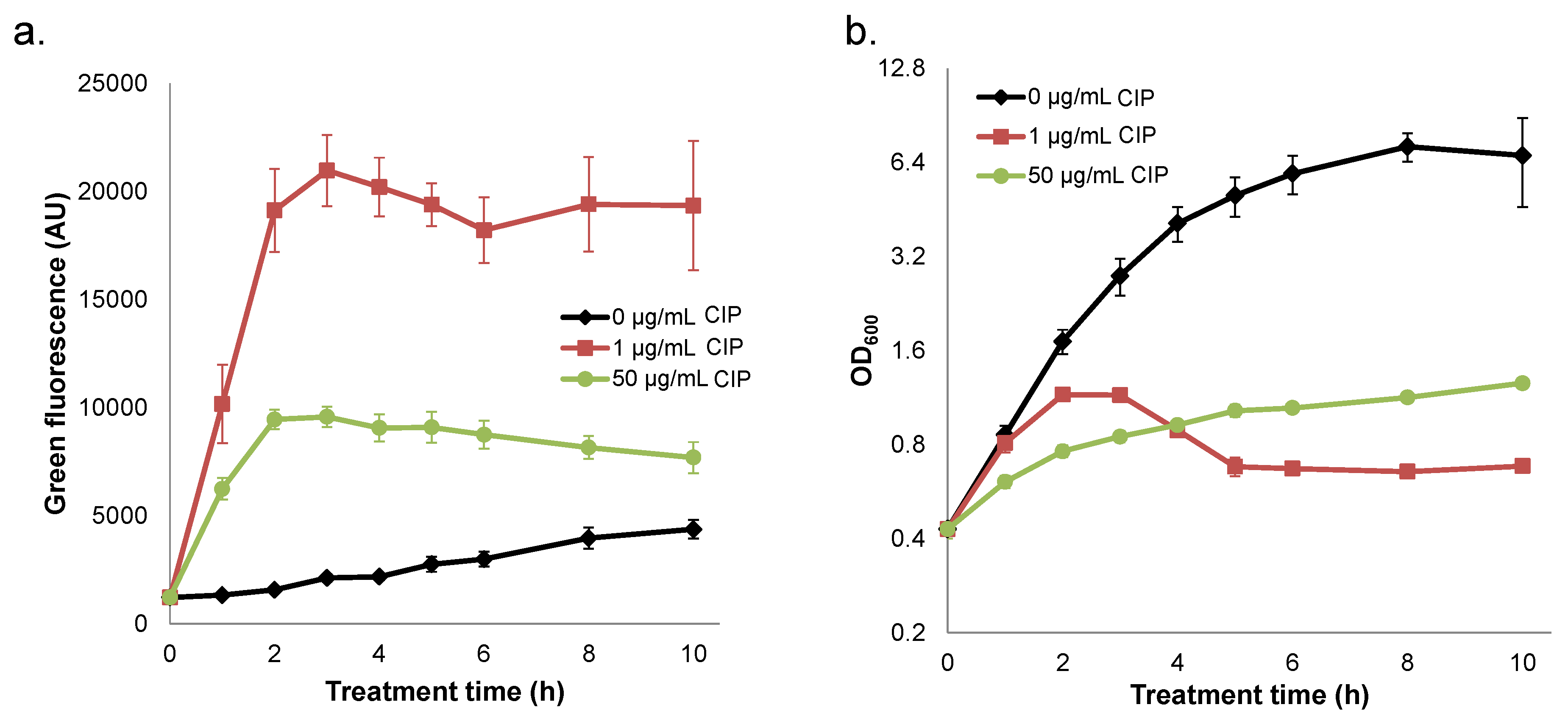
2.5. Interplay between Prophage Induction and Translational Inhibition by Ciprofloxacin Yields Non-Monotonic Survival
3. Experimental Section
3.1. Bacterial Strains, Chemicals, and Media
| Strains/Plasmids | Description | Source |
|---|---|---|
| Bacterial Strains: | ||
| S. aureus 8325 | Wild-type strain | [46] |
| S. aureus 8325 pES10 | Wild-type strain harboring pES10 | This study |
| S. aureus RN4220 | Cloning strain derivative of S. aureus NCTC 8325 | Lab of Thomas Muir |
| S. aureus RN10395 | 80α lysogen used for phage transduction to facilitate plasmid transformation | Lab of Richard Novick |
| E. coli SA08B | Cloning strain for plasmid construction | Lucigen |
| Plasmids: | ||
| pCN51 | pCN shuttle vector (Pcad-cadC), ermC (S. aureus), amp ColE1 ori (E. coli) | Lab of Thomas Muir |
| pUA66 | Plasmid source of gfpmut2 | [47] |
| pES1 | pCN51 with gfpmut2 (gene 10 RBS) | This study |
| pES4 | pES1 with MCS replacing Pcad-cadC | This study |
| pES5 | pES4 with gfpmut2 (TIR RBS) | This study |
| pES10 | PrecA-TIR-gfpmut2 | This study |
3.2. Fluorescent Reporter Construction
| Primer | Sequence |
|---|---|
| gfpmut2_FW_KpnI + 3stop_2 | 5′-CTGACAGGTACCGTTAACTAATTAATTTAAGAAGGAGATATACATATGAG-3′ |
| gfpmut2 + EcoRI_REV | 5′-AGCATAGAATTCTTATTTGTATAGTTCATCCATGCCA-3′ |
| MCS_FW | 5′-TCATCATGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGGCATGC-3′ |
| MCS_REV | 5′-CTAGTAGGCATGCCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACC-3′ |
| gfpmut2_KpnI_TIR_FW | 5′-CATCATGGTACCTGATTAACTTTATAAGGAGGAAAAACATATGAGTAAAGGAGAAGA-3′ |
| SbfI_PrecA_FW | 5′-TAGTAGCCTGCAGGATGATGGTATTACTAATGGTGC-3′ |
| KpnI_PrecA_REV | 5′-TAGTAGGGTACCAGCGAGACCTCCTAATTGAAATTGCTA-3′ |
3.3. Measurement of MIC
3.4. Antibiotic Treatment Assays
3.5. Plaque Assays
3.6. Fluorescence Assays
3.7. Statistical Analyses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Valaperta, R.; Tejada, M.R.; Frigerio, M.; Moroni, A.; Ciulla, E.; Cioffi, S.; Capelli, P.; Costa, E. Staphylococcus aureus nosocomial infections: The role of a rapid and low-cost characterization for the establishment of a surveillance system. New Microbiol. 2010, 33, 223–232. [Google Scholar] [PubMed]
- Valentino, M.D.; Foulston, L.; Sadaka, A.; Kos, V.N.; Villet, R.A.; Santa Maria, J., Jr.; Lazinski, D.W.; Camilli, A.; Walker, S.; Hooper, D.C.; et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 2014, 5, e01729-14. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 2011, 473, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.J.; Levin, B.R. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Lechner, S.; Lewis, K.; Bertram, R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 2012, 22, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: An in vitro study. J. Med. Microbiol. 2009, 58, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 2011, 14, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.M.; Fazen, C.H.; Henry, T.C.; Mok, W.W.; Orman, M.A.; Sandvik, E.L.; Volzing, K.G.; Brynildsen, M.P. The role of metabolism in bacterial persistence. Front. Microbiol. 2014, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Kint, C.I.; Verstraeten, N.; Fauvart, M.; Michiels, J. New-found fundamentals of bacterial persistence. Trends Microbiol. 2012, 20, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Gerdes, K.; Lewis, K.; McKinney, J.D. A problem of persistence: Still more questions than answers? Nat. Rev. Microbiol. 2013, 11, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Donegan, N.P.; Memmi, G.; Cheung, A.L. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8871–8879. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, S.; Zhang, Y.; Yamaguchi, Y.; Chen, L.; Kreiswirth, B.N.; Inouye, M. Staphylococcus aureus yoeB homologues inhibit translation initiation. J. Bacteriol. 2009, 191, 5868–5872. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Thompson, E.T.; Fu, Z.; Cheung, A.L. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 2010, 192, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Donegan, N.P.; Cheung, A.L. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 2009, 191, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Mechler, L.; Nolle, N.; Krismer, B.; Zelder, M.E.; Götz, F.; Bertram, R. The MazEF toxin-antitoxin system alters the β-lactam susceptibility of Staphylococcus aureus. PloS ONE 2015, 10, e0126118. [Google Scholar] [CrossRef] [PubMed]
- Gefen, O.; Balaban, N.Q. The importance of being persistent: Heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 2009, 33, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Fauvart, M.; De Groote, V.N.; Michiels, J. Role of persister cells in chronic infections: Clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 2011, 60, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Froshauer, S.; Silvia, A.M.; Chidambaram, M.; Sharma, B.; Weinstock, G.M. Sensitization of bacteria to danofloxacin by temperate prophages. Antimicrob. Agents Chemother. 1996, 40, 1561–1563. [Google Scholar] [PubMed]
- Piddock, L.J.; Walters, R.N.; Diver, J.M. Correlation of quinolone MIC and inhibition of DNA, RNA, and protein synthesis and induction of the SOS response in Escherichia coli. Antimicrob. Agents Chemother. 1990, 34, 2331–2336. [Google Scholar] [CrossRef] [PubMed]
- Selva, L.; Viana, D.; Regev-Yochay, G.; Trzcinski, K.; Corpa, J.M.; Lasa, I.; Novick, R.P.; Penades, J.R. Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. USA 2009, 106, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Crumplin, G.C.; Smith, J.T. Nalidixic acid: An antibacterial paradox. Antimicrob. Agents Chemother. 1975, 8, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Cirz, R.T.; Jones, M.B.; Gingles, N.A.; Minogue, T.D.; Jarrahi, B.; Peterson, S.N.; Romesberg, F.E. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 2007, 189, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Orman, M.A.; Mok, W.W.; Brynildsen, M.P. Aminoglycoside-enabled elucidation of bacterial persister metabolism. Curr. Protoc. Microbiol. 2015, 36. [Google Scholar] [CrossRef]
- Lewin, C.S.; Amyes, S.G. The bactericidal activity of DR-3355, an optically active isomer of ofloxacin. J. Med. Microbiol. 1989, 30, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Lewin, C.S.; Morrissey, I.; Smith, J.T. The mode of action of quinolones: The paradox in activity of low and high concentrations and activity in the anaerobic environment. Eur. J. Clin. Microbiol. Infect. Dis. 1991, 10, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Goerke, C.; Koller, J.; Wolz, C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Maiques, E.; Knecht, E.; Lasa, I.; Novick, R.P.; Penades, J.R. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 2005, 56, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Novick, R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 1967, 33, 155–166. [Google Scholar] [CrossRef]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [PubMed]
- Amato, S.M.; Brynildsen, M.P. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PloS ONE 2014, 9, e93110. [Google Scholar] [CrossRef] [PubMed]
- Orman, M.A.; Brynildsen, M.P. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Pearl, S.; Gabay, C.; Kishony, R.; Oppenheim, A.; Balaban, N.Q. Nongenetic individuality in the host-phage interaction. PLoS Biol. 2008, 6, e120. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.V.; Kokjohn, T.A. General microbiology of recA: Environmental and evolutionary significance. Annu. Rev. Microbiol. 1990, 44, 365–394. [Google Scholar] [CrossRef] [PubMed]
- Bae, T.; Baba, T.; Hiramatsu, K.; Schneewind, O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 2006, 62, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Genetic systems in staphylococci. Method. Enzymol. 1991, 204, 587–636. [Google Scholar]
- Cirz, R.T.; Jones, M.B.; Gingles, N.A.; Minogue, T.D.; Jarrahi, B.; Peterson, S.N.; Romesberg, F.E. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 2007, 189, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Zaslaver, A.; Bren, A.; Ronen, M.; Itzkovitz, S.; Kikoin, I.; Shavit, S.; Liebermeister, W.; Surette, M.G.; Alon, U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 2006, 3, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Anton, A.I.; Barry, P.; Alfonso, B.; Fang, Y.; Novick, R.P. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70, 6076–6085. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.L.; Boles, B.R.; Lauderdale, K.J.; Thoendel, M.; Kavanaugh, J.S.; Horswill, A.R. Fluorescent reporters for Staphylococcus aureus. J. Microbiol. Meth. 2009, 77, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; Lindow, S.E. An improved gfp cloning cassette designed for prokaryotic transcriptional fusions. Gene 1997, 191, 149–153. [Google Scholar] [CrossRef]
- Bose, J.L.; Fey, P.D.; Bayles, K.W. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 2013, 79, 2218–2224. [Google Scholar] [CrossRef] [PubMed]
- Monk, I.R.; Shah, I.M.; Xu, M.; Tan, M.-W.; Foster, T.J. Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 2012, 3, e00277-11. [Google Scholar] [CrossRef] [PubMed]
- Thompson, N.E.; Pattee, P.A. Transformation in Staphylococcus aureus: Role of bacteriophage and incidence of competence among strains. J. Bacteriol. 1977, 129, 778–788. [Google Scholar] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandvik, E.L.; Fazen, C.H.; Henry, T.C.; Mok, W.W.K.; Brynildsen, M.P. Non-Monotonic Survival of Staphylococcus aureus with Respect to Ciprofloxacin Concentration Arises from Prophage-Dependent Killing of Persisters. Pharmaceuticals 2015, 8, 778-792. https://doi.org/10.3390/ph8040778
Sandvik EL, Fazen CH, Henry TC, Mok WWK, Brynildsen MP. Non-Monotonic Survival of Staphylococcus aureus with Respect to Ciprofloxacin Concentration Arises from Prophage-Dependent Killing of Persisters. Pharmaceuticals. 2015; 8(4):778-792. https://doi.org/10.3390/ph8040778
Chicago/Turabian StyleSandvik, Elizabeth L., Christopher H. Fazen, Theresa C. Henry, Wendy W.K. Mok, and Mark P. Brynildsen. 2015. "Non-Monotonic Survival of Staphylococcus aureus with Respect to Ciprofloxacin Concentration Arises from Prophage-Dependent Killing of Persisters" Pharmaceuticals 8, no. 4: 778-792. https://doi.org/10.3390/ph8040778
APA StyleSandvik, E. L., Fazen, C. H., Henry, T. C., Mok, W. W. K., & Brynildsen, M. P. (2015). Non-Monotonic Survival of Staphylococcus aureus with Respect to Ciprofloxacin Concentration Arises from Prophage-Dependent Killing of Persisters. Pharmaceuticals, 8(4), 778-792. https://doi.org/10.3390/ph8040778





