The Expanding Role of Non-Coding RNAs in Neurodegenerative Diseases: From Biomarkers to Therapeutic Targets
Abstract
1. Introduction
2. The Biological Basis of ncRNAs in the Nervous System
2.1. miRNA and the Nervous System
2.2. lncRNAs and the Nervous System
2.3. circRNAs and the Nervous System
2.4. piRNAs and the Nervous System
2.5. RNA-Binding Proteins (RBPs) and Epitranscriptome
3. ncRNAs in Specific Neurodegenerative Diseases
3.1. ncRNAs in AD: Regulation of Amyloid-Beta Production, Tau Pathology, and Synaptic Dysfunction
3.2. ncRNAs in PD: Modulation of Alpha-Synuclein Aggregation, Autophagy-Lysosomal Pathways, and Mitochondrial Homeostasis
3.3. ncRNAs in HD: RNA Toxicity from CAG Repeats, Splicing Dysregulation, and RNA-Targeted Therapeutic Interventions
3.4. ncRNA Dysregulation in ALS and Frontotemporal Dementia (FTD): Impacts on Protein Aggregation, miRNA Biogenesis, and RNA Homeostasis
4. ncRNAs as Biomarkers
4.1. Sample Types and Preprocessing: Biofluid Sources, Handling Protocols, and Quality Control for Reliable ncRNA Detection
4.2. Detection Platforms: Comparative Analysis of qPCR, ddPCR, and NGS for ncRNA Quantification in Neurodegenerative Diseases
4.3. Machine Learning Approaches: Integration of ncRNA Data with Multimodal Models for Predictive Diagnostics and Mechanistic Insights
5. From Mechanism to Therapy: RNA Interventions and Delivery
5.1. Therapeutic Modalities: The RNA Intervention Spectrum and Mechanisms from ASO/siRNA to CRISPRi/a
5.2. Clinical and Preclinical Evidence: Translational Progress Across ALS, Tauopathy, and HD
5.3. Brain Delivery Strategies: Comparing Intrathecal/Intraventricular Routes, Receptor-Mediated BBB Shuttles, and Exosome Carriers
5.4. Safety and Quality—Innate Immune Activation, Organ Burden, CMC Consistency, and Biomarker Monitoring
6. Key Challenges and Future Prospects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3′UTR | 3′ untranslated region |
| AAV | adeno-associated virus |
| AAV-miRNAs | adeno-associated virus vectors expressing artificial microRNAs |
| AD | Alzheimer’s disease |
| ADAR | adenosine deaminase acting on RNA |
| AI | artificial intelligence |
| ALS | amyotrophic lateral sclerosis |
| ALSFRS-R | ALS Functional Rating Scale–Revised |
| AMT-130 | AAV5-miHTT gene therapy AMT-130 for Huntington’s disease |
| APP | amyloid precursor protein |
| ASO | antisense oligonucleotide |
| ASOs | antisense oligonucleotides |
| AUC | area under the receiver operating characteristic curve |
| Aβ | amyloid-β peptide |
| BBB | blood–brain barrier |
| C9ORF72 | chromosome 9 open reading frame 72 |
| CMA | chaperone-mediated autophagy |
| CNS | central nervous system |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| CRISPRi | CRISPR interference |
| CSF | cerebrospinal fluid |
| ddPCR | droplet digital polymerase chain reaction |
| DNA | deoxyribonucleic acid |
| EV | extracellular vesicle |
| EVs | extracellular vesicles |
| FTD | frontotemporal dementia |
| GPCR | G protein–coupled receptor |
| HD | Huntington’s disease |
| HTT | huntingtin |
| ICV | intracerebroventricular |
| ISR | integrated stress response |
| IT | intrathecal |
| LAMP2A | lysosome-associated membrane protein 2A |
| LLPS | liquid–liquid phase separation |
| LTP | long-term potentiation |
| MAPT | microtubule-associated protein tau |
| MAPTRx | tau-targeting antisense oligonucleotide MAPT(Rx) |
| MISEV2023 | Minimal Information for Studies of Extracellular Vesicles 2023 |
| MRI | magnetic resonance imaging |
| NDD | neurodegenerative disease |
| NDDs | neurodegenerative diseases |
| NDEVs | neuron-derived extracellular vesicles |
| NfL | neurofilament light chain |
| NGS | next-generation sequencing |
| NMDA | N-methyl-D-aspartate |
| NRI | net reclassification index |
| PD | Parkinson’s disease |
| PET | positron emission tomography |
| PIWI | P-element-induced wimpy testis protein family |
| PSP | progressive supranuclear palsy |
| QC | quality control |
| RAN | repeat-associated non-ATG |
| RBP | RNA-binding protein |
| RBPs | RNA-binding proteins |
| RISC | RNA-induced silencing complex |
| RNA | ribonucleic acid |
| RNAi | RNA interference |
| RVG | rabies virus glycoprotein |
| SNP | single-nucleotide polymorphism |
| SNPs | single-nucleotide polymorphisms |
| SNCA | alpha-synuclein |
| SOD1-ALS | amyotrophic lateral sclerosis with SOD1 mutations |
| TfR1 | transferrin receptor 1 |
| XAI | explainable artificial intelligence |
| ceRNA | competing endogenous RNA |
| circRNAs | circular RNAs |
| iLTP | inhibitory long-term potentiation |
| lncRNA | long non-coding RNA |
| lncRNAs | long non-coding RNAs |
| m(6)A | N6-methyladenosine |
| m6A | N6-methyladenosine |
| mHTT | mutant huntingtin |
| miRNA | microRNA |
| miRNAs | microRNAs |
| mRNA | messenger RNA |
| ncRNA | non-coding RNA |
| ncRNAs | non-coding RNAs |
| nt | nucleotide |
| piRNA | PIWI-interacting RNA |
| piRNAs | PIWI-interacting RNAs |
| pre-miRNAs | precursor microRNAs |
| pri-miRNAs | primary microRNAs |
| qPCR | quantitative polymerase chain reaction |
References
- Alzheimer’s Association. 2024 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2024, 20, 3708–3821. [Google Scholar]
- Steinmetz, J.D.; Seeher, K.M.; Schiess, N.; Nichols, E.; Cao, B.; Servili, C.; Cavallera, V.; Cousin, E.; Hagins, H.; Moberg, M.E.; et al. Global, regional, and national burden of disorders affecting the nervous system, 1990–2021: A systematic analysis for the Global Burden of Disease Study 2021. Lancet Neurol. 2024, 23, 344–381. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, S. RNA dysregulation in neurodegenerative diseases. EMBO J. 2025, 44, 613–638. [Google Scholar] [CrossRef]
- Alzarea, S.I. Non-coding RNA-mediated gene regulation in Alzheimer’s disease pathogenesis: Molecular insights and emerging innovations. Saudi Pharm. J. 2025, 33, 33. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, A.K.; Vij, P.; Lopez, S.; Leslie, S.M.; Doxtater, K.; Khan, M.M.; Yallapu, M.M.; Chauhan, S.C.; Maestre, G.E.; Tripathi, M.K. Long Non-Coding RNAs: New Insights in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2268. [Google Scholar] [CrossRef]
- Ilieva, M.S. Non-Coding RNAs in Neurological and Neuropsychiatric Disorders: Unraveling the Hidden Players in Disease Pathogenesis. Cells 2024, 13, 1063. [Google Scholar] [CrossRef] [PubMed]
- Talebi Taheri, A.; Golshadi, Z.; Zare, H.; Alinaghipour, A.; Faghihi, Z.; Dadgostar, E.; Tamtaji, Z.; Aschner, M.; Mirzaei, H.; Tamtaji, O.R.; et al. The Potential of Targeting Autophagy-Related Non-coding RNAs in the Treatment of Alzheimer’s and Parkinson’s Diseases. Cell. Mol. Neurobiol. 2024, 44, 28. [Google Scholar] [CrossRef]
- Li, J.; Wang, X. Functional roles of conserved lncRNAs and circRNAs in eukaryotes. Noncoding RNA Res. 2024, 9, 1271–1279. [Google Scholar] [CrossRef]
- Song, G.; Yang, Z.; Guo, J.; Zheng, Y.; Su, X.; Wang, X. Interactions Among lncRNAs/circRNAs, miRNAs, and mRNAs in Neuropathic Pain. Neurotherapeutics 2020, 17, 917–931. [Google Scholar] [CrossRef]
- Li, M.L.; Wang, W.; Jin, Z.B. Circular RNAs in the Central Nervous System. Front. Mol. Biosci. 2021, 8, 629593. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.P. MicroRNAs in microglia: Deciphering their role in neurodegenerative diseases. Front. Cell. Neurosci. 2024, 18, 1391537. [Google Scholar] [CrossRef]
- Rege, S.D.; Geetha, T.; Pondugula, S.R.; Zizza, C.A.; Wernette, C.M.; Babu, J.R. Noncoding RNAs in Neurodegenerative Diseases. ISRN Neurol. 2013, 2013, 375852. [Google Scholar] [CrossRef] [PubMed]
- Mo, M. Editorial: Non-coding RNAs in neurodegenerative diseases. Front. Neurosci. 2023, 17, 1241737. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Rajgor, D.; Fiuza, M.; Parkinson, G.T.; Hanley, J.G. The PICK1 Ca2+ sensor modulates N-methyl-d-aspartate (NMDA) receptor-dependent microRNA-mediated translational repression in neurons. J. Biol. Chem. 2017, 292, 9774–9786. [Google Scholar] [CrossRef]
- Hebert, S.S.; Papadopoulou, A.S.; Smith, P.; Galas, M.C.; Planel, E.; Silahtaroglu, A.N.; Sergeant, N.; Buee, L.; De Strooper, B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 2010, 19, 3959–3969. [Google Scholar] [CrossRef]
- Cheng, T.L.; Wang, Z.; Liao, Q.; Zhu, Y.; Zhou, W.H.; Xu, W.; Qiu, Z. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev. Cell 2014, 28, 547–560. [Google Scholar] [CrossRef]
- Dalla Costa, I.; Buchanan, C.N.; Zdradzinski, M.D.; Sahoo, P.K.; Smith, T.P.; Thames, E.; Kar, A.N.; Twiss, J.L. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2021, 22, 77–91. [Google Scholar] [CrossRef]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef]
- Sotoudeh Anvari, M.; Vasei, H.; Najmabadi, H.; Badv, R.S.; Golipour, A.; Mohammadi-Yeganeh, S.; Salehi, S.; Mohamadi, M.; Goodarzynejad, H.; Mowla, S.J. Identification of microRNAs associated with human fragile X syndrome using next-generation sequencing. Sci. Rep. 2022, 12, 5011. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, M.J.; Landry, M.; Favereaux, A. MicroRNA and chronic pain: From mechanisms to therapeutic potential. Pharmacol. Ther. 2017, 180, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Wu, Z.J.; Zhu, L.J.; Niu, T.X.; Liu, B.; Li, J. Emerging roles of miRNAs in neuropathic pain: From new findings to novel mechanisms. Front. Mol. Neurosci. 2023, 16, 1110975. [Google Scholar] [CrossRef] [PubMed]
- Dubes, S.; Favereaux, A.; Thoumine, O.; Letellier, M. miRNA-Dependent Control of Homeostatic Plasticity in Neurons. Front. Cell. Neurosci. 2019, 13, 536. [Google Scholar] [CrossRef] [PubMed]
- Dubes, S.; Soula, A.; Benquet, S.; Tessier, B.; Poujol, C.; Favereaux, A.; Thoumine, O.; Letellier, M. miR-124-dependent tagging of synapses by synaptopodin enables input-specific homeostatic plasticity. EMBO J. 2022, 41, e109012. [Google Scholar] [CrossRef]
- Aslani, M.; Mortazavi-Jahromi, S.S.; Mirshafiey, A. Efficient roles of miR-146a in cellular and molecular mechanisms of neuroinflammatory disorders: An effectual review in neuroimmunology. Immunol. Lett. 2021, 238, 1–20. [Google Scholar] [CrossRef]
- Fan, W.; Liang, C.; Ou, M.; Zou, T.; Sun, F.; Zhou, H.; Cui, L. MicroRNA-146a Is a Wide-Reaching Neuroinflammatory Regulator and Potential Treatment Target in Neurological Diseases. Front. Mol. Neurosci. 2020, 13, 90. [Google Scholar] [CrossRef]
- Barbosa, M.; Gomes, C.; Sequeira, C.; Goncalves-Ribeiro, J.; Pina, C.C.; Carvalho, L.A.; Moreira, R.; Vaz, S.H.; Vaz, A.R.; Brites, D. Recovery of Depleted miR-146a in ALS Cortical Astrocytes Reverts Cell Aberrancies and Prevents Paracrine Pathogenicity on Microglia and Motor Neurons. Front. Cell Dev. Biol. 2021, 9, 634355. [Google Scholar] [CrossRef]
- Long, J.M.; Ray, B.; Lahiri, D.K. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 2012, 287, 31298–31310. [Google Scholar] [CrossRef]
- Liang, C.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Deng, W.; Liu, Y.; Qin, C. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012, 1455, 103–113. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef]
- Lellahi, S.M.; Rosenlund, I.A.; Hedberg, A.; Kiaer, L.T.; Mikkola, I.; Knutsen, E.; Perander, M. The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 2018, 293, 18965–18976. [Google Scholar] [CrossRef]
- Sunwoo, J.S.; Lee, S.T.; Im, W.; Lee, M.; Byun, J.I.; Jung, K.H.; Park, K.I.; Jung, K.Y.; Lee, S.K.; Chu, K.; et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol. Neurobiol. 2017, 54, 1577–1586. [Google Scholar] [CrossRef]
- Yadav, M.; Harding, R.J.; Li, T.; Xu, X.; Gall-Duncan, T.; Khan, M.; Bardile, C.F.; Sequiera, G.L.; Duan, S.; Chandrasekaran, R.; et al. Huntingtin is an RNA binding protein and participates in NEAT1-mediated paraspeckles. Sci. Adv. 2024, 10, eado5264. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, D.; Todoerti, K.; Nobili, L.; Tassone, P.; Amodio, N.; Neri, A. LncRNA NEAT1 in Paraspeckles: A Structural Scaffold for Cellular DNA Damage Response Systems? Non-Coding RNA 2020, 6, 26. [Google Scholar] [CrossRef]
- Li, K.; Wang, Z. lncRNA NEAT1: Key player in neurodegenerative diseases. Ageing Res. Rev. 2023, 86, 101878. [Google Scholar] [CrossRef]
- Xie, S.P.; Zhou, F.; Li, J.; Duan, S.J. NEAT1 regulates MPP+-induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci. Lett. 2019, 708, 134340. [Google Scholar] [CrossRef]
- Esmaeili, A.; Yazdanpanah, N.; Rezaei, N. LncRNAs Orchestrating Neuroinflammation: A Comprehensive Review. Cell. Mol. Neurobiol. 2025, 45, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Yang, H.; Xu, Y.; Zhou, X.; Zhang, X.; Xie, Z.; Bi, J. The effect of BACE1-AS on beta-amyloid generation by regulating BACE1 mRNA expression. BMC Mol. Biol. 2019, 20, 23. [Google Scholar] [CrossRef] [PubMed]
- Sayad, A.; Najafi, S.; Hussen, B.M.; Abdullah, S.T.; Movahedpour, A.; Taheri, M.; Hajiesmaeili, M. The Emerging Roles of the beta-Secretase BACE1 and the Long Non-coding RNA BACE1-AS in Human Diseases: A Focus on Neurodegenerative Diseases and Cancer. Front. Aging Neurosci. 2022, 14, 853180. [Google Scholar] [CrossRef]
- Rani, N.; Nowakowski, T.J.; Zhou, H.; Godshalk, S.E.; Lisi, V.; Kriegstein, A.R.; Kosik, K.S. A Primate lncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron 2016, 90, 1174–1188. [Google Scholar] [CrossRef]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e7. [Google Scholar] [CrossRef]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Mao, Y.; Li, B.; Zhu, Y.; Zhang, S.; Wang, S.; Jiang, Y.; Xu, N.; Xie, Y.; et al. NEAT1 regulates microtubule stabilization via FZD3/GSK3beta/P-tau pathway in SH-SY5Y cells and APP/PS1 mice. Aging 2020, 12, 23233–23250. [Google Scholar]
- Li, Y.; Fan, H.; Ni, M.; Zhang, W.; Fang, F.; Sun, J.; Lyu, P.; Ma, P. Targeting lncRNA NEAT1 Hampers Alzheimer’s Disease Progression. Neuroscience 2023, 529, 88–98. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, H.H. Circular RNAs: Diagnostic and Therapeutic Perspectives in CNS Diseases. Curr. Med. Sci. 2023, 43, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Gruner, H.; Cortes-Lopez, M.; Cooper, D.A.; Bauer, M.; Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016, 6, 38907. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Zajaczkowski, E.L.; Bredy, T.W. Circular RNAs in the Brain: A Possible Role in Memory? Neuroscientist 2021, 27, 473–486. [Google Scholar] [CrossRef]
- Zajaczkowski, E.L.; Zhao, Q.; Liau, W.S.; Gong, H.; Madugalle, S.U.; Periyakaruppiah, A.; Leighton, L.J.; Musgrove, M.; Ren, H.; Davies, J.; et al. Localised Cdr1as activity is required for fear extinction memory. Neurobiol. Learn. Mem. 2023, 203, 107777. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Loubalova, Z.; Konstantinidou, P.; Haase, A.D. Themes and variations on piRNA-guided transposon control. Mob. DNA 2023, 14, 10. [Google Scholar] [CrossRef]
- Andersen, P.R.; Tirian, L.; Vunjak, M.; Brennecke, J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 2017, 549, 54–59. [Google Scholar] [CrossRef]
- Li, Z.; Tang, X.; Shen, E.Z. How mammalian piRNAs instruct de novo DNA methylation of transposons. Signal Transduct. Target. Ther. 2020, 5, 190. [Google Scholar] [CrossRef]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef]
- Ochoa, E.; Ramirez, P.; Gonzalez, E.; De Mange, J.; Ray, W.J.; Bieniek, K.F.; Frost, B. Pathogenic tau-induced transposable element-derived dsRNA drives neuroinflammation. Sci. Adv. 2023, 9, eabq5423. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyo, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef]
- Abdelhamid, R.F.; Ogawa, K.; Beck, G.; Ikenaka, K.; Takeuchi, E.; Yasumizu, Y.; Jinno, J.; Kimura, Y.; Baba, K.; Nagai, Y.; et al. piRNA/PIWI Protein Complex as a Potential Biomarker in Sporadic Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2022, 59, 1693–1705. [Google Scholar] [CrossRef]
- Kim, K.W.; Tang, N.H.; Andrusiak, M.G.; Wu, Z.; Chisholm, A.D.; Jin, Y. A Neuronal piRNA Pathway Inhibits Axon Regeneration in C. elegans. Neuron 2018, 97, 511–519.e6. [Google Scholar] [CrossRef]
- Kawahara, Y.; Mieda-Sato, A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. USA 2012, 109, 3347–3352. [Google Scholar] [CrossRef]
- Morlando, M.; Dini Modigliani, S.; Torrelli, G.; Rosa, A.; Di Carlo, V.; Caffarelli, E.; Bozzoni, I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012, 31, 4502–4510. [Google Scholar] [CrossRef]
- Kim, H.J.; Taylor, J.P. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 2017, 96, 285–297. [Google Scholar] [CrossRef]
- Rosenthal, J.J.; Seeburg, P.H. A-to-I RNA editing: Effects on proteins key to neural excitability. Neuron 2012, 74, 432–439. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Ries, R.J.; Pickering, B.F.; Poh, H.X.; Namkoong, S.; Jaffrey, S.R. m6A governs length-dependent enrichment of mRNAs in stress granules. Nat. Struct. Mol. Biol. 2023, 30, 1525–1535. [Google Scholar] [CrossRef]
- Miller, T.M.; Cudkowicz, M.E.; Genge, A.; Shaw, P.J.; Sobue, G.; Bucelli, R.C.; Chio, A.; Van Damme, P.; Ludolph, A.C.; Glass, J.D.; et al. Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2022, 387, 1099–1110. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, H.; Dong, W.; Quan, X.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Qin, C. miR-29c regulates BACE1 protein expression. Brain Res. 2011, 1395, 108–115. [Google Scholar] [CrossRef]
- Lei, X.; Lei, L.; Zhang, Z.; Zhang, Z.; Cheng, Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015, 8, 1565–1574. [Google Scholar]
- Liu, Z.; Zhang, H.; Sun, L.; Zhu, K.; Lang, W. miR-29c-3p Increases Cell Viability and Suppresses Apoptosis by Regulating the TNFAIP1/NF-kappaB Signaling Pathway via TNFAIP1 in Abeta-Treated Neuroblastoma Cells. Neurochem. Res. 2020, 45, 2375–2384. [Google Scholar] [CrossRef]
- El Fatimy, R.; Li, S.; Chen, Z.; Mushannen, T.; Gongala, S.; Wei, Z.; Balu, D.T.; Rabinovsky, R.; Cantlon, A.; Elkhal, A.; et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 2018, 136, 537–555. [Google Scholar] [CrossRef]
- Walgrave, H.; Balusu, S.; Snoeck, S.; Vanden Eynden, E.; Craessaerts, K.; Thrupp, N.; Wolfs, L.; Horre, K.; Fourne, Y.; Ronisz, A.; et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell 2021, 28, 1805–1821.e8. [Google Scholar] [CrossRef]
- Wong, H.K.; Veremeyko, T.; Patel, N.; Lemere, C.A.; Walsh, D.M.; Esau, C.; Vanderburg, C.; Krichevsky, A.M. De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer’s disease. Hum. Mol. Genet. 2013, 22, 3077–3092. [Google Scholar] [CrossRef]
- Bazrgar, M.; Khodabakhsh, P.; Prudencio, M.; Mohagheghi, F.; Ahmadiani, A. The role of microRNA-34 family in Alzheimer’s disease: A potential molecular link between neurodegeneration and metabolic disorders. Pharmacol. Res. 2021, 172, 105805. [Google Scholar] [CrossRef]
- Dong, Z.; Gu, H.; Guo, Q.; Liang, S.; Xue, J.; Yao, F.; Liu, X.; Li, F.; Liu, H.; Sun, L.; et al. Profiling of Serum Exosome MiRNA Reveals the Potential of a MiRNA Panel as Diagnostic Biomarker for Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 3084–3094. [Google Scholar] [CrossRef]
- Song, S.; Lee, J.U.; Jeon, M.J.; Kim, S.; Sim, S.J. Detection of multiplex exosomal miRNAs for clinically accurate diagnosis of Alzheimer’s disease using label-free plasmonic biosensor based on DNA-Assembled advanced plasmonic architecture. Biosens. Bioelectron. 2022, 199, 113864. [Google Scholar] [CrossRef]
- Alhenaky, A.; Alhazmi, S.; Alamri, S.H.; Alkhatabi, H.A.; Alharthi, A.; Alsaleem, M.A.; Abdelnour, S.A.; Hassan, S.M. Exosomal MicroRNAs in Alzheimer’s Disease: Unveiling Their Role and Pioneering Tools for Diagnosis and Treatment. J. Clin. Med. 2024, 13, 6960. [Google Scholar] [CrossRef]
- Yu, X.; Sun, X.; Wei, M.; Deng, S.; Zhang, Q.; Guo, T.; Shao, K.; Zhang, M.; Jiang, J.; Han, Y.; et al. Innovative Multivariable Model Combining MRI Radiomics and Plasma Indexes Predicts Alzheimer’s Disease Conversion: Evidence from a 2-Cohort Longitudinal Study. Research 2024, 7, 0354. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.J.; Cheng, Y.; Xie, Y.; Cheng, Y.; Zhao, S.; Jiang, Y.; Bai, T.; Huo, Y.; Wang, K.; et al. Multimodal integration of plasma biomarkers, MRI, and genetic risk to predict cerebral amyloid burden in Alzheimer’s disease. Neuroimage 2025, 322, 121550. [Google Scholar] [CrossRef]
- Gascon, E.; Gao, F.B. Cause or Effect: Misregulation of microRNA Pathways in Neurodegeneration. Front. Neurosci. 2012, 6, 48. [Google Scholar] [CrossRef]
- Shaheen, N.; Shaheen, A.; Osama, M.; Nashwan, A.J.; Bharmauria, V.; Flouty, O. MicroRNAs regulation in Parkinson’s disease, and their potential role as diagnostic and therapeutic targets. NPJ Park. Dis. 2024, 10, 186. [Google Scholar] [CrossRef]
- Henrich, M.T.; Oertel, W.H.; Surmeier, D.J.; Geibl, F.F. Mitochondrial dysfunction in Parkinson’s disease—A key disease hallmark with therapeutic potential. Mol. Neurodegener. 2023, 18, 83. [Google Scholar] [CrossRef]
- McMillan, K.J.; Murray, T.K.; Bengoa-Vergniory, N.; Cordero-Llana, O.; Cooper, J.; Buckley, A.; Wade-Martins, R.; Uney, J.B.; O’Neill, M.J.; Wong, L.F.; et al. Loss of MicroRNA-7 Regulation Leads to alpha-Synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo. Mol. Ther. 2017, 25, 2404–2414. [Google Scholar] [CrossRef]
- Sala, G.; Marinig, D.; Arosio, A.; Ferrarese, C. Role of Chaperone-Mediated Autophagy Dysfunctions in the Pathogenesis of Parkinson’s Disease. Front. Mol. Neurosci. 2016, 9, 157. [Google Scholar] [CrossRef]
- Sang, Q.; Liu, X.; Wang, L.; Qi, L.; Sun, W.; Wang, W.; Sun, Y.; Zhang, H. CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 2018, 10, 1281–1293. [Google Scholar] [CrossRef]
- Liao, J.; Zhang, Q.; Huang, J.; He, H.; Lei, J.; Shen, Y.; Wang, J.; Xiao, Y. The emerging role of circular RNAs in Parkinson’s disease. Front. Neurosci. 2023, 17, 1137363. [Google Scholar] [CrossRef]
- Minones-Moyano, E.; Porta, S.; Escaramis, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Marti, E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. [Google Scholar] [CrossRef]
- Citterio, L.A.; Mancuso, R.; Agostini, S.; Meloni, M.; Clerici, M. Serum and Exosomal miR-7-1-5p and miR-223-3p as Possible Biomarkers for Parkinson’s Disease. Biomolecules 2023, 13, 865. [Google Scholar] [CrossRef]
- Vaitkiene, P.; Pranckeviciene, A.; Radziunas, A.; Miseikaite, A.; Miniotaite, G.; Belickiene, V.; Laucius, O.; Deltuva, V. Association of Serum Extracellular Vesicle miRNAs with Cognitive Functioning and Quality of Life in Parkinson’s Disease. Biomolecules 2024, 14, 1000. [Google Scholar] [CrossRef]
- Dos Santos, M.C.T.; Barreto-Sanz, M.A.; Correia, B.R.S.; Bell, R.; Widnall, C.; Perez, L.T.; Berteau, C.; Schulte, C.; Scheller, D.; Berg, D.; et al. miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 2018, 9, 17455–17465. [Google Scholar] [CrossRef]
- Wu, L.; Xu, Q.; Zhou, M.; Chen, Y.; Jiang, C.; Jiang, Y.; Lin, Y.; He, Q.; Zhao, L.; Dong, Y.; et al. Plasma miR-153 and miR-223 Levels as Potential Biomarkers in Parkinson’s Disease. Front. Neurosci. 2022, 16, 865139. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, M.; Yan, R.; Liu, J.; Maddila, S.; Junn, E.; Mouradian, M.M. MicroRNA-7 Protects Against Neurodegeneration Induced by α-Synuclein Preformed Fibrils in the Mouse Brain. Neurotherapeutics 2021, 18, 2529–2540. [Google Scholar] [CrossRef]
- Azadfar, P.; Noormohammadi, Z.; Noroozian, M.; Eidi, A.; Mortazavi, P. Effect of memantine on expression of Bace1-as and Bace1 genes in STZ-induced Alzheimeric rats. Mol. Biol. Rep. 2020, 47, 5737–5745. [Google Scholar] [CrossRef]
- Choi, D.C.; Yoo, M.; Kabaria, S.; Junn, E. MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci. Lett. 2018, 678, 118–123. [Google Scholar] [CrossRef]
- Fan, Z.; Lu, M.; Qiao, C.; Zhou, Y.; Ding, J.H.; Hu, G. MicroRNA-7 Enhances Subventricular Zone Neurogenesis by Inhibiting NLRP3/Caspase-1 Axis in Adult Neural Stem Cells. Mol. Neurobiol. 2016, 53, 7057–7069. [Google Scholar] [CrossRef]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Kay, C.; Leavitt, B.R.; Nance, M.; Ross, C.A.; Scahill, R.I.; Wetzel, R.; et al. Huntington disease. Nat. Rev. Dis. Primers 2015, 1, 15005. [Google Scholar] [CrossRef]
- de Mezer, M.; Wojciechowska, M.; Napierala, M.; Sobczak, K.; Krzyzosiak, W.J. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011, 39, 3852–3863. [Google Scholar] [CrossRef]
- Nalavade, R.; Griesche, N.; Ryan, D.P.; Hildebrand, S.; Krauss, S. Mechanisms of RNA-induced toxicity in CAG repeat disorders. Cell Death Dis. 2013, 4, e752. [Google Scholar] [CrossRef]
- Ayyildiz, D.; Bergonzoni, G.; Monziani, A.; Tripathi, T.; Doring, J.; Kerschbamer, E.; Di Leva, F.; Pennati, E.; Donini, L.; Kovalenko, M.; et al. CAG repeat expansion in the Huntington’s disease gene shapes linear and circular RNAs biogenesis. PLoS Genet. 2023, 19, e1010988. [Google Scholar] [CrossRef]
- Sonmez, A.; Mustafa, R.; Ryll, S.T.; Tuorto, F.; Wacheul, L.; Ponti, D.; Litke, C.; Hering, T.; Kojer, K.; Koch, J.; et al. Nucleolar stress controls mutant Huntington toxicity and monitors Huntington’s disease progression. Cell Death Dis. 2021, 12, 1139. [Google Scholar] [CrossRef]
- Aviner, R.; Lee, T.T.; Masto, V.B.; Li, K.H.; Andino, R.; Frydman, J. Polyglutamine-mediated ribotoxicity disrupts proteostasis and stress responses in Huntington’s disease. Nat. Cell Biol. 2024, 26, 892–902. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H.; et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef]
- Conroy, F.; Miller, R.; Alterman, J.F.; Hassler, M.R.; Echeverria, D.; Godinho, B.; Knox, E.G.; Sapp, E.; Sousa, J.; Yamada, K.; et al. Chemical engineering of therapeutic siRNAs for allele-specific gene silencing in Huntington’s disease models. Nat. Commun. 2022, 13, 5802. [Google Scholar] [CrossRef]
- Hirunagi, T.; Sahashi, K.; Tachikawa, K.; Leu, A.I.; Nguyen, M.; Mukthavaram, R.; Karmali, P.P.; Chivukula, P.; Tohnai, G.; Iida, M.; et al. Selective suppression of polyglutamine-expanded protein by lipid nanoparticle-delivered siRNA targeting CAG expansions in the mouse CNS. Mol. Ther. Nucleic Acids 2021, 24, 1–10. [Google Scholar] [CrossRef]
- Keskin, S.; Brouwers, C.C.; Sogorb-Gonzalez, M.; Martier, R.; Depla, J.A.; Valles, A.; van Deventer, S.J.; Konstantinova, P.; Evers, M.M. AAV5-miHTT Lowers Huntingtin mRNA and Protein without Off-Target Effects in Patient-Derived Neuronal Cultures and Astrocytes. Mol. Ther. Methods Clin. Dev. 2019, 15, 275–284. [Google Scholar] [CrossRef]
- Cheng, P.H.; Li, C.L.; Chang, Y.F.; Tsai, S.J.; Lai, Y.Y.; Chan, A.W.; Chen, C.M.; Yang, S.H. miR-196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am. J. Hum. Genet. 2013, 93, 306–312. [Google Scholar] [CrossRef]
- Kunkanjanawan, T.; Carter, R.L.; Prucha, M.S.; Yang, J.; Parnpai, R.; Chan, A.W. miR-196a Ameliorates Cytotoxicity and Cellular Phenotype in Transgenic Huntington’s Disease Monkey Neural Cells. PLoS ONE 2016, 11, e0162788. [Google Scholar] [CrossRef]
- Hawley, Z.C.E.; Campos-Melo, D.; Strong, M.J. Evidence of A Negative Feedback Network Between TDP-43 and miRNAs Dependent on TDP-43 Nuclear Localization. J. Mol. Biol. 2020, 432, 166695. [Google Scholar] [CrossRef]
- Hawley, Z.C.E.; Campos-Melo, D.; Strong, M.J. MiR-105 and miR-9 regulate the mRNA stability of neuronal intermediate filaments. Implications for the pathogenesis of amyotrophic lateral sclerosis (ALS). Brain Res. 2019, 1706, 93–100. [Google Scholar] [CrossRef]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Waller, R.; Goodall, E.F.; Milo, M.; Cooper-Knock, J.; Da Costa, M.; Hobson, E.; Kazoka, M.; Wollff, H.; Heath, P.R.; Shaw, P.J.; et al. Serum miRNAs miR-206, 143–3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 2017, 55, 123–131. [Google Scholar] [CrossRef]
- Ash, P.E.; Bieniek, K.F.; Gendron, T.F.; Caulfield, T.; Lin, W.L.; Dejesus-Hernandez, M.; van Blitterswijk, M.M.; Jansen-West, K.; Paul, J.W., 3rd; Rademakers, R.; et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 2013, 77, 639–646. [Google Scholar] [CrossRef]
- Cooper-Knock, J.; Walsh, M.J.; Higginbottom, A.; Robin Highley, J.; Dickman, M.J.; Edbauer, D.; Ince, P.G.; Wharton, S.B.; Wilson, S.A.; Kirby, J.; et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 2014, 137, 2040–2051. [Google Scholar] [CrossRef]
- Cooper-Knock, J.; Bury, J.J.; Heath, P.R.; Wyles, M.; Higginbottom, A.; Gelsthorpe, C.; Highley, J.R.; Hautbergue, G.; Rattray, M.; Kirby, J.; et al. C9ORF72 GGGGCC Expanded Repeats Produce Splicing Dysregulation which Correlates with Disease Severity in Amyotrophic Lateral Sclerosis. PLoS ONE 2015, 10, e0127376. [Google Scholar] [CrossRef]
- Parameswaran, J.; Zhang, N.; Braems, E.; Tilahun, K.; Pant, D.C.; Yin, K.; Asress, S.; Heeren, K.; Banerjee, A.; Davis, E.; et al. Antisense, but not sense, repeat expanded RNAs activate PKR/eIF2alpha-dependent ISR in C9ORF72 FTD/ALS. Elife 2023, 12, e85902. [Google Scholar] [CrossRef]
- Smith, P.Y.; Hernandez-Rapp, J.; Jolivette, F.; Lecours, C.; Bisht, K.; Goupil, C.; Dorval, V.; Parsi, S.; Morin, F.; Planel, E.; et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015, 24, 6721–6735. [Google Scholar] [CrossRef]
- Donnelly, C.J.; Zhang, P.W.; Pham, J.T.; Haeusler, A.R.; Mistry, N.A.; Vidensky, S.; Daley, E.L.; Poth, E.M.; Hoover, B.; Fines, D.M.; et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 2013, 80, 415–428. [Google Scholar] [CrossRef]
- Sareen, D.; O’Rourke, J.G.; Meera, P.; Muhammad, A.K.; Grant, S.; Simpkinson, M.; Bell, S.; Carmona, S.; Ornelas, L.; Sahabian, A.; et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013, 5, 208ra149. [Google Scholar] [CrossRef]
- Basso, D.; Padoan, A.; Laufer, T.; Aneloni, V.; Moz, S.; Schroers, H.; Pelloso, M.; Saiz, A.; Krapp, M.; Fogar, P.; et al. Relevance of pre-analytical blood management on the emerging cardiovascular protein biomarkers TWEAK and HMGB1 and on miRNA serum and plasma profiling. Clin. Biochem. 2017, 50, 186–193. [Google Scholar] [CrossRef]
- Shah, J.S.; Soon, P.S.; Marsh, D.J. Comparison of Methodologies to Detect Low Levels of Hemolysis in Serum for Accurate Assessment of Serum microRNAs. PLoS ONE 2016, 11, e0153200. [Google Scholar] [CrossRef]
- Sorensen, S.S.; Nygaard, A.B.; Christensen, T. miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia—An exploratory study. Transl. Neurodegener. 2016, 5, 6. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- Norman, M.; Ter-Ovanesyan, D.; Trieu, W.; Lazarovits, R.; Kowal, E.J.K.; Lee, J.H.; Chen-Plotkin, A.S.; Regev, A.; Church, G.M.; Walt, D.R. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat. Methods 2021, 18, 631–634. [Google Scholar] [CrossRef]
- Fotuhi, S.N.; Khalaj-Kondori, M.; Hoseinpour Feizi, M.A.; Talebi, M. Long Non-coding RNA BACE1-AS May Serve as an Alzheimer’s Disease Blood-Based Biomarker. J. Mol. Neurosci. 2019, 69, 351–359. [Google Scholar] [CrossRef]
- Khodayi, M.; Khalaj-Kondori, M.; Hoseinpour Feizi, M.A.; Jabarpour Bonyadi, M.; Talebi, M. Plasma lncRNA profiling identified BC200 and NEAT1 lncRNAs as potential blood-based biomarkers for late-onset Alzheimer’s disease. EXCLI J. 2022, 21, 772–785. [Google Scholar]
- Ma, W.; Li, Y.; Wang, C.; Xu, F.; Wang, M.; Liu, Y. Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem. Funct. 2016, 34, 511–515. [Google Scholar] [CrossRef]
- Schulz, J.; Takousis, P.; Wohlers, I.; Itua, I.O.G.; Dobricic, V.; Rucker, G.; Binder, H.; Middleton, L.; Ioannidis, J.P.A.; Perneczky, R.; et al. Meta-analyses identify differentially expressed micrornas in Parkinson’s disease. Ann. Neurol. 2019, 85, 835–851. [Google Scholar] [CrossRef]
- Chunhui, G.; Yanqiu, Y.; Jibing, C.; Ning, L.; Fujun, L. Exosomes and non-coding RNAs: Bridging the gap in Alzheimer’s pathogenesis and therapeutics. Metab. Brain Dis. 2025, 40, 84. [Google Scholar] [CrossRef]
- Feng, L.; Liao, Y.T.; He, J.C.; Xie, C.L.; Chen, S.Y.; Fan, H.H.; Su, Z.P.; Wang, Z. Plasma long non-coding RNA BACE1 as a novel biomarker for diagnosis of Alzheimer disease. BMC Neurol. 2018, 18, 4. [Google Scholar] [CrossRef]
- Soliman, H.M.; Ghonaim, G.A.; Gharib, S.M.; Chopra, H.; Farag, A.K.; Hassanin, M.H.; Nagah, A.; Emad-Eldin, M.; Hashem, N.E.; Yahya, G.; et al. Exosomes in Alzheimer’s Disease: From Being Pathological Players to Potential Diagnostics and Therapeutics. Int. J. Mol. Sci. 2021, 22, 10794. [Google Scholar] [CrossRef]
- Gutierrez-Tordera, L.; Papandreou, C.; Novau-Ferre, N.; Garcia-Gonzalez, P.; Rojas, M.; Marquie, M.; Chapado, L.A.; Papagiannopoulos, C.; Fernandez-Castillo, N.; Valero, S.; et al. Exploring small non-coding RNAs as blood-based biomarkers to predict Alzheimer’s disease. Cell Biosci. 2024, 14, 8. [Google Scholar] [CrossRef]
- Park, C.; Weerakkody, J.S.; Schneider, R.; Miao, S.; Pitt, D. CNS cell-derived exosome signatures as blood-based biomarkers of neurodegenerative diseases. Front. Neurosci. 2024, 18, 1426700. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Edelman, J.J.; Kao, S.C.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Xu, Y.Z.; Cheng, M.G.; Wang, X.; Hu, Y. The emerging role of non-coding RNAs from extracellular vesicles in Alzheimer’s disease. J. Integr. Neurosci. 2021, 20, 239–245. [Google Scholar] [CrossRef]
- Sun, Y.M.; Chen, Y.Q. Principles and innovative technologies for decrypting noncoding RNAs: From discovery and functional prediction to clinical application. J. Hematol. Oncol. 2020, 13, 109. [Google Scholar] [CrossRef]
- Almohaimeed, H.M.; Assiri, R.; Althubaiti, E.H.; Aggad, W.S.; Shaheen, S.; Shaheen, M.Y.; Batarfi, M.A.; Alharbi, N.A.; Alshehri, A.M.; Alkhudhairy, B.S.M. Non-coding RNAs as key players in the neurodegenerative diseases: Multi-platform strategies and approaches for exploring the Genome’s dark matter. J. Chem. Neuroanat. 2023, 129, 102236. [Google Scholar] [CrossRef]
- Mahmud, T.; Barua, K.; Habiba, S.U.; Sharmen, N.; Hossain, M.S.; Andersson, K. An Explainable AI Paradigm for Alzheimer’s Diagnosis Using Deep Transfer Learning. Diagnostics 2024, 14, 345. [Google Scholar] [CrossRef]
- Shobeiri, P.; Alilou, S.; Jaberinezhad, M.; Zare, F.; Karimi, N.; Maleki, S.; Teixeira, A.L.; Perry, G.; Rezaei, N. Circulating long non-coding RNAs as novel diagnostic biomarkers for Alzheimer’s disease (AD): A systematic review and meta-analysis. PLoS ONE 2023, 18, e0281784. [Google Scholar] [CrossRef]
- Hao, Y.; Xie, B.; Fu, X.; Xu, R.; Yang, Y. New Insights into lncRNAs in Abeta Cascade Hypothesis of Alzheimer’s Disease. Biomolecules 2022, 12, 1802. [Google Scholar] [CrossRef]
- Zhao, M.Y.; Wang, G.Q.; Wang, N.N.; Yu, Q.Y.; Liu, R.L.; Shi, W.Q. The long-non-coding RNA NEAT1 is a novel target for Alzheimer’s disease progression via miR-124/BACE1 axis. Neurol. Res. 2019, 41, 489–497. [Google Scholar] [CrossRef]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef]
- Wang, F.; Liang, Y.; Wang, Q.W. Interpretable machine learning-driven biomarker identification and validation for Alzheimer’s disease. Sci. Rep. 2024, 14, 30770. [Google Scholar] [CrossRef]
- Bloch, L.; Friedrich, C.M.; Alzheimer’s Disease Neuroimaging, I. Data analysis with Shapley values for automatic subject selection in Alzheimer’s disease data sets using interpretable machine learning. Alzheimer’s Res. Ther. 2021, 13, 155. [Google Scholar] [CrossRef]
- Lin, R.; Wichadakul, D. Interpretable Deep Learning Model Reveals Subsequences of Various Functions for Long Non-Coding RNA Identification. Front. Genet. 2022, 13, 876721. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Ao, X.; Yu, W.; Zhang, L.; Wang, Y.; Chang, W. The Role of Non-coding RNAs in Alzheimer’s Disease: From Regulated Mechanism to Therapeutic Targets and Diagnostic Biomarkers. Front. Aging Neurosci. 2021, 13, 654978. [Google Scholar] [CrossRef]
- Wang, D.; Wang, P.; Bian, X.; Xu, S.; Zhou, Q.; Zhang, Y.; Ding, M.; Han, M.; Huang, L.; Bi, J.; et al. Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 227–238. [Google Scholar] [CrossRef]
- Ding, H.; Huang, Z.; Chen, M.; Wang, C.; Chen, X.; Chen, J.; Zhang, J. Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Park. Relat. Disord. 2016, 22, 68–73. [Google Scholar] [CrossRef]
- Ghit, A.; Deeb, H.E. Cytokines, miRNAs, and Antioxidants as Combined Non-invasive Biomarkers for Parkinson’s Disease. J. Mol. Neurosci. 2022, 72, 1133–1140. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef]
- Bennett, C.F.; Kordasiewicz, H.B.; Cleveland, D.W. Antisense Drugs Make Sense for Neurological Diseases. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 831–852. [Google Scholar] [CrossRef]
- Mummery, C.J.; Borjesson-Hanson, A.; Blackburn, D.J.; Vijverberg, E.G.B.; De Deyn, P.P.; Ducharme, S.; Jonsson, M.; Schneider, A.; Rinne, J.O.; Ludolph, A.C.; et al. Tau-targeting antisense oligonucleotide MAPT(Rx) in mild Alzheimer’s disease: A phase 1b, randomized, placebo-controlled trial. Nat. Med. 2023, 29, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Rook, M.E.; Southwell, A.L. Antisense Oligonucleotide Therapy: From Design to the Huntington Disease Clinic. BioDrugs 2022, 36, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.S.; Keiser, M.S.; Davidson, B.L. Recent advances in RNA interference therapeutics for CNS diseases. Neurotherapeutics 2013, 10, 473–485. [Google Scholar] [CrossRef]
- Martier, R.; Konstantinova, P. Gene Therapy for Neurodegenerative Diseases: Slowing Down the Ticking Clock. Front. Neurosci. 2020, 14, 580179. [Google Scholar] [CrossRef]
- Seyhan, A.A. Trials and Tribulations of MicroRNA Therapeutics. Int. J. Mol. Sci. 2024, 25, 1469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, P.; Bian, Z. Long Noncoding RNAs in Neurodegenerative Diseases: Pathogenesis and Potential Implications as Clinical Biomarkers. Front. Mol. Neurosci. 2021, 14, 685143. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, eaah7111. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Moriyama, H.; Yokota, T. Recent Progress of Antisense Oligonucleotide Therapy for Superoxide-Dismutase-1-Mutated Amyotrophic Lateral Sclerosis: Focus on Tofersen. Genes 2024, 15, 1342. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef]
- Everett, W.H.; Bucelli, R.C. Tofersen for SOD1 ALS. Neurodegener. Dis. Manag. 2024, 14, 149–160. [Google Scholar] [CrossRef]
- McGuigan, A.; Blair, H.A. Tofersen: A Review in Amyotrophic Lateral Sclerosis Associated with SOD1 Mutations. CNS Drugs 2025, 39, 903–912. [Google Scholar] [CrossRef]
- McColgan, P.; Thobhani, A.; Boak, L.; Schobel, S.A.; Nicotra, A.; Palermo, G.; Trundell, D.; Zhou, J.; Schlegel, V.; Sanwald Ducray, P.; et al. Tominersen in Adults with Manifest Huntington’s Disease. N. Engl. J. Med. 2023, 389, 2203–2205. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Estevez-Fraga, C.; van Roon-Mom, W.M.C.; Flower, M.D.; Scahill, R.I.; Wild, E.J.; Munoz-Sanjuan, I.; Sampaio, C.; Rosser, A.E.; Leavitt, B.R. Potential disease-modifying therapies for Huntington’s disease: Lessons learned and future opportunities. Lancet Neurol. 2022, 21, 645–658. [Google Scholar] [CrossRef]
- Spronck, E.A.; Valles, A.; Lampen, M.H.; Montenegro-Miranda, P.S.; Keskin, S.; Heijink, L.; Evers, M.M.; Petry, H.; Deventer, S.J.V.; Konstantinova, P.; et al. Intrastriatal Administration of AAV5-miHTT in Non-Human Primates and Rats Is Well Tolerated and Results in miHTT Transgene Expression in Key Areas of Huntington Disease Pathology. Brain Sci. 2021, 11, 129. [Google Scholar] [CrossRef]
- Valles, A.; Evers, M.M.; Stam, A.; Sogorb-Gonzalez, M.; Brouwers, C.; Vendrell-Tornero, C.; Acar-Broekmans, S.; Paerels, L.; Klima, J.; Bohuslavova, B.; et al. Widespread and sustained target engagement in Huntington’s disease minipigs upon intrastriatal microRNA-based gene therapy. Sci. Transl. Med. 2021, 13, eabb8920. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.; Lee, M.; Kim, M. Gene Therapy for Huntington’s Disease: The Final Strategy for a Cure? J. Mov. Disord. 2022, 15, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, H. Innovative Therapeutic Approaches for Huntington’s Disease: From Nucleic Acids to GPCR-Targeting Small Molecules. Front. Cell. Neurosci. 2021, 15, 785703. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The blood-brain barrier: Structure, regulation, and drug delivery. Signal Transduct. Target. Ther. 2023, 8, 217. [Google Scholar] [CrossRef]
- Mazur, C.; Powers, B.; Zasadny, K.; Sullivan, J.M.; Dimant, H.; Kamme, F.; Hesterman, J.; Matson, J.; Oestergaard, M.; Seaman, M.; et al. Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI Insight 2019, 4, e129240. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Bak, M.; Kempen, P.J.; Melander, F.; Burkhart, A.; Thomsen, M.S.; Nielsen, M.S.; Moos, T.; Andresen, T.L. Antibody affinity and valency impact brain uptake of transferrin receptor-targeted gold nanoparticles. Theranostics 2018, 8, 3416–3436. [Google Scholar] [CrossRef]
- Pornnoppadol, G.; Bond, L.G.; Lucas, M.J.; Zupancic, J.M.; Kuo, Y.H.; Zhang, B.; Greineder, C.F.; Tessier, P.M. Bispecific antibody shuttles targeting CD98hc mediate efficient and long-lived brain delivery of IgGs. Cell Chem. Biol. 2024, 31, 361–372.e8. [Google Scholar] [CrossRef]
- Kingwell, K. A new shuttle for drug delivery across the blood-brain barrier. Nat. Rev. Drug Discov. 2023, 22, 952. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-scale production, isolation, drug loading efficiency, and biodistribution and uptake. J. Control. Release 2022, 347, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.M.; Wiklander, P.B.; Nordin, J.Z.; Al-Shawi, R.; Wood, M.J.; Vithlani, M.; Schapira, A.H.; Simons, J.P.; El-Andaloussi, S.; Alvarez-Erviti, L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord. 2014, 29, 1476–1485. [Google Scholar] [CrossRef]
- Sumner, C.J.; Miller, T.M. The expanding application of antisense oligonucleotides to neurodegenerative diseases. J. Clin. Investig. 2024, 134, e186116. [Google Scholar] [CrossRef]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X.H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2021, 20, 427–453. [Google Scholar] [CrossRef]
- Henry, S.P.; Arfvidsson, C.; Arrington, J.; Canadi, J.; Crowe, D.; Gupta, S.; Lohmann, S.; Massonnet, B.; Mytych, D.; Rogers, T.; et al. Assessment of the Immunogenicity Potential for Oligonucleotide-Based Drugs. Nucleic Acid. Ther. 2022, 32, 369–377. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Teunissen, C.E.; Otto, M.; Piehl, F.; Sormani, M.P.; Gattringer, T.; Barro, C.; Kappos, L.; Comabella, M.; Fazekas, F.; et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 2018, 14, 577–589. [Google Scholar] [CrossRef]
- Liu, Z.; Song, S.Y. Genomic and Transcriptomic Approaches Advance the Diagnosis and Prognosis of Neurodegenerative Diseases. Genes 2025, 16, 135. [Google Scholar] [CrossRef]
- Bu, F.T.; Wang, H.Y.; Xu, C.; Song, K.L.; Dai, Z.; Wang, L.T.; Ying, J.; Chen, J. The role of m6A-associated membraneless organelles in the RNA metabolism processes and human diseases. Theranostics 2024, 14, 4683–4700. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Azam, H.M.H.; Rossling, R.I.; Geithe, C.; Khan, M.M.; Dinter, F.; Hanack, K.; Pruss, H.; Husse, B.; Roggenbuck, D.; Schierack, P.; et al. MicroRNA biomarkers as next-generation diagnostic tools for neurodegenerative diseases: A comprehensive review. Front. Mol. Neurosci. 2024, 17, 1386735. [Google Scholar] [CrossRef]
- Wilton-Clark, H.; Yan, E.; Yokota, T. Preparing for Patient-Customized N-of-1 Antisense Oligonucleotide Therapy to Treat Rare Diseases. Genes 2024, 15, 821. [Google Scholar] [CrossRef]
- Alhamadani, F.; Zhang, K.; Parikh, R.; Wu, H.; Rasmussen, T.P.; Bahal, R.; Zhong, X.B.; Manautou, J.E. Adverse Drug Reactions and Toxicity of the Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. 2022, 50, 879–887. [Google Scholar] [CrossRef]
- Kim, K.R.; Kang, J.H.; Thai, H.B.D.; Back, J.H.; Mao, C.; Lee, J.E.; Ko, Y.T.; Ahn, D.R. Systemic Brain Delivery of Oligonucleotide Therapeutics Enhanced by Protein Corona-Assisted DNA Cubes. Small Methods 2025, 9, e2400902. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Ding, L.; Jia, F.; Zhang, Z.; Long, C. Integrated Transcriptome Analysis Reveals Molecular Subtypes and ceRNA Networks in Multiple Sclerosis. Degener. Neurol. Neuromuscul. Dis. 2024, 14, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.; Hu, W.; Zhang, B.; Narykov, O.; Diekhans, M.; Marrocco, J.; Balacco, J.; Ndhlovu, L.C.; Milner, T.A.; Fedrigo, O.; et al. Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain. Nat. Neurosci. 2024, 27, 1051–1063. [Google Scholar] [CrossRef] [PubMed]

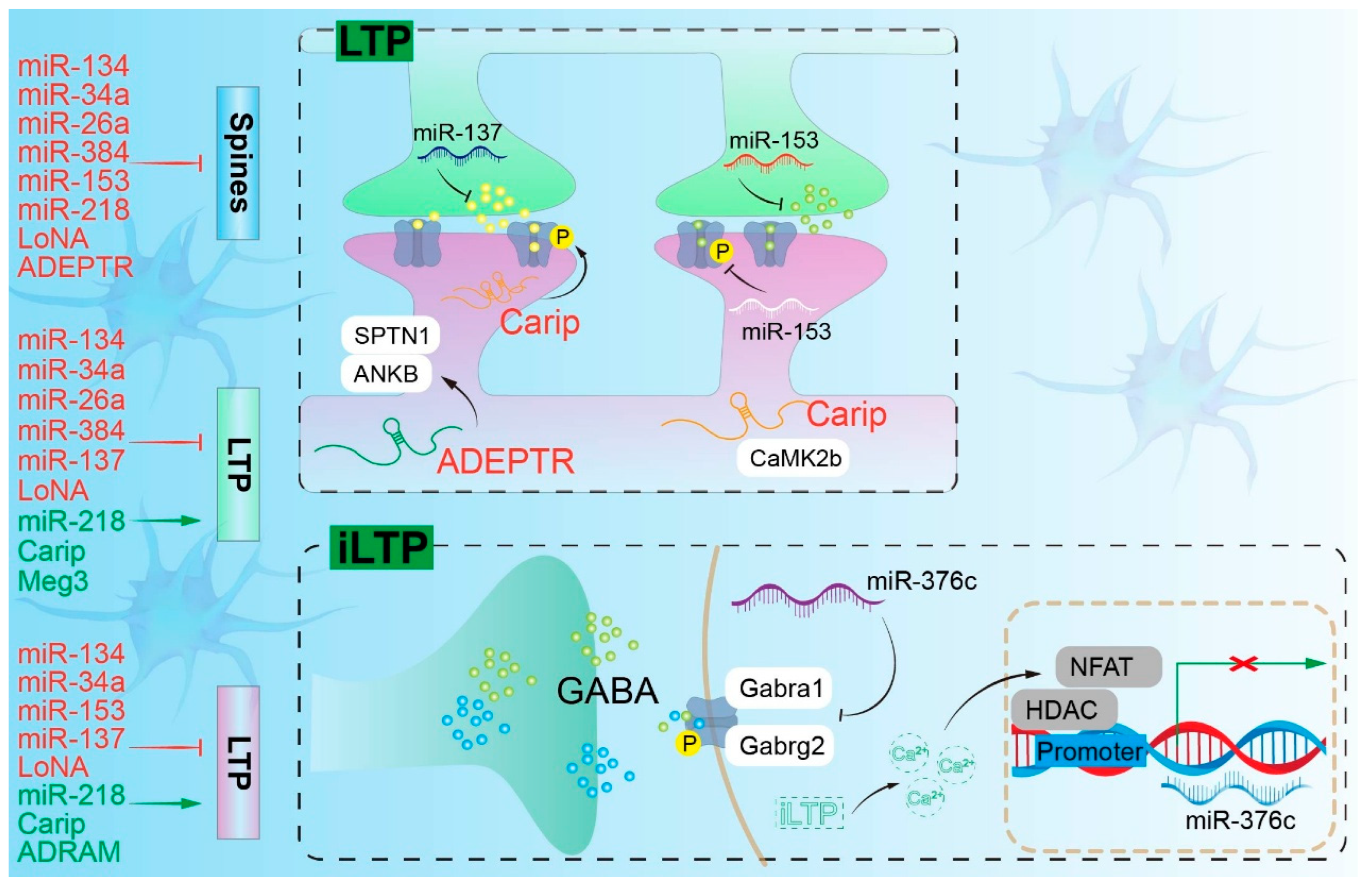

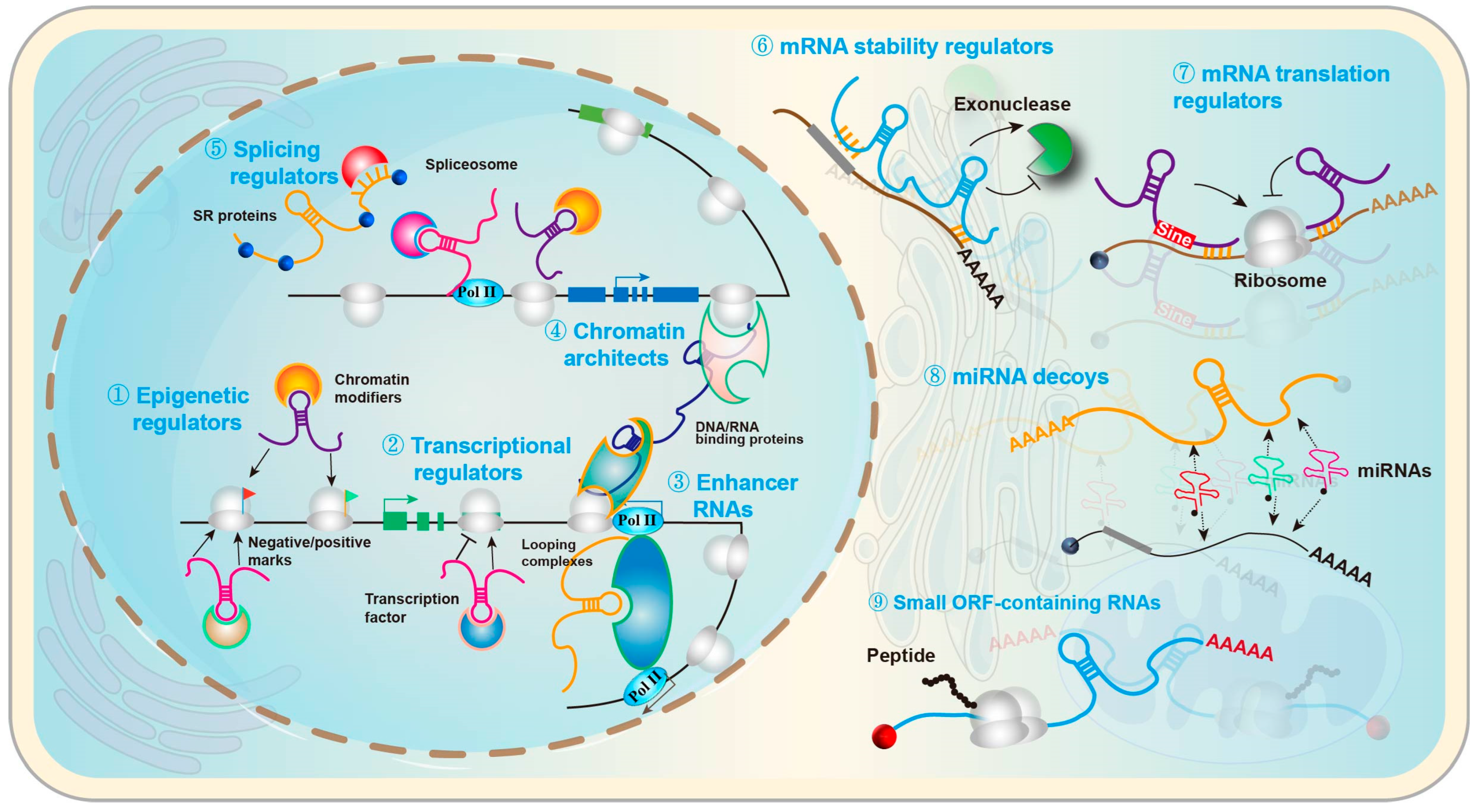
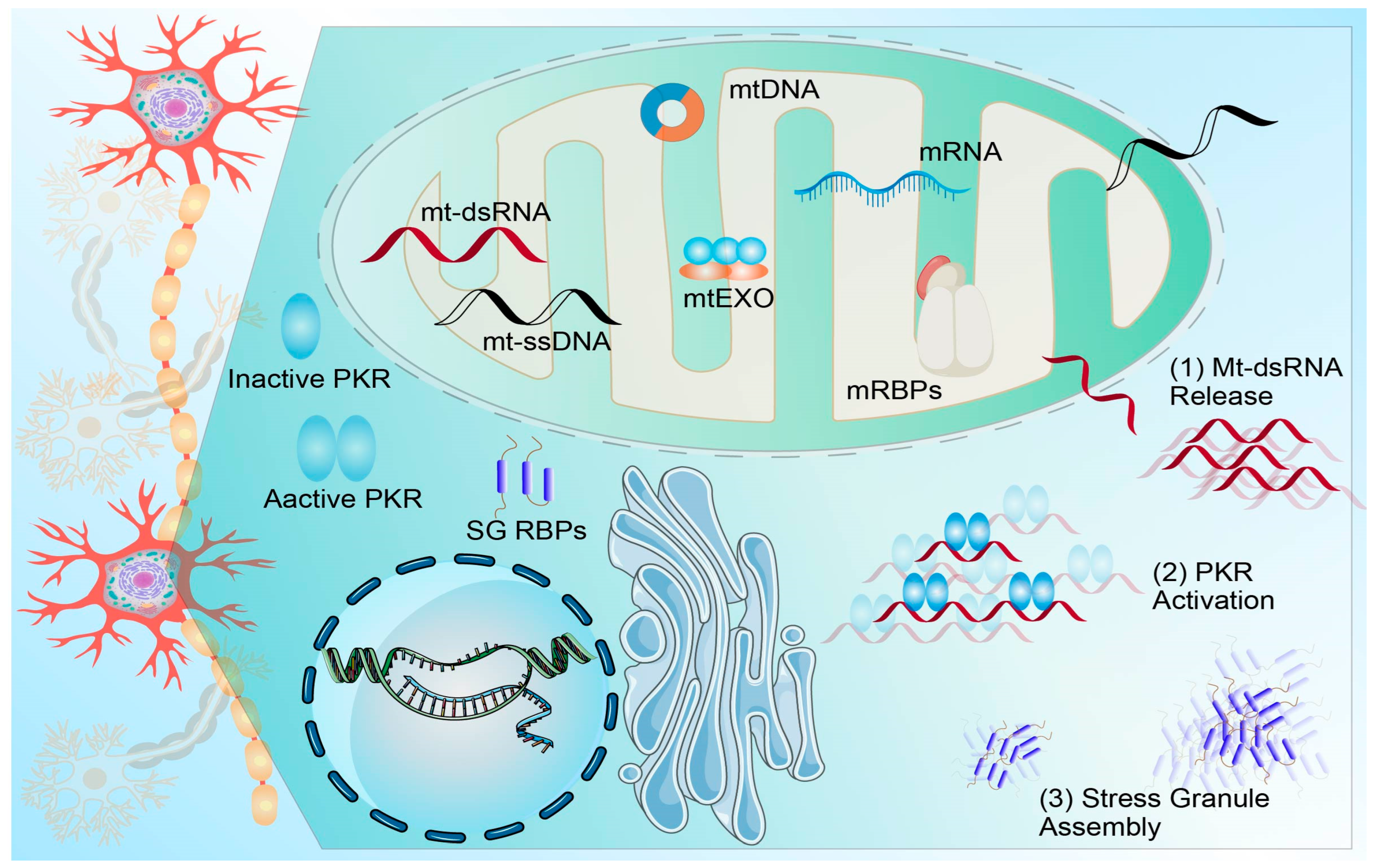
| Disease | ncRNA (Type) | Primary Target/Pathway | Change in Disease Context | Key Functional Effect | Model/Sample | Evidence Strength | Key Knowledge Gaps | Refs. |
|---|---|---|---|---|---|---|---|---|
| Alzheimer’s disease | miR-132 (microRNA) | Regulates networks controlling adult hippocampal neurogenesis and synaptic plasticity | Consistently downregulated in hippocampus of AD patients and mouse models | miR-132 replacement restores adult hippocampal neurogenesis and rescues memory deficits in AD mouse models | Multiple AD mouse models, human neural stem cells, post-mortem human hippocampus | Strong | Optimal delivery method for clinical translation undefined; long-term safety of miR-132 restoration unknown | [82] |
| Alzheimer’s disease | BACE1-AS (lncRNA, antisense to BACE1) | BACE1 mRNA/BACE1 protein and amyloidogenic processing | BACE1-AS expression is altered in STZ-induced AD rats and further increased after memantine treatment | Changes in BACE1-AS in brain and blood parallel alterations in BACE1 protein, suggesting roles in AD pathogenesis and treatment monitoring | STZ-induced AD rat model, brain and blood tissues | Moderate | Causality vs. correlation unclear; human validation limited to postmortem tissue | [103] |
| Alzheimer’s disease | miR-29a/b/c (microRNA) | BACE1 3′UTR/β-secretase activity | Downregulated in sporadic AD brain tissue | Reduced miR-29 leads to elevated BACE1, increased Aβ production, and plaque formation | Human postmortem brain, cell lines, AD mouse models | Strong | Specificity of miR-29 targeting; potential off-target effects of miR-29 mimics | [78,79] |
| Alzheimer’s disease | miR-34c (microRNA) | SIRT1, SYT1/survival and synaptic pathways | Upregulated in AD hippocampus | Promotes apoptosis, mitochondrial stress, and memory impairment | AD mouse models, human postmortem tissue | Moderate | Direction of causality unclear; functional redundancy with miR-34a/b | [80,81] |
| Alzheimer’s disease | NEAT1 (lncRNA) | FZD3/GSK3β/p-tau axis; paraspeckle formation | Upregulated in AD models | Modulates microtubule stability and tau phosphorylation; context-dependent neuroprotective or pathogenic roles | SH-SY5Y cells, APP/PS1 mice | Moderate | Conflicting reports on protective vs. deleterious effects; cell type-specific functions undefined | [47,48] |
| Parkinson’s disease | miR-7 (microRNA) | SNCA (α-synuclein) and autophagy machinery | miR-7 downregulation contributes to α-synuclein accumulation | miR-7 overexpression promotes autophagic clearance of α-synuclein monomers and aggregates, protecting against α-synuclein-induced toxicity | Differentiated human ReNcell VM cells overexpressing α-synuclein | Moderate | No clinical trial data; optimal delivery route to substantia nigra undefined | [104] |
| Parkinson’s disease | miR-7 (microRNA) | NLRP3/caspase-1 inflammasome and neuroinflammation | α-Synuclein overexpression reduces miR-7 and activates NLRP3 inflammasome | Stereotactic delivery of miR-7 mimics inhibits NLRP3/caspase-1 activation and improves subventricular zone neurogenesis | A53T α-synuclein transgenic mice; adult neural stem cells; intracerebroventricular miR-7 mimics | Moderate | Intracerebroventricular delivery not practical for chronic therapy; systemic delivery challenges | [105] |
| Parkinson’s disease | miR-34b/c (microRNA) | DJ-1, Parkin/mitophagy and mitochondrial quality control | Downregulated in PD substantia nigra | Compromises mitochondrial quality control and produces energetic deficits | Human postmortem substantia nigra, cell lines | Preliminary | Single postmortem study; functional validation limited | [97] |
| Parkinson’s disease | circSNCA (circRNA) | miR-7 sponging/SNCA expression | Elevated in PD models | circSNCA elevation suppresses miR-7, increases SNCA, promotes autophagy suppression and redox stress | Cell models, pramipexole treatment studies | Preliminary | Limited to cell culture; in vivo validation needed | [95,96] |
| Disease | ncRNA(s) | Sample Type | Direction of Change | Diagnostic Performance/Clinical Association | Study Population/Notes | Validation Status | Methodological Quality | Key Limitations | Refs. |
|---|---|---|---|---|---|---|---|---|---|
| Alzheimer’s disease | BACE1 (lncRNA) | Plasma | Increased in AD versus non-AD patients | Plasma BACE1 level showed high specificity (≈88%) for discriminating AD from non-AD subjects | Case–control study comparing four lncRNAs in plasma of AD and non-AD individuals; BACE1 showed the most robust separation | Discovery only | Moderate (QUADAS-2) | Single-center; no external validation; small sample size | [135] |
| Alzheimer’s disease | BACE1-AS (lncRNA) | Plasma and plasma-derived exosomes | Overall AD vs. control difference modest; but BACE1-AS levels differ between pre-AD and full AD subgroups | Plasma BACE1-AS discriminates pre-AD and controls, full AD and controls, and pre-AD and full AD with high specificity in ROC analyses | Cross-sectional cohort including AD, pre-AD and cognitively normal controls; BACE1-AS quantified in plasma and exosomes | Internal validation | Moderate (QUADAS-2) | Cross-sectional design; exosome isolation method variability | [134] |
| Alzheimer’s disease | Exosomal BACE1-AS (lncRNA) | Plasma exosomes | Elevated in AD compared with controls | Exosomal BACE1-AS alone yields AUC ≈ 0.76; combining exosomal BACE1-AS with right entorhinal cortex volume and thickness increases AUC and improves both sensitivity and specificity | AD patients and controls with plasma exosome profiling and 3D MRI of entorhinal cortex; combined molecular–imaging signature proposed as detection biomarker | Internal validation | Moderate (QUADAS-2) | Requires MRI integration; exosome isolation not standardized | [158] |
| Alzheimer’s disease | miR-29a, miR-125b panel (microRNA) | Serum/plasma exosomes | Altered in AD | Multi-analyte panels combining exosomal miRNAs improve diagnostic accuracy with reported sensitivities >80% | Multiple cohort studies | Discovery + partial validation | Low-Moderate (QUADAS-2) | Heterogeneous methods across studies; no consensus panel | [85,86,87] |
| Alzheimer’s disease | miR-135a (microRNA) | CSF, plasma | Altered in AD progression | Tracks disease progression in longitudinal studies | Longitudinal cohort studies | Internal validation | Moderate (QUADAS-2) | Limited sample sizes; normalization variability | [141] |
| Parkinson’s disease | miR-221 (microRNA) | Serum | Decreased in PD relative to healthy controls | Serum miR-221 shows ROC AUC ≈ 0.79 and correlates positively with UPDRS-III and UPDRS-V scores, suggesting association with motor severity | 138 PD patients and 112 controls; qRT–PCR panel of 16 PD-related miRNAs measured in serum | Internal validation | Moderate (QUADAS-2) | Single-center; no external validation cohort | [136] |
| Parkinson’s disease | miR-195, miR-185, miR-15b, miR-221, miR-181a (microRNA panel) | Serum | miR-195 upregulated; miR-185, miR-15b, miR-221 and miR-181a downregulated in PD | Five-miRNA signature accurately distinguishes PD from healthy individuals and is proposed as a serum-based diagnostic panel | Discovery and validation cohorts of PD patients and controls; high-throughput qRT–PCR and ROC analysis | External validation | High (QUADAS-2) | Validation cohort from same geographic region; generalizability uncertain | [159] |
| Parkinson’s disease | miR-214, miR-221, miR-141 (microRNAs, combined with cytokines and antioxidants) | Serum | miR-214 decreased; miR-221 and miR-141 decreased together with altered cytokines and antioxidant markers | Combined panel of miRNAs, cytokines, α-synuclein and antioxidant markers improves discrimination between PD patients and controls compared with single markers | 20 PD patients and 15 controls; integrated analysis of serum cytokines, α-synuclein, miRNAs and antioxidant enzymes | Discovery only | Low (QUADAS-2) | Very small sample size (n = 35); no validation | [160] |
| Parkinson’s disease | EV-miRNAs (miR-7, miR-153, miR-19b) | CSF, serum EVs | Altered levels correlating with PD severity | Panels integrating EV-miRNAs with circRNAs show promising early diagnostic performance | Longitudinal and cohort studies | Internal validation | Moderate (QUADAS-2) | EV isolation heterogeneity; study-specific panels | [99,100,101] |
| Disease/Indication | Therapeutic Strategy | Target RNA/Pathway | Delivery Approach | Key Preclinical or Clinical Outcome | Development Stage | Evidence Level | Key Limitations/Failures | Refs. |
|---|---|---|---|---|---|---|---|---|
| Alzheimer’s disease | miR-132 replacement therapy | miR-132 deficiency affecting adult hippocampal neurogenesis and memory circuits | Hippocampal delivery of miR-132 (e.g., viral vectors or mimics) in AD mouse models | Restoring miR-132 expression rescues adult hippocampal neurogenesis and ameliorates memory deficits in AD mouse models, supporting miR-132 as a therapeutic candidate | Preclinical (mouse models, human neural stem cells) | Preclinical (in vivo) | No human safety data; delivery to human hippocampus technically challenging; long-term expression stability unknown | [82] |
| Alzheimer’s disease | BACE1-AS targeting | BACE1-AS lncRNA/BACE1 stabilization | ASO or siRNA targeting BACE1-AS | Knockdown reduces BACE1 levels and Aβ production in cell models | Preclinical (cell culture) | Preclinical (in vitro) | Cell culture only; in vivo validation lacking; potential off-target effects on BACE1 regulation | [42,43] |
| Alzheimer’s disease | Anti-miR-34c therapy | miR-34c/SIRT1, synaptic targets | Antagomir delivery | Inhibition of miR-34c rescues memory deficits in AD mouse models | Preclinical (mouse models) | Preclinical (in vivo) | Specificity concerns (miR-34 family redundancy); no human data | [84] |
| Parkinson’s disease | miR-7 overexpression to reduce α-synuclein burden | SNCA (α-synuclein) and autophagy pathway | Lentiviral miR-7 overexpression in neuron-like cells (ReNcell VM) expressing α-synuclein | miR-7 reduces monomeric and aggregated α-synuclein and promotes autophagic clearance, highlighting miR-7 as a potential disease-modifying approach | Preclinical (human neuron-like cell model) | Preclinical (in vitro) | Cell model only; no in vivo PD model testing; delivery to substantia nigra undefined | [104] |
| Parkinson’s disease | miR-7 mimic administration to modulate neuroinflammation and neurogenesis | NLRP3/caspase-1 inflammasome and α-synuclein–induced neuroinflammation | Stereotactic intracerebroventricular injection of synthetic miR-7 mimics in A53T α-synuclein transgenic mice | miR-7 mimics suppress NLRP3/caspase-1 activation and improve subventricular zone neurogenesis, suggesting potential to restore regenerative capacity in PD | Preclinical (transgenic mouse model) | Preclinical (in vivo) | Invasive delivery (ICV injection); chronic dosing not evaluated; translation to human unclear | [105] |
| Parkinson’s disease | circSNCA modulation | circSNCA/miR-7 sponging | Pharmacological (pramipexole) | Pramipexole downregulates circSNCA, restores miR-7, and attenuates apoptosis | Preclinical (cell models) | Preclinical (in vitro) | Indirect mechanism; pramipexole’s circSNCA effects may be secondary to dopaminergic action | [95,96] |
| SOD1-mutant ALS | Antisense oligonucleotide tofersen | SOD1 mRNA (reducing mutant SOD1 protein) | Repeated intrathecal administration of ASO (100 mg) | Phase 3 trial shows tofersen lowers CSF SOD1 and plasma neurofilament light; early-start treatment in the open-label extension is associated with more favorable functional outcomes despite primary endpoint not being met | FDA Approved (April 2023) | Phase 3 + Approved | Primary clinical endpoint not met in initial analysis; requires repeated lumbar punctures; limited to SOD1 mutation carriers (~2% of ALS) | [77,173] |
| Tauopathy/mild AD | MAPT-targeting ASO (BIIB080/MAPTRx) | MAPT mRNA/tau protein | Intrathecal administration | ~50% reduction in CSF total tau and p-tau181; reductions persist ≥12 weeks post-dosing; numerical tau-PET reductions | Phase 1b completed; Phase 2 ongoing | Phase 2 | Not powered for clinical outcomes; long-term safety unknown; requires repeated IT dosing | [164] |
| Huntington’s disease | HTT-lowering ASO tominersen | HTT mRNA (both mutant and wild-type) | Intrathecal administration | Phase 1/2a showed dose-dependent CSF mHTT reduction; Phase 3 halted for futility with worse outcomes in some treatment groups | Phase 3 (terminated) | Phase 3 (Failed) | CRITICAL FAILURE: Non-allele-selective; possible over-suppression of wild-type HTT; younger patients showed worse outcomes; dosing interval may have been suboptimal | [112,176] |
| Huntington’s disease | AAV5-miHTT (AMT-130) gene therapy | HTT mRNA via artificial microRNA | Single neurosurgical intrastriatal injection | Preclinical: 60–80% HTT mRNA/protein reduction sustained ≥12 months in rodents and NHPs with functional rescue; Phase 1/2 ongoing | Phase 1/2 (ongoing) | Phase 1/2 | Irreversible; requires neurosurgery; long-term safety of permanent HTT suppression unknown; phase 3 tominersen failure raises caution | [178,179] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhao, X.; Zheng, Y.; Cai, X.; Yao, Y.; Qin, D. The Expanding Role of Non-Coding RNAs in Neurodegenerative Diseases: From Biomarkers to Therapeutic Targets. Pharmaceuticals 2026, 19, 92. https://doi.org/10.3390/ph19010092
Zhao X, Zheng Y, Cai X, Yao Y, Qin D. The Expanding Role of Non-Coding RNAs in Neurodegenerative Diseases: From Biomarkers to Therapeutic Targets. Pharmaceuticals. 2026; 19(1):92. https://doi.org/10.3390/ph19010092
Chicago/Turabian StyleZhao, Xuezhi, Yongquan Zheng, Xiaoyu Cai, Yao Yao, and Dongxu Qin. 2026. "The Expanding Role of Non-Coding RNAs in Neurodegenerative Diseases: From Biomarkers to Therapeutic Targets" Pharmaceuticals 19, no. 1: 92. https://doi.org/10.3390/ph19010092
APA StyleZhao, X., Zheng, Y., Cai, X., Yao, Y., & Qin, D. (2026). The Expanding Role of Non-Coding RNAs in Neurodegenerative Diseases: From Biomarkers to Therapeutic Targets. Pharmaceuticals, 19(1), 92. https://doi.org/10.3390/ph19010092








