The SPINK Protein Family in Cancer: Emerging Roles in Tumor Progression, Therapeutic Resistance, and Precision Oncology
Abstract
1. Introduction
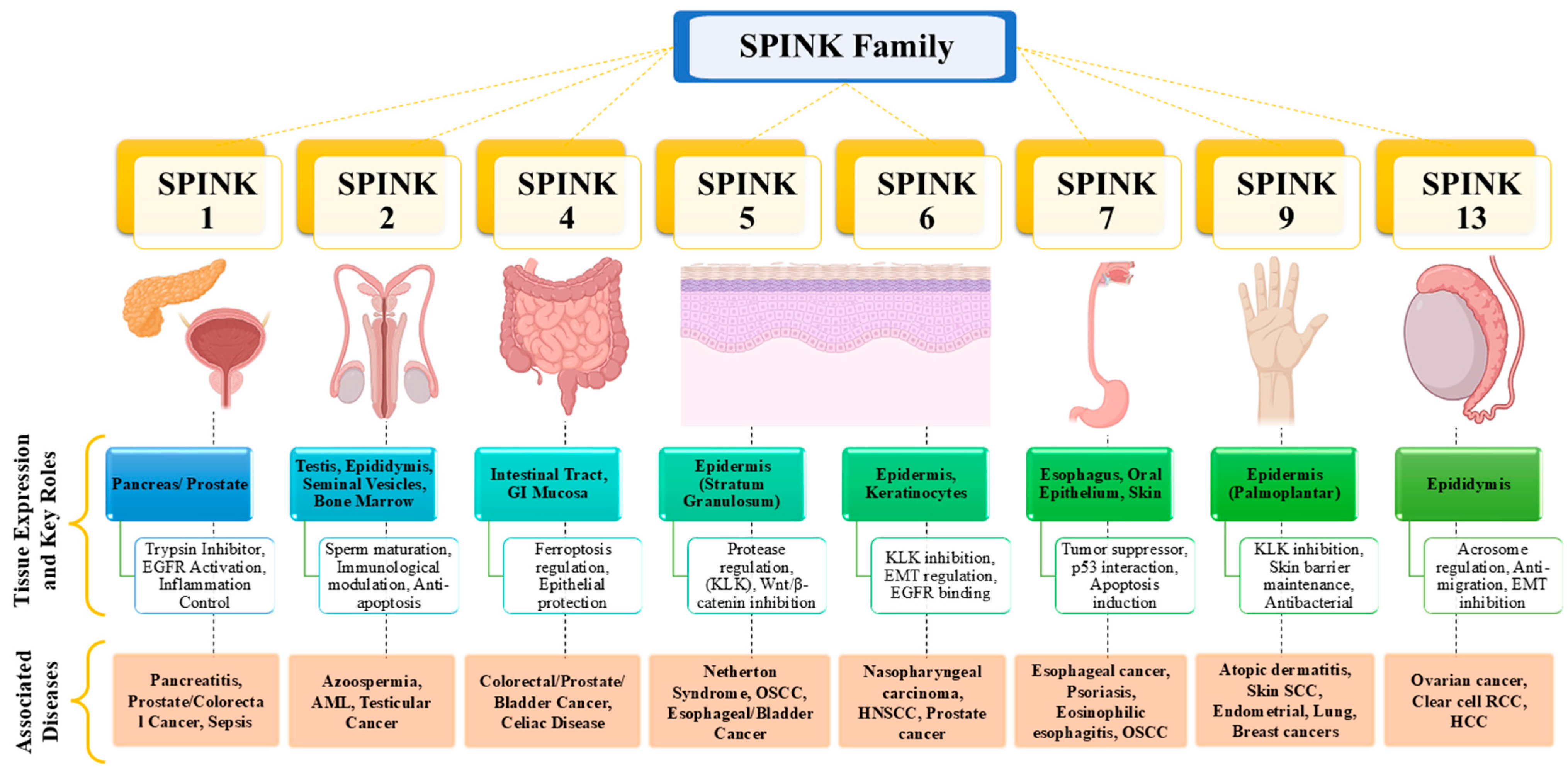
2. SPINK Family and Cancer: Classification and Functional Roles
2.1. Overview of SPINK Family Proteins
| S. No. | SPINK Member | Target Protease | Known Substrate | Functional Role | References |
|---|---|---|---|---|---|
| 1 | SPINK1 | Trypsin, KLK5, 7 | Pancreatic zymogens, Desmoglein-1 (KLK5/7) | Pancreatitis, cancer cell proliferation, Skin (Inflammation) | [25,26,27] |
| 2 | SPINK2 | Acrosin, Trypsin-like serine proteases | Acrosomal proteins, Apoptotic regulators (Unknown) | Spermatogenesis, Apoptotic resistance in bone marrow | [22,28] |
| 3 | SPINK4 | Trypsin, Elastase | Intestinal epithelial proteins, mucins | Colonic inflammation, intestinal homeostasis | [29] |
| 4 | SPINK5 (LEKTI) | KLK5, 7, 14 | Desmogleins, Corneo-desmosomal proteins | Skin barrier integrity, Netherton syndrome | [30,31] |
| 5 | SPINK6 | KLK5, 7, 14 | Fibronectin, Desmosomal proteins (via KLKs) | Skin desquamation, anti-inflammatory response | [32,33] |
| 6 | SPINK7 | Predicted trypsin-like serine protases | Unknown | Esophageal epithelial protection | [34,35] |
| 7 | SPINK9 | KLK5 | Corneo-desmosomal proteins | Palmoplantar skin barrier protection | [36] |
| 8 | SPINK13 | Predicted trypsin-like proteases | Unknown | Tumor suppression in HCC | [10,37] |
2.2. Role of SPINK Pathways and Different Isoforms in Cancer
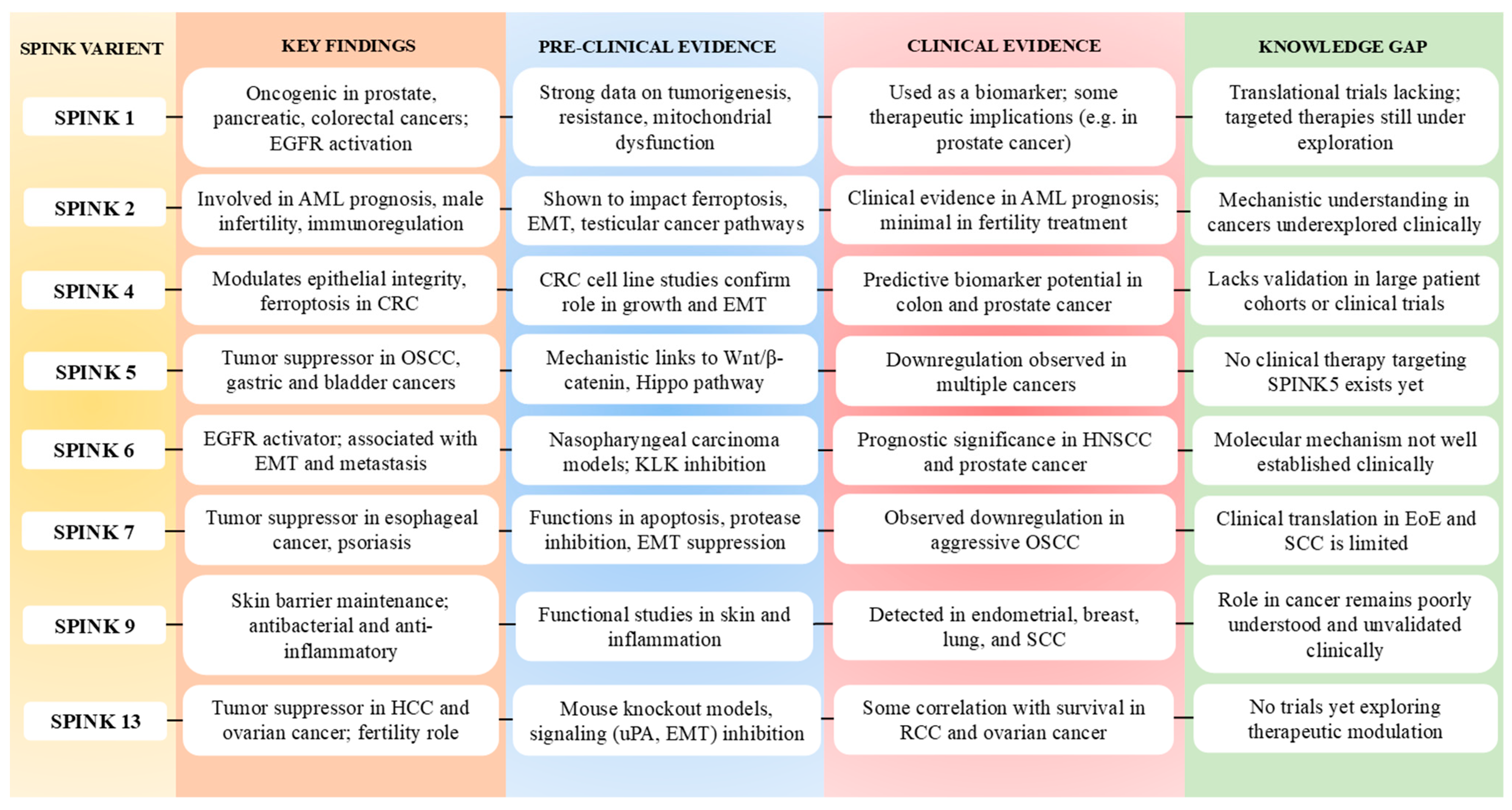
2.2.1. SPINK1
2.2.2. SPINK2
2.2.3. SPINK4
2.2.4. SPINK5
2.2.5. SPINK6
2.2.6. SPINK7
2.2.7. SPINK9
2.2.8. SPINK13
2.3. SPINK as a Biomarker and Therapeutic Target
| S. No. | Aspect | Details | Cancer Type | Reference |
|---|---|---|---|---|
| 1 | Diagnostic Biomarker | Elevated SPINK1 expression correlates with poor prognosis and tumor grade. | Prostate, Pancreatic | [106,107] |
| 2 | Prognostic Biomarker | High SPINK expression lined to aggressive tumor behavior and recurrence. | Colorectal, Ovarian | [96,108] |
| 3 | Predictive Biomarker | SPINK mutations predict resistance to chemotherapy and poor outcomes. | Lung, Gastric | [109,110] |
| 4 | Therapeutic Target | Inhibiting SPINK1 reduces tumor growth and enhances chemosensitivity. | Prostate, HCC | [10] |
| 5 | Role in Metabolic Pathways | SPINK—mediated mitochondrial dysfunction promotes metabolic reprogramming. | Breast, Pancreatic | [11] |
| 6 | Immune Modulation | SPINK overexpression facilitates immune evasion by modulating TME | Colorectal, Lung | [7] |
| 7 | Potential Therapeutics | SPINK inhibitors and mitochondrial modulators are under preclinical testing | Multiple Cancer Types | [19] |
2.4. SPINK and Its Relation to Various Diseases
2.4.1. Acute and Chronic Pancreatitis
2.4.2. Azoospermia
2.4.3. Celiac Disease
2.4.4. Netherton Syndrome
2.4.5. Eosinophilic Esophagitis
2.4.6. Psoriasis and Eczema
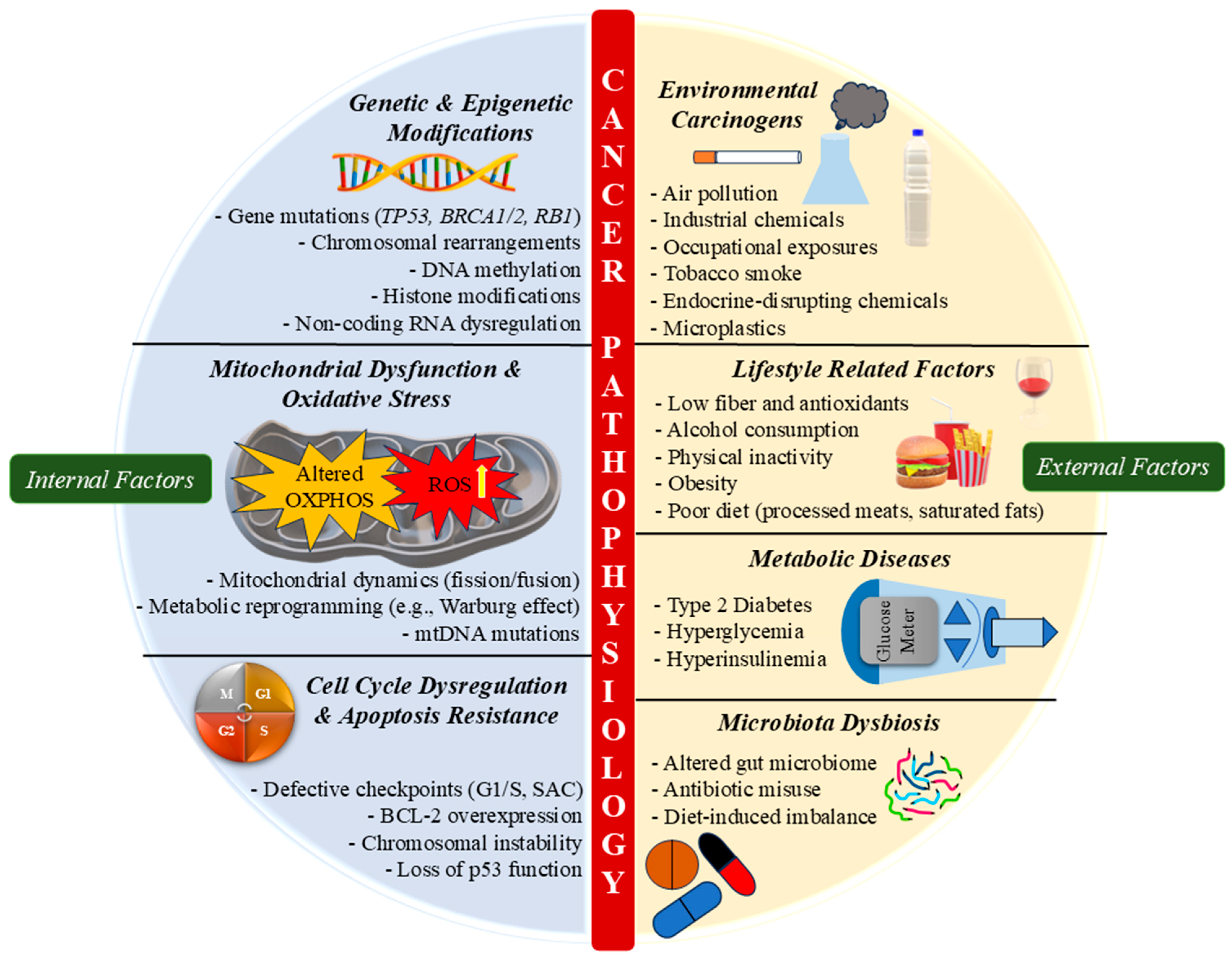
3. Cancer Pathophysiology: Role of Internal and External Factors
3.1. Genetic and Epigenetic Alterations
3.2. Environmental and Lifestyle Influences
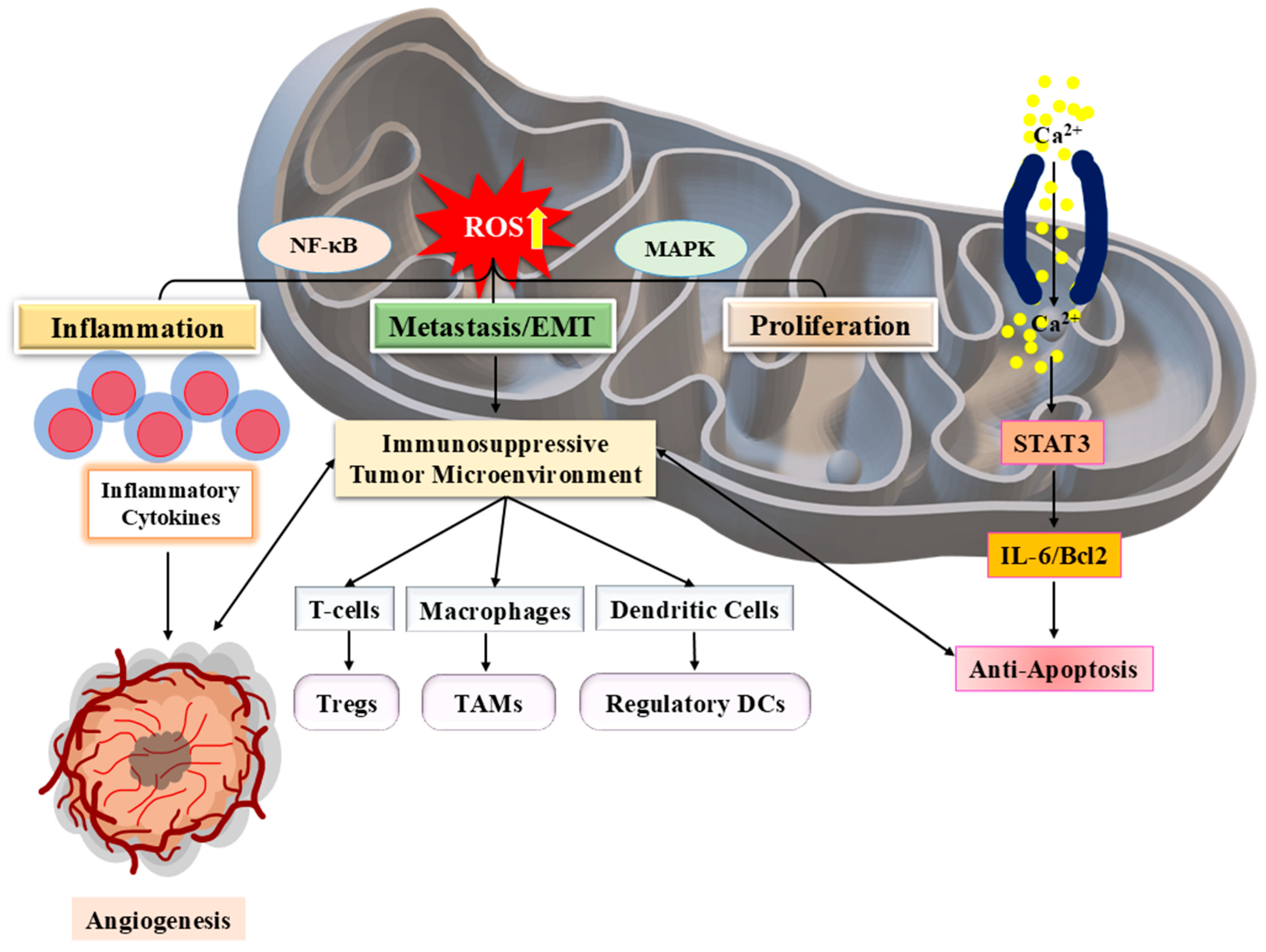
3.3. Inflammatory Responses and Tumor Microenvironment
3.4. Dysregulation of Cell Cycle and Apoptosis
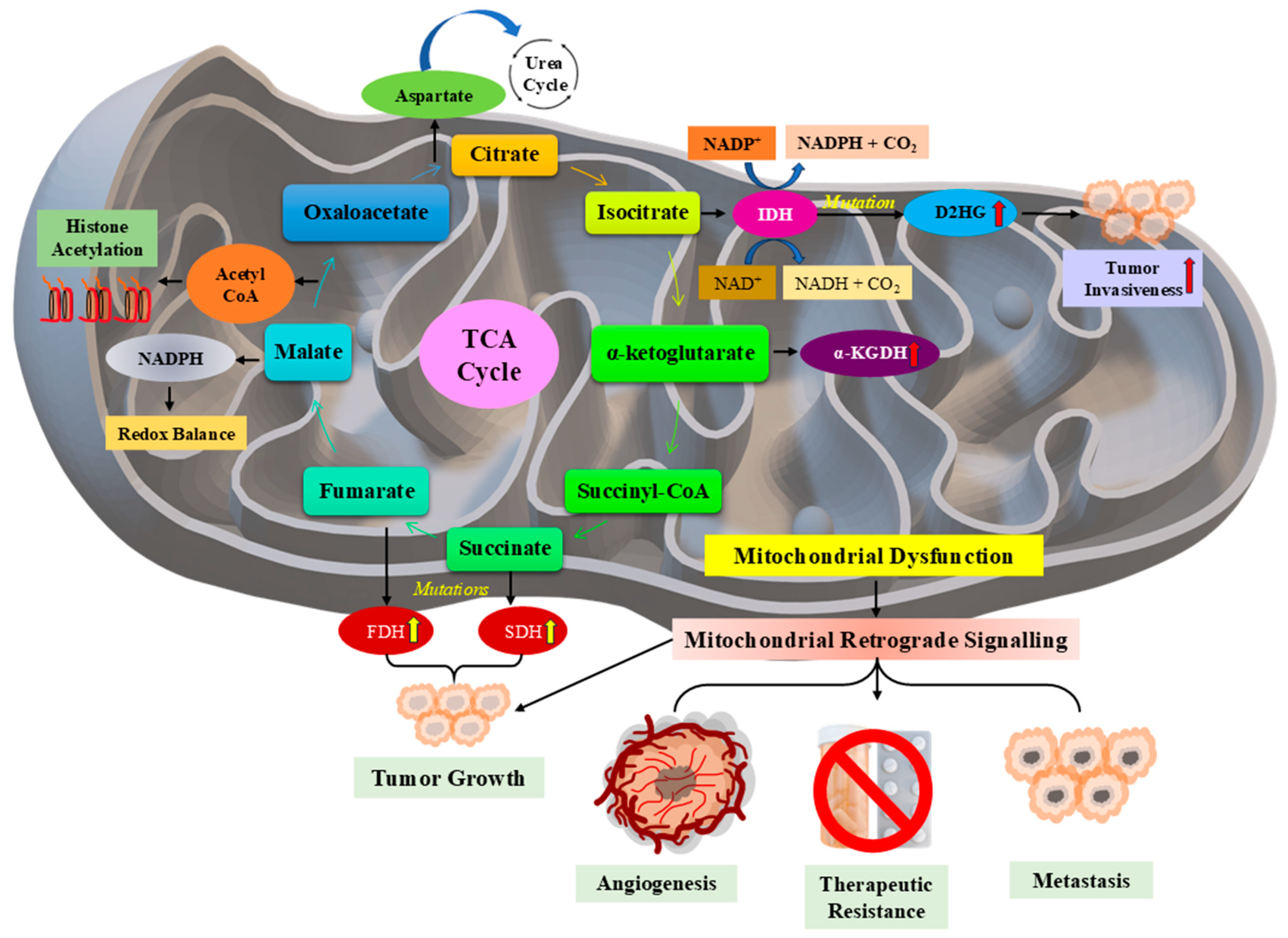
3.5. Mitochondrial Dysfunction in Cancer Progression
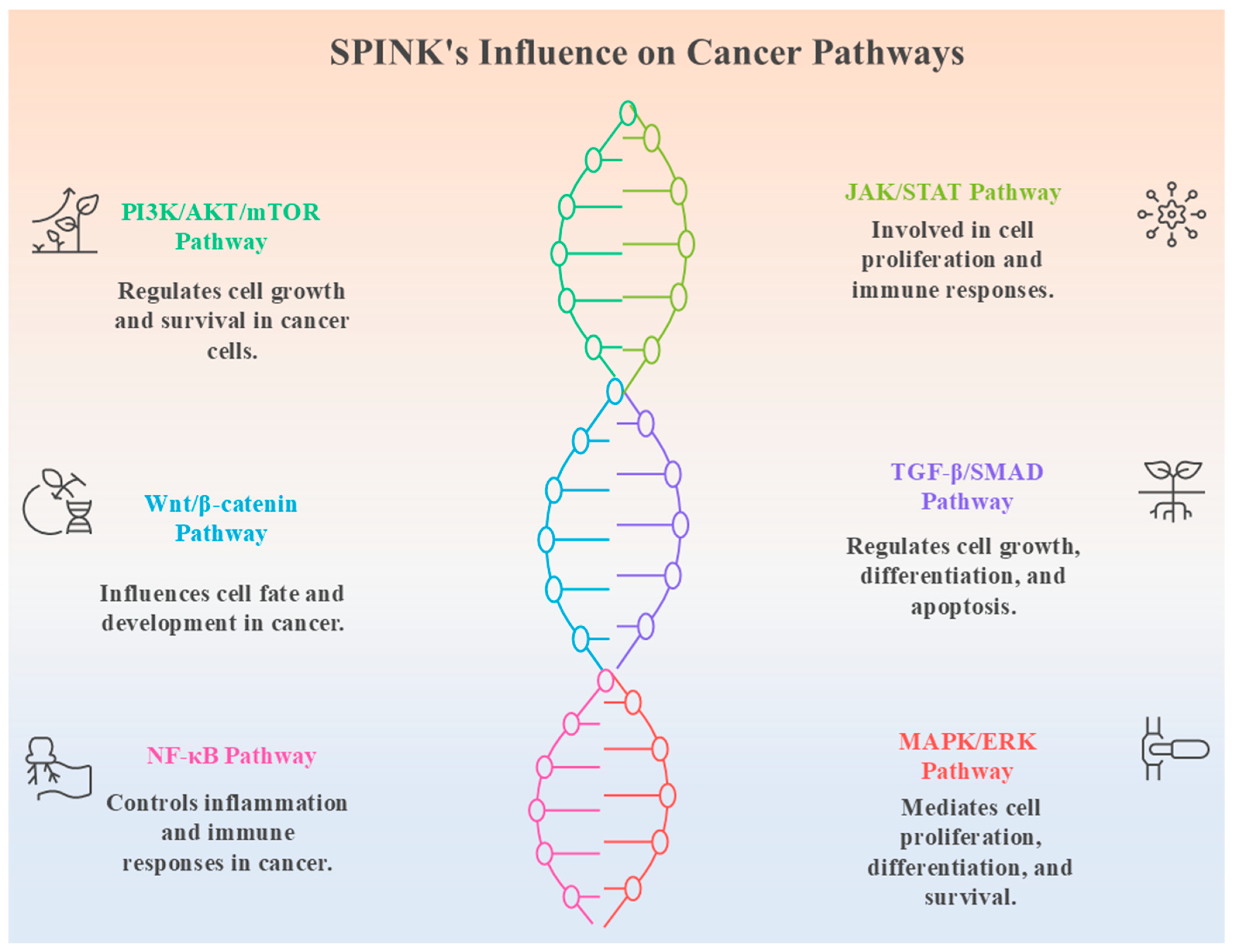
4. SPINK-Mediated Signalling Mechanisms in Cancer
4.1. SPINK Interactions with Growth Factor Receptors
4.2. Impact on Cell Proliferation, Metastasis, and Therapy Resistance
4.3. Crosstalk with Other Oncogenic Pathways
5. Therapeutic Implications and Future Directions
5.1. Potential Strategies for Targeting SPINK in Cancer Therapy
| S. No. | Strategy | Mechanism of Action | Therapeutic Approach | Cancer Type | References |
|---|---|---|---|---|---|
| 1 | SPINK Inhibitors | Direct inhibition of SPINK proteins to suppress tumor growth | Small-molecule inhibitors | Prostate, Pancreatic | [10] |
| 2 | RNA Interference (RNAi) | Silencing SPINK gene expression to inhibit oncogenic activity | siRNA and shRNA-based therapies | Hepatocellular, Colorectal | [219] |
| 3 | CRISPR—Cas9 Gene Editing | Targeted deletion or correction of SPINK gene mutation | Genome-editing technology | Breast, Lung | [220,221] |
| 4 | Mitochondrial Modulators | Restoration of mitochondrial function altered by SPINK dysregulation | Antioxidants OXPHOS inhibitors | Breast, Pancreatic | [11] |
| 5 | Immune Modulation | Enhance anti—tumor immunity by targeting SPINK-induced immune evasion | Immune checkpoint inhibitors | Colorectal, Lung | [222,223] |
| 6 | Combination Therapy | SPINK inhibition alongside chemotherapy or targeted therapy | Dual drug regimens | Prostate, Ovarian | [224,225] |
| 7 | Nutritional Modulation | Diet-based approaches to reduce SPINK-mediated inflammation | Antioxidant-rich diets, supplements | Multiple Cancer Types | [226] |
5.2. Challenges in Developing SPINK Inhibitors
5.3. Emerging Trends in Cancer Therapy
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADT | Androgen Deprivation Therapy |
| AHR | Aryl Hydrocarbon Receptor |
| AML | Acute Myeloid Leukemia |
| AR | Androgen Receptor |
| CD | Celiac Disease |
| CIN | Chromosomal Instability |
| CRC | Colorectal Cancer |
| CRPC | Castration Resistant Prostate Cancer |
| ECRG2 | Esophageal Cancer-related Gene 2 |
| EGFR | Epidermal Growth Factor Receptor |
| EHMT2 | Euchromatic Histone Lysine Methyltransferase 2 |
| EMT | Epithelial–Mesenchymal Transition |
| EoE | Eosinophilic Esophagitis |
| GI | Gastrointestinal |
| GSK3β | Glycogen Synthase Kinase 3 beta |
| HCC | Hepatocellular Carcinoma |
| HDAC | Histone Deacetylase |
| HIF | Hypoxia Inducible Factor |
| HMT | Histone Methyltransferase |
| HNSCC | Head and Neck Squamous Cell Carcinoma |
| HO1 | Heme Oxygenase 1 |
| IL | Interleukin |
| KLK | Kallikrein |
| LEKTI | Lymphoepithelial Kazal-type-related Inhibitor |
| MMP | Matrix Metalloproteinase |
| mtDNA | mitochondrial DNA |
| mTOR | Mechanistic Target of Rapamycin |
| NAFLD | Non-Alcoholic Fatty Liver Disease |
| NOA | Non-Obstructive Azoospermia |
| NS | Netherton Syndrome |
| OMTKY3 | Turkey Ovomucoid Third Domain |
| OS | Oxidative Stress |
| OSCC | Oral Squamous Cell Carcinoma |
| PC | Prostate Cancer |
| PST1 | Pancreatic Secretory Trypsin Inhibitor |
| PTEN | Phosphatase and Tensin Homolog |
| RCC | Renal Cell Carcinoma |
| ROS | Reactive Oxygen Species |
| SAC | Spindle Assembly Checkpoints |
| SASP | Senescence Associated Secretory Phenotype |
| SCC | Squamous Cell Carcinoma |
| shRNA | Short Hairpin RNA |
| SPINK | Serine Protease Inhibitor Kazal Type |
| TERT | Telomerase Reverse Transcriptase |
| TIG1 | Tazarotene-induced Gene 1 |
| TME | Tumor Microenvironment |
| TNF | Tumor Necrosis Factor |
| uPA | Urokinase-type Plasminogen Activator |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Sathishkumar, K.; Chaturvedi, M.; Das, P.; Stephen, S.; Mathur, P. Cancer incidence estimates for 2022 & projection for 2025: Result from National Cancer Registry Programme, India. Indian J. Med. Res. 2022, 156, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.M. Epidemiology of Cancer. Clin. Chem. 2024, 70, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, S.M.; Al Doghaither, H.A.; Al-Ghafar, A.B. General insight into cancer: An overview of colorectal cancer (review). Mol. Clin. Oncol. 2021, 15, 271. [Google Scholar] [CrossRef]
- Swain, N.; Hosalkar, R.; Thakur, M.; Prabhu, A.H. Hallmarks of Cancer: Its Concept and Critique. In Microbes and Oral Squamous Cell Carcinoma: A Network Spanning Infection and Inflammation; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar] [CrossRef]
- Roy, M.; Datta, A. Cancer: Types and Hallmarks. In Cancer Genetics and Therapeutics; Springer: Singapore, 2019; pp. 1–26. [Google Scholar] [CrossRef]
- Hanahan, D.; Monje, M. Cancer hallmarks intersect with neuroscience in the tumor microenvironment. Cancer Cell 2023, 41, 573–580. [Google Scholar] [CrossRef]
- Chae, H.-S.; Hong, S.-T. Overview of Cancer Metabolism and Signaling Transduction. Int. J. Mol. Sci. 2022, 24, 12. [Google Scholar] [CrossRef]
- Al-Ostoot, F.H.; Salah, S.; Khanum, S.A. An Overview of Cancer Biology, Pathophysiological Development and It’s Treatment Modalities: Current Challenges of Cancer anti-Angiogenic Therapy. Cancer Investig. 2024, 42, 559–604. [Google Scholar] [CrossRef]
- Lun, Y.; Sun, J.; Wei, L.; Liu, B.; Li, Z.; Dong, W.; Zhao, W. SPINK13 acts as a tumor suppressor in hepatocellular carcinoma by inhibiting Akt phosphorylation. Cell Death Dis. 2024, 15, 822. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Guo, Y.; Shi, X.; Chen, X.; Feng, W.; Wu, L.-L.; Zhang, J.; Yu, S.; Wang, Y.; et al. An Overview: The Diversified Role of Mitochondria in Cancer Metabolism. Int. J. Biol. Sci. 2023, 19, 897–915. [Google Scholar] [CrossRef]
- Radzak, S.M.A.; Khair, S.Z.M.; Ahmad, F.; Patar, A.; Idris, Z.; Yusoff, A.M. Insights regarding mitochondrial DNA copy number alterations in human cancer (Review). Int. J. Mol. Med. 2022, 50, 104. [Google Scholar] [CrossRef]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA variation and cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef]
- Shadhu, K.; Xi, C. Inflammation and pancreatic cancer: An updated review. Saudi J. Gastroenterol. 2019, 25, 3. [Google Scholar] [CrossRef]
- Gukovsky, I.; Li, N.; Todoric, J.; Gukovskaya, A.; Karin, M. Inflammation, Autophagy, and Obesity: Common Features in the Pathogenesis of Pancreatitis and Pancreatic Cancer. Gastroenterology 2013, 144, 1199–1209.e4. [Google Scholar] [CrossRef]
- Szabo, I.; Zoratti, M.; Biasutto, L. Targeting mitochondrial ion channels for cancer therapy. Redox Biol. 2021, 42, 101846. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Babuharisankar, A.P.; Lin, Y.-C.; Lien, H.-W.; Lo, Y.K.; Chou, H.-Y.; Tangeda, V.; Cheng, L.-C.; Cheng, A.N.; Lee, A.Y.-L. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: Foe or friend? J. Biomed. Sci. 2022, 29, 74. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Lim, S.-N.; Chen, C.-Y.; Chi, H.-C.; Yeh, C.-T.; Lin, W.-R. Functional Role of Mitochondrial DNA in Cancer Progression. Int. J. Mol. Sci. 2022, 23, 1659. [Google Scholar] [CrossRef]
- Kumar, S.; Dhamija, B.; Attrish, D.; Sawant, V.; Sengar, M.; Thorat, J.; Shet, T.; Jain, H.; Purwar, R. Genetic alterations and oxidative stress in T cell lymphomas. Pharmacol. Ther. 2022, 236, 108109. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.C.; Wang, W.P.; Chi, Y.H. AKT phosphorylation as a predictive biomarker for PI3K/mTOR dual inhibition-induced proteolytic cleavage of mTOR companion proteins in small cell lung cancer. Cell Biosci. 2022, 12, 122. [Google Scholar] [CrossRef]
- Chen, D.; Shi, Z.; Gao, X.; Yang, Y.; Lei, X.; Hu, Y. SPINK1 is a Potential Diagnostic and Prognostic Biomarker for Sepsis. Infect. Drug Resist. 2024, 17, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Di Bella, V.; Nigro, L.L.; Privitera, A.P.; Bonaccorso, P.; Scuderi, C.; Condorelli, D.F. Temporary serine protease inhibition and the role of SPINK2 in human bone marrow. iScience 2023, 26, 106949. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.R.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef]
- Lilly, A.C.; Astsaturov, I.; Golemis, E.A. Intrapancreatic fat, pancreatitis and pancreatic cancer. Cell. Mol. Life Sci. 2023, 80, 206. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, Y.; Shao, D.; Pan, Y.; Gao, X.; Zhao, P.; Liu, Q.; Shang, G.; Shang, W.; Fu, Z.; et al. High expression of serine protease inhibitor kazal type 1 predicts poor prognosis and promotes the progression and invasion of oral tongue squamous cell carcinoma. Arch. Oral Biol. 2024, 164, 106003. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Shi, Z.; Johnson, J.J.; Liu, Y.; Stack, M.S. Kallikrein-5 promotes cleavage of desmoglein-1 and loss of cell-cell cohesion in oral squamous cell carcinoma. J. Biol. Chem. 2011, 286, 9127–9135. [Google Scholar] [CrossRef]
- Kind, S.; Castillo, C.; Uhlig, R.; Gorbokon, N.; Lennartz, M.; Rico, S.D.; Reiswich, V.; Viehweger, F.; Kluth, M.; Hube-Magg, C.; et al. Abstract 3302: KLK7 expression in human tumors: A tissue microarray study on 13,447 tumors. Cancer Res. 2023, 83, 794. [Google Scholar] [CrossRef]
- Kherraf, Z.; Christou-Kent, M.; Karaouzene, T.; Amiri-Yekta, A.; Martinez, G.; Vargas, A.S.; Lambert, E.; Borel, C.; Dorphin, B.; Aknin-Seifer, I.; et al. SPINK 2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol. Med. 2017, 9, 1132–1149. [Google Scholar] [CrossRef]
- Wang, Y.; Han, J.; Yang, G.; Zheng, S.; Zhou, G.; Liu, X.; Cao, X.; Li, G.; Zhang, B.; Xie, Z.; et al. Therapeutic potential of the secreted Kazal-type serine protease inhibitor SPINK4 in colitis. Nat. Commun. 2024, 15, 5874. [Google Scholar] [CrossRef]
- Deraison, C.; Bonnart, C.; Lopez, F.; Besson, C.; Robinson, R.; Jayakumar, A.; Wagberg, F.; Brattsand, M.; Hachem, J.P.; Leonardsson, G.; et al. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol. Biol. Cell 2007, 18, 3607–3619. [Google Scholar] [CrossRef]
- Zani, M.B.; Sant’Ana, A.M.; Tognato, R.C.; Chagas, J.R.; Puzer, L. Human Tissue Kallikreins-Related Peptidases Are Targets for the Treatment of Skin Desquamation Diseases. Front. Med. 2022, 8, 777619. [Google Scholar] [CrossRef]
- Meyer-Hoffert, U.; Wu, Z.; Kantyka, T.; Fischer, J.; Latendorf, T.; Hansmann, B.; Bartels, J.; He, Y.; Gläser, R.; Schröder, J.M. Isolation of SPINK6 in human skin: Selective inhibitor of kallikrein-related peptidases. J. Biol. Chem. 2010, 285, 32174–32181. [Google Scholar] [CrossRef]
- Fischer, J.; Wu, Z.; Kantyka, T.; Sperrhacke, M.; Dimitrieva, O.; Koblyakova, Y.; Ahrens, K.; Graumann, N.; Baurecht, H.; Reiss, K.; et al. Characterization of Spink6 in Mouse Skin: The Conserved Inhibitor of Kallikrein-Related Peptidases Is Reduced by Barrier Injury. J. Investig. Dermatol. 2013, 134, 1305–1312. [Google Scholar] [CrossRef]
- Azouz, N.P.; Michael, D.; Furio, L.; Hovnanian, A.; Rothenberg, M.E. Loss of SPINK7 in Esophageal Epithelial Cells Unleashes a Pro-Inflammatory Response Characterized by Excessive Cytokine Production and Loss of Barrier Function. J. Allergy Clin. Immunol. 2016, 137, AB280. [Google Scholar] [CrossRef]
- Azouz, N.P.; Ynga-Durand, M.A.; Caldwell, J.M.; Jain, A.; Rochman, M.; Fischesser, D.M.; Ray, L.M.; Bedard, M.C.; Mingler, M.K.; Forney, C.; et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci. Transl. Med. 2018, 10, eaap9736. [Google Scholar] [CrossRef] [PubMed]
- Brännström, K.; Öhman, A.; Von Pawel Rammingen, U.; Olofsson, A.; Brattsand, M. Characterization of SPINK9, a KLK5-specific inhibitor expressed in palmo-plantar epidermis. Biol. Chem. 2012, 393, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Lun, Y.; Zhou, X.; He, S.; Gao, L.; Liu, Y.; He, Z.; Li, B.; Wang, C. Novel urokinase-plasminogen activator inhibitor SPINK13 inhibits growth and metastasis of hepatocellular carcinoma in vivo. Pharmacol. Res. 2019, 143, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yu, H.; Ni, Z.; Hu, S.; Ma, W.; Chu, C.; Liu, Q.; Zhang, Y. Spink13, an epididymis-specific gene of the kazal-type serine protease inhibitor (SPINK) family, is essential for the acrosomal integrity and male fertility. J. Biol. Chem. 2013, 288, 10154–10165. [Google Scholar] [CrossRef]
- Christeller, J.T. Evolutionary mechanisms acting on proteinase inhibitor variability. FEBS J. 2005, 272, 5710–5722. [Google Scholar] [CrossRef]
- Mehner, C.; Radisky, E.S. Bad Tumors Made Worse: SPINK1. Front. Cell Dev. Biol. 2019, 7, 10. [Google Scholar] [CrossRef]
- Chen, F.; Long, Q.; Fu, D.; Zhu, D.; Ji, Y.; Han, L.; Zhang, B.; Xu, Q.; Liu, B.; Li, Y.; et al. Targeting SPINK1 in the damaged tumour microenvironment alleviates therapeutic resistance. Nat. Commun. 2018, 9, 4315. [Google Scholar] [CrossRef]
- Kereszturi, É.; Sahin-Tóth, M. Pancreatic cancer cell lines heterozygous for the SPINK1 p.N34S haplotype exhibit diminished expression of the variant allele. Pancreas 2017, 46, e54–e55. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Su, B.; Lu, N.; Song, J.; Yang, X.; Fu, W.; Tan, W.; Han, B. Serine protease inhibitor Kazal type 1 promotes epithelial-mesenchymal transition through EGFR signaling pathway in prostate cancer. Prostate 2014, 74, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-C. Functional Roles of SPINK1 in Cancers. Int. J. Mol. Sci. 2021, 22, 3814. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Garcia, V.; Tawil, Y.; Wise, H.M.; Leslie, N.R. Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin. Cancer Biol. 2019, 59, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Ateeq, B. Molecular Underpinnings Governing Genetic Complexity of ETS-Fusion-Negative Prostate Cancer. Trends Mol. Med. 2019, 25, 1024–1038. [Google Scholar] [CrossRef]
- Pu, N.; Masson, E.; Cooper, D.N.; Génin, E.; Férec, C.; Chen, J.M. Chronic pancreatitis: The true pathogenic culprit within the spink1 n34s-containing haplotype is no longer at large. Genes 2021, 12, 1683. [Google Scholar] [CrossRef]
- Wang, Q.-W.; Zou, W.-B.; Masson, E.; Férec, C.; Liao, Z.; Chen, J.-M. Genetics and clinical implications of SPINK1 in the pancreatitis continuum and pancreatic cancer. Hum. Genom. 2025, 19, 32. [Google Scholar] [CrossRef]
- Gezer, S.; Emrence, Z.; Elverdi, T.; Ar, M.C.; Yaylaz, B.S.; Paçal, F.; Ünüvar, A.; Sarlman, M.; Eşkazan, A.E.; Karaman, S.; et al. Elevación de SPINK2 en leucemia mieloide aguda. Adv. Lab. Med. 2023, 4, 98–104. [Google Scholar] [CrossRef]
- Pitts, H.A.; Cheng, C.K.; Cheung, J.S.; Sun, M.K.H.; Yung, Y.L.; Chan, H.Y.; Wong, R.S.M.; Yip, S.F.; Lau, K.N.; Wong, W.S.; et al. SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response. Int. J. Mol. Sci. 2023, 24, 9696. [Google Scholar] [CrossRef]
- Nagel, F.; Susemihl, A.; Eulberg, T.; Delcea, M. Identification of Kazal Inhibitor Scaffolds with Identical Canonical Binding Loops and Their Effects on Binding Properties. Biochemistry 2023, 62, 535–542. [Google Scholar] [CrossRef]
- Barresi, V.; Di Bella, V.; Andriano, N.; Privitera, A.P.; Bonaccorso, P.; La Rosa, M.; Iachelli, V.; Spampinato, G.; Pulvirenti, G.; Scuderi, C.; et al. Nup-98 rearrangements led to the identification of candidate biomarkers for primary induction failure in pediatric acute myeloid leukemia. Int. J. Mol. Sci. 2021, 22, 4575. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, J.; Zhang, G.; Xue, Y.; Zhang, G.; Wu, X. Elevated SPINK2 gene expression is a predictor of poor prognosis in acute myeloid leukemia. Oncol. Lett. 2019, 18, 2877–2884. [Google Scholar] [CrossRef] [PubMed]
- Köberle, B.; Schoch, S. Platinum complexes in colorectal cancer and other solid tumors. Cancers 2021, 13, 2073. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, K.; Li, J.; Lu, D.; Hu, B. Association and diagnostic value of serum SPINK4 in colorectal cancer. PeerJ 2019, 7, e6679. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.; Yaron, J.R.; Zhang, L.; Macaulay, C.; McFadden, G. Serpins: Development for therapeutic applications. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef]
- Chen, T.J.; Tian, Y.F.; Chou, C.L.; Chan, T.C.; He, H.L.; Li, W.S.; Tsai, H.H.; Li, C.F.; Lai, H.Y. High spink4 expression predicts poor outcomes among rectal cancer patients receiving ccrt. Curr. Oncol. 2021, 28, 2373–2384. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, Y.; Ren, H.; Lei, Y. Identification of Prognosis-Related Genes in Bladder Cancer Microenvironment across TCGA Database. Biomed. Res. Int. 2020, 2020, 9143695. [Google Scholar] [CrossRef]
- Wang, Q.; Lv, Q.; Bian, H.; Yang, L.; Guo, K.L.; Ye, S.S.; Dong, X.F.; Tao, L.L. A novel tumor suppressor SPINK5 targets Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med. 2019, 8, 2360–2371. [Google Scholar] [CrossRef]
- Chen, S.H.; Hsiao, S.Y.; Chang, K.Y.; Chang, J.Y. New insights into oral squamous cell carcinoma: From clinical aspects to molecular tumorigenesis. Int. J. Mol. Sci. 2021, 22, 2252. [Google Scholar] [CrossRef]
- Wu, H.-T.; Chen, W.-T.; Chen, W.-J.; Li, C.-L.; Liu, J. Bioinformatics Analysis Reveals That ANXA1 and SPINK5 Are Novel Tumor Suppressor Genes in Patients with Oral Squamous Cell Carcinoma. Transl. Cancer Res. 2021, 10, 1682–1694. [Google Scholar] [CrossRef]
- Sun, S.; Su, G.; Zheng, X. Inhibition of the Tumor Suppressor Gene SPINK5 via EHMT2 Induces the Oral Squamous Cell Carcinoma Development. Mol. Biotechnol. 2024, 66, 208–221. [Google Scholar] [CrossRef]
- Alves, M.G.; Kodama, M.H.; da Silva, E.Z.M.; Gomes, B.B.M.; da Silva, R.A.A.; Vieira, G.V.; Alves, V.M.; da Fonseca, C.K.; Santana, A.C.; Cecílio, N.T.; et al. Relative expression of KLK5 to LEKTI is associated with aggressiveness of oral squamous cell carcinoma. Transl. Oncol. 2021, 14, 100970. [Google Scholar] [CrossRef] [PubMed]
- da Silva, E.Z.M.; Fraga-Silva, T.F.d.C.; Yuan, Y.; Alves, M.G.; Publio, G.A.; da Fonseca, C.K.; Kodama, M.H.; Vieira, G.V.; Candido, M.F.; Innocentini, L.M.A.R.; et al. Kallikrein 5 Inhibition by the Lympho-Epithelial Kazal-Type Related Inhibitor Hinders Matriptase-Dependent Carcinogenesis. Cancers 2021, 13, 4395. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Bong, S.K.; Lee, S.; Jung, Y.; Jegal, H.; Kim, J.; Kim, S.K.; Kim, Y.K.; Kim, S.N. Compound K improves skin barrier function by increasing SPINK5 expression. J. Ginseng Res. 2020, 44, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Wu, K.; Qin, X.; Yuan, J.; Yan, M.; Zhang, J.; Wang, L.; Ji, T.; Cao, W.; Chen, W. A novel tumor suppressor spink5 serves as an independent prognostic predictor for patients with head and neck squamous cell carcinoma. Cancer Manag. Res. 2020, 12, 4855–4869. [Google Scholar] [CrossRef]
- Li, R.G.; Deng, H.; Liu, X.H.; Chen, Z.Y.; Wan, S.S.; Wang, L. Histone Methyltransferase G9a Promotes the Development of Renal Cancer through Epigenetic Silencing of Tumor Suppressor Gene SPINK5. Oxid. Med. Cell Longev. 2021, 2021, 6650781. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Shen, Y.; He, P.; Ding, J.; Chen, Y. G9a stimulates CRC growth by inducing p53 Lys373 dimethylation-dependent activation of Plk1. Theranostics 2018, 8, 2884–2895. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The hippo pathway: Biology; pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Jung, S.; Fischer, J.; Spudy, B.; Kerkow, T.; Sönnichsen, F.D.; Xue, L.; Bonvin, A.M.J.J.; Goettig, P.; Magdolen, V.; Meyer-Hoffert, U.; et al. The solution structure of the kallikrein-related peptidases inhibitor SPINK6. Biochem. Biophys. Res. Commun. 2016, 471, 103–108. [Google Scholar] [CrossRef]
- Plaza, K.; Kalinska, M.; Bochenska, O.; Meyer-Hoffert, U.; Wu, Z.; Fischer, J.; Falkowski, K.; Sasiadek, L.; Bielecka, E.; Potempa, B.; et al. Gingipains of porphyromonas gingivalis affect the stability and function of serine protease inhibitor of Kazal-type 6(SPINK6), a tissue inhibitor of human kallikreins. J. Biol. Chem. 2016, 291, 18753–18764. [Google Scholar] [CrossRef]
- Zheng, L.S.; Yang, J.P.; Cao, Y.; Peng, L.X.; Sun, R.; Xie, P.; Wang, M.Y.; Meng, D.F.; Luo, D.H.; Zou, X.; et al. SPINK6 promotes metastasis of nasopharyngeal carcinoma via binding and activation of epithelial growth factor receptor. Cancer Res. 2017, 77, 579–589. [Google Scholar] [CrossRef]
- Liao, C.; Wang, Q.; An, J.; Long, Q.; Wang, H.; Xiang, M.; Xiang, M.; Zhao, Y.; Liu, Y.; Liu, J.; et al. Partial EMT in squamous cell carcinoma: A snapshot. Int. J. Biol. Sci. 2021, 17, 3036–3047. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, L.; Li, H.; Jin, C.; Yu, Y.; Hou, L.; Liu, X.; Yu, Y.; Yan, R.; Xue, F. Identification of Microenvironment Related Potential Biomarkers of Biochemical Recurrence at 3 Years after Prostatectomy in Prostate Adenocarcinoma. Aging 2021, 13, 16024–16042. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Meng, G.; Zhang, W. A six-mRNA prognostic model to predict survival in head and neck squamous cell carcinoma. Cancer Manag. Res. 2019, 11, 131–142. [Google Scholar] [CrossRef]
- Weber, C.; Fischer, J.; Redelfs, L.; Rademacher, F.; Harder, J.; Weidinger, S.; Wu, Z.; Meyer-Hoffert, U. The serine protease inhibitor of Kazal-type 7 (SPINK7) is expressed in human skin. Arch. Dermatol. Res. 2017, 309, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Pennacchiotti, G.; Valdés-Gutiérrez, F.; González-Arriagada, W.A.; Montes, H.F.; Parra, J.M.R.; Guida, V.A.; Gómez, S.E.; Guerrero-Gimenez, M.E.; Fernandez-Muñoz, J.M.; Zoppino, F.C.M.; et al. SPINK7 expression changes accompanied by HER2, P53 and RB1 can be relevant in predicting oral squamous cell carcinoma at a molecular level. Sci. Rep. 2021, 11, 6939. [Google Scholar] [CrossRef]
- Azouz, N.P.; Klingler, A.M.; Pathre, P.; Besse, J.A.; Ben Baruch-Morgenstern, N.; Ballaban, A.Y.; Osswald, G.A.; Brusilovsky, M.; Habel, J.E.; Caldwell, J.M.; et al. Functional role of kallikrein 5 and proteinase-activated receptor 2 in eosinophilic esophagitis. Sci. Transl. Med. 2020, 12, eaaz7773. [Google Scholar] [CrossRef]
- Zhao, N.; Wang, G.; Long, S.; Liu, D.; Gao, J.; Xu, Y.; Wang, C.; Wang, A.; Wang, F.; Hao, Y.; et al. Neutrophils-derived Spink7 as one safeguard against experimental murine colitis. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166125. [Google Scholar] [CrossRef]
- Patel, H.; Sheikh, M.S.; Huang, Y. ECRG2/SPINK7 Tumor Suppressor as Modulator of DNA Damage Response. Int. J. Mol. Sci. 2024, 25, 5854. [Google Scholar] [CrossRef]
- Kanapathipillai, M. Treating p53 mutant aggregation-associated cancer. Cancers 2018, 10, 154. [Google Scholar] [CrossRef]
- Hou, X.F.; Xu, L.P.; Song, H.Y.; Li, S.; Wu, C.; Wang, J.F. ECRG2 enhances the anti-cancer effects of cisplatin in cisplatin-resistant esophageal cancer cells via upregulation of p53 and downregulation of PCNA. World J. Gastroenterol. 2017, 23, 1796–1803. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, W.; Yan, H.; Hu, Y.; He, Q.; Luo, P. Regulation of p53 stability as a therapeutic strategy for cancer. Biochem. Pharmacol. 2021, 185, 114407. [Google Scholar] [CrossRef]
- Li, X.; Xiao, X.; Chang, R.; Zhang, C. Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a migration inhibitor in esophageal squamous cell carcinoma. J. Cell Biochem. 2020, 121, 468–481. [Google Scholar] [CrossRef]
- Daneva, G.N.; Tsiakanikas, P.; Adamopoulos, P.G.; Scorilas, A. Kallikrein-related peptidases: Mechanistic understanding for potential therapeutic targeting in cancer. Expert. Opin. Ther. Targets 2024, 28, 875–894. [Google Scholar] [CrossRef]
- Pampalakis, G. Anti-KLK5/KLK7 Antibody-based Strategies for the Treatment of Epidermal Diseases. Curr. Pharm. Des. 2023, 29, 2354–2357. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, Y.; Fischer, J.; Bartels, J.; Schröder, J.M.; Meyer-Hoffert, U. Skin-Derived SPINK9 Kills Escherichia coli. J. Investig. Dermatol. 2019, 139, 1135–1142. [Google Scholar] [CrossRef]
- Ortloff, A.; Bustamante, F.A.; Molina, L.; Ojeda, J.; Figueroa, C.D.; Ehrenfeld, P. Kallikrein-related Peptidase 5 (KLK5) Expression and Distribution in Canine Cutaneous Squamous Cell Carcinoma. J. Comp. Pathol. 2020, 174, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Redelfs, L.; Fischer, J.; Weber, C.; Wu, Z.; Meyer-Hoffert, U. The serine protease inhibitor of Kazal-type 9 (SPINK9) is expressed in lichen simplex chronicus, actinic keratosis and squamous cell carcinoma. Arch. Dermatol. Res. 2016, 308, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-M.; Shang, H.; Feng, X.-L.; Qi, C.; Zhang, S.-E.; Sun, Y.-C.; Gou, C.-Y.; Sun, Y.-J.; Zhang, G.-L. Epididymis May Be More Important in Male Fertility than Our Cognition. Explor. Immunol. 2023, 4, 309–324. [Google Scholar] [CrossRef]
- Björkgren, I.; Sipilä, P. The impact of epididymal proteins on sperm function. Reproduction 2019, 158, R155–R167. [Google Scholar] [CrossRef]
- Jin, M.; Fujiwara, E.; Kakiuchi, Y.; Okabe, M.; Satouh, Y.; Baba, S.A.; Chiba, K.; Hirohashi, N. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc. Natl. Acad. Sci. USA 2011, 108, 4892–4896. [Google Scholar] [CrossRef]
- Ou, C.M.; Tang, J.B.; Huang, M.S.; Gandhi, P.S.S.; Geetha, S.; Li, S.H.; Chen, Y.H. The mode of reproductive-derived Spink (serine protease inhibitor Kazal-type) action in the modulation of mammalian sperm activity. Int. J. Androl. 2012, 35, 52–62. [Google Scholar] [CrossRef]
- Cai, S.Y.; Yang, T.; Chen, Y.; Wang, J.W.; Li, L.; Xu, M.J. Gene expression profiling of ovarian carcinomas and prognostic analysis of outcome. J. Ovarian Res. 2015, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Zhang, P.; Dong, S.; Li, L.; Cai, J.; Xu, M. Downregulation of SPINK13 Promotes Metastasis by Regulating uPA in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2018, 45, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Lu, Y.; Lu, B. Genomic/epigenomic alterations in ovarian carcinoma: Translational insight into clinical practice. J. Cancer 2016, 7, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.H.; Shi, S.N.; Wang, J.; Xu, Y.; Tian, X.; Wan, F.N.; Cao, D.L.; Qu, Y.Y.; Zhang, H.L.; Ye, D.W. The role of serine peptidase inhibitor kazal type 13 (SPINK13) as a clinicopathological and prognostic biomarker in patients with clear cell renal cell carcinoma. Med. Sci. Monit. 2019, 25, 9458–9470. [Google Scholar] [CrossRef]
- Hsieh, S.C.; Tsai, J.P.; Yang, S.F.; Tang, M.J.; Hsieh, Y.H. Metformin inhibits the invasion of human hepatocellular carcinoma cells and enhances the chemosensitivity to sorafenib through a downregulation of the ERK/JNK-mediated NF-κB-dependent pathway that reduces uPA and MMP-9 expression. Amino Acids 2014, 46, 2809–2822. [Google Scholar] [CrossRef]
- Yang, C.; Guo, L.; Du, J.; Zhang, Q.; Zhang, L. SPINK1 Overexpression Correlates with Hepatocellular Carcinoma Treatment Resistance Revealed by Single Cell RNA-Sequencing and Spatial Transcriptomics. Biomolecules 2024, 14, 265. [Google Scholar] [CrossRef]
- Ma, C.; Li, H. Prognostic and diagnostic value of SPINK mRNAs expression in head and neck squamous cell carcinoma based on genome-wide analysis. Explor. Med. 2024, 5, 912–925. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, L.; Yu, T.; Zeng, J.; Chen, M. SPINK2 is a prognostic biomarker related to immune infiltration in acute myeloid leukemia. Am. J. Transl. Res. 2022, 14, 197–210. [Google Scholar]
- Cao, X.; Luo, N.; Liu, X.; Guo, K.; Deng, M.; Lv, C. Crosstalk of SPINK4 Expression with Patient Mortality, Immunotherapy and Metastasis in Pan-Cancer Based on Integrated Multi-Omics Analyses. Onco Targets Ther. 2025, 18, 161–177. [Google Scholar] [CrossRef]
- Huo, J.T.; Tuersun, A.; Yu, S.Y.; Zhang, Y.C.; Feng, W.Q.; Xu, Z.Q.; Zhao, J.K.; Zong, Y.P.; Lu, A.G. Leveraging a KRAS-based signature to predict the prognosis and drug sensitivity of colon cancer and identifying SPINK4 as a new biomarker. Sci. Rep. 2023, 13, 22230. [Google Scholar] [CrossRef]
- Zhao, C.; Zhu, H.; Tian, Y.; Sun, Y.; Zhang, Z. SPINK5 is a key regulator of eosinophil extracellular traps in head and neck squamous cell carcinoma. Discov. Oncol. 2024, 15, 627. [Google Scholar] [CrossRef]
- Yun, S.J.; Kim, S.K.; Kim, J.; Cha, E.J.; Kim, J.S.; Kim, S.J.; Ha, Y.S.; Kim, Y.H.; Jeong, P.; Kang, H.W.; et al. Transcriptomic features of primary prostate cancer and their prognostic relevance to castration-resistant prostate cancer. Oncotarget 2017, 8, 114845–114855. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Hu, C.; Zheng, S.; Zhang, X.; Chen, R.; Zhou, Q. A novel gene signature for prognosis prediction and chemotherapy response in patients with pancreatic cancer. Aging 2021, 13, 12493–12513. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jia, Z.; Liu, J.; Xu, X.; Wang, H.; Li, D.; Qiu, Z. Transcription activation of SPINK4 by ELF-1 augments progression of colon cancer by regulating biological behaviors. Tissue Cell 2023, 84, 102190. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Q.; Yu, X.; Lu, L.; Zhou, Z.; Li, M.; Xia, R.; Gan, X.; Hu, Y.; Guo, G.; et al. The microprotein HDSP promotes gastric cancer progression through activating the MECOM-SPINK1-EGFR signaling axis. Nat. Commun. 2024, 15, 8381. [Google Scholar] [CrossRef]
- Rochman, S. New Initiative Takes Fresh Approach to Increase Value in Cancer Care. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Demcsák, A.; Sahin-Tóth, M. Heterozygous Spink1 Deficiency Promotes Trypsin-dependent Chronic Pancreatitis in Mice. Cell Mol. Gastroenterol. Hepatol. 2024, 18, 101361. [Google Scholar] [CrossRef]
- Liu, M.; Ma, L.; An, W.; Yang, Y.; Liu, J.; Jiang, H.; Yuan, J.; Sun, X.; Zhu, J.; Yan, M.; et al. Heterozygous Spink1 c.194+2T>C mutation promotes chronic pancreatitis after acute attack in mice. Pancreatology 2024, 24, 677–689. [Google Scholar] [CrossRef]
- Piseddu, I.; Vielhauer, J.; Mayerle, J. Genetic Testing in Acute and Chronic Pancreatitis. Curr. Treat. Options Gastroenterol. 2022, 20, 429–444. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Mao, X.-T.; Sun, C.; Wang, Y.-H.; Zheng, Y.-Z.; Xiong, S.-H.; Liu, M.-Y.; Mao, S.-H.; Wang, Q.-W.; Ma, G.-X.; et al. Pancreas-directed AAV8-hSPINK1 gene therapy safely and effectively protects against pancreatitis in mice. Gut 2024, 73, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Ghieh, F.; Mitchell, V.; Mandon-Pepin, B.; Vialard, F. Genetic defects in human azoospermia. Basic. Clin. Androl. 2019, 29, 4. [Google Scholar] [CrossRef] [PubMed]
- Cioppi, F.; Rosta, V.; Krausz, C. Genetics of azoospermia. Int. J. Mol. Sci. 2021, 22, 3264. [Google Scholar] [CrossRef]
- Wu, F.T.; Chen, C.P.; Chen, S.W.; Chern, S.R.; Chen, P.T.; Chiu, C.L.; Lee, C.C.; Chen, W.L.; Wang, W. Concomitance of, a balanced reciprocal translocation of t(4; 17)(q12; q11.2) encompassing SPINK2 at 4q12 and NOS at 17q11.2 and an AZFa sY86 deletion in an infertile male. Taiwan. J. Obstet. Gynecol. 2023, 62, 336–342. [Google Scholar] [CrossRef]
- Wapenaar, M.C.; Monsuur, A.J.; Poell, J.; Van ’t Slot, R.; Meijer, J.W.R.; Meijer, G.A.; Mulder, C.J.; Mearin, M.L.; Wijmenga, C. The SPINK gene family and celiac disease susceptibility. Immunogenetics 2007, 59, 349–357. [Google Scholar] [CrossRef]
- Pietz, G.; De, R.; Hedberg, M.; Sjöberg, V.; Sandström, O.; Hernell, O.; Hammarström, S.; Hammarström, M.L. Immunopathology of childhood celiac disease—Key role of intestinal epithelial cells. PLoS ONE 2017, 12, e0185025. [Google Scholar] [CrossRef]
- Sarri, C.A.; Roussaki-Schulze, A.; Vasilopoulos, Y.; Zafiriou, E.; Patsatsi, A.; Stamatis, C.; Gidarokosta, P.; Sotiriadis, D.; Sarafidou, T.; Mamuris, Z. Netherton Syndrome: A Genotype-Phenotype Review. Mol. Diagn. Ther. 2017, 21, 137–152. [Google Scholar] [CrossRef]
- Xu, M.; Shi, Y.; Lin, L.; Wang, L.; Zhu, X.; Xiong, J.; Yin, J.; Qi, Q.; Yang, W. The role of SPINK5 mutation distribution in phenotypes of Netherton syndrome. Front. Genet. 2025, 16, 1475054. [Google Scholar] [CrossRef]
- Hannula-Jouppi, K.; Laasanen, S.-L.; Ilander, M.; Furio, L.; Tuomiranta, M.; Marttila, R.; Jeskanen, L.; Häyry, V.; Kanerva, M.; Kivirikko, S.; et al. Intrafamily and Interfamilial Phenotype Variation and Immature Immunity in Patients with Netherton Syndrome and Finnish SPINK5 Founder Mutation. JAMA Dermatol. 2016, 152, 435. [Google Scholar] [CrossRef]
- Petrova, E.; López-Gay, J.M.; Fahrner, M.; Leturcq, F.; de Villartay, J.P.; Barbieux, C.; Gonschorek, P.; Tsoi, L.C.; Gudjonsson, J.E.; Schilling, O.; et al. Comparative analyses of Netherton syndrome patients and Spink5 conditional knock-out mice uncover disease-relevant pathways. Commun. Biol. 2024, 7, 152. [Google Scholar] [CrossRef]
- Furio, L.; Pampalakis, G.; Michael, I.P.; Nagy, A.; Sotiropoulou, G.; Hovnanian, A. KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome. PLoS Genet. 2015, 11, e1005389. [Google Scholar] [CrossRef]
- Morrison, H.A.; Hoyt, K.J.; Mounzer, C.; Ivester, H.M.; Barnes, B.H.; Sauer, B.; McGowan, E.C.; Allen, I.C. Expression profiling identifies key genes and biological functions associated with eosinophilic esophagitis in human patients. Front. Allergy 2023, 4, 1239273. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Oshima, T.; Huang, X.; Tomita, T.; Fukui, H.; Miwa, H. Esophageal Mucosal Permeability as a Surrogate Measure of Cure in Eosinophilic Esophagitis. J. Clin. Med. 2022, 11, 4246. [Google Scholar] [CrossRef] [PubMed]
- Azouz, N.P.; Klingler, A.M.; Rochman, M.; Paul, M.; Caldwell, J.M.; Brusilovsky, M.; Dwyer, A.T.; Chen, X.; Miller, D.; Lynch, A.; et al. Aryl Hydrocarbon Receptor Suppresses Eosinophilic Esophagitis Responses through OVOL1 and SPINK7. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sotiropoulou, G.; Zingkou, E.; Pampalakis, G. Reconstructing the epidermal proteolytic cascades in health and disease. J. Pathol. 2022, 257, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, G.; Long, S.; Lv, X.; Ran, X.; Wang, J.; Su, Y.; Wang, T. The antiprotease Spink7 promotes inflammation resolution by modulating multiple proteases activities during wound healing. Clin. Transl. Med. 2025, 15, e70291. [Google Scholar] [CrossRef]
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Front. Biosci.-Landmark 2020, 25, 1058–1109. [Google Scholar] [CrossRef]
- Leão, R.; Lee, D.; Figueiredo, A.; Hermanns, T.; Wild, P.; Komosa, M.; Lau, I.; Mistry, M.; Nunes, N.M.; Price, A.J.; et al. Combined genetic and epigenetic alterations of the TERT promoter affect clinical and biological behavior of bladder cancer. Int. J. Cancer 2019, 144, 1676–1684. [Google Scholar] [CrossRef]
- Grady, W.M. Epigenetic alterations in the gastrointestinal tract: Current and emerging use for biomarkers of cancer. Adv. Cancer Res. 2021, 151, 425–468. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef]
- Fath, M.K.; Azargoonjahromi, A.; Kiani, A.; Jalalifar, F.; Osati, P.; Oryani, M.A.; Shakeri, F.; Nasirzadeh, F.; Khalesi, B.; Nabi-Afjadi, M.; et al. The role of epigenetic modifications in drug resistance and treatment of breast cancer. Cell Mol. Biol. Lett. 2022, 27, 52. [Google Scholar] [CrossRef]
- Miranda-Galvis, M.; Loveless, R.; Kowalski, L.P.; Teng, Y. Impacts of environmental factors on head and neck cancer pathogenesis and progression. Cells 2021, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Mbemi, A.; Khanna, S.; Njiki, S.; Yedjou, C.G.; Tchounwou, P.B. Impact of gene–environment interactions on cancer development. Int. J. Environ. Res. Public Health 2020, 17, 8089. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Chan, A.T. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Pati, S.; Irfan, W.; Jameel, A.; Ahmed, S.; Shahid, R.K. Obesity and Cancer: A Current Overview of Epidemiology, Pathogenesis, Outcomes, and Management. Cancers 2023, 15, 485. [Google Scholar] [CrossRef]
- Lega, I.C.; Lipscombe, L.L. Review: Diabetes, Obesity, and Cancer-Pathophysiology and Clinical Implications. Endocr. Rev. 2020, 41, 33–52. [Google Scholar] [CrossRef]
- Lin, X.; Li, H. Obesity: Epidemiology, Pathophysiology, and Therapeutics. Front. Endocrinol. 2021, 12, 706978. [Google Scholar] [CrossRef]
- Choi, J.-W.; Hua, T.N.M. Impact of Lifestyle Behaviors on Cancer Risk and Prevention. J. Lifestyle Med. 2021, 11, 1–7. [Google Scholar] [CrossRef]
- Deshpande, R.P.; Sharma, S.; Watabe, K. The confounders of cancer immunotherapy: Roles of lifestyle, metabolic disorders and sociological factors. Cancers 2020, 12, 2983. [Google Scholar] [CrossRef]
- Jardim, S.R.; de Souza, L.M.P.; de Souza, H.S.P. The Rise of Gastrointestinal Cancers as a Global Phenomenon: Unhealthy Behavior or Progress? Int. J. Environ. Res. Public Health 2023, 20, 3640. [Google Scholar] [CrossRef]
- Denk, D.; Greten, F.R. Inflammation: The incubator of the tumor microenvironment. Trends Cancer 2022, 8, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Grant, R.; Mishra, P.; Nilubol, N. The Role of Tumor Necrosis Factor in Manipulating the Immunological Response of Tumor Microenvironment. Front. Immunol. 2021, 12, 656908. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Xue, H.; Sun, Y.; Zhang, C.; Song, Y.; Qi, Y. The Role of Tumor Inflammatory Microenvironment in Lung Cancer. Front. Pharmacol. 2021, 12, 688625. [Google Scholar] [CrossRef]
- Sherman, M.H.; Beatty, G.L. Tumor Microenvironment in Pancreatic Cancer Pathogenesis and Therapeutic Resistance. Annu. Rev. Pathol. Mech. Dis. 2023, 18, 123–148. [Google Scholar] [CrossRef]
- Habanjar, O.; Bingula, R.; Decombat, C.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. Crosstalk of Inflammatory Cytokines within the Breast Tumor Microenvironment. Int. J. Mol. Sci. 2023, 24, 4002. [Google Scholar] [CrossRef]
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor microenvironment in breast cancer—Updates on therapeutic implications and pathologic assessment. Cancers 2021, 13, 4233. [Google Scholar] [CrossRef]
- Maiorino, L.; Daßler-Plenker, J.; Sun, L.; Egeblad, M. Innate Immunity and Cancer Pathophysiology. Annu. Rev. Pathol. Mech. Dis. 2021, 17, 425–457. [Google Scholar] [CrossRef]
- Ene, C.V.; Nicolae, I.; Geavlete, B.; Geavlete, P.; Ene, C.D. IL-6 Signaling Link between Inflammatory Tumor Microenvironment and Prostatic Tumorigenesis. Anal. Cell. Pathol. 2022, 2022, 5980387. [Google Scholar] [CrossRef]
- Jang, J.H.; Kim, D.H.; Surh, Y.J. Dynamic roles of inflammasomes in inflammatory tumor microenvironment. NPJ Precis. Oncol. 2021, 5, 18. [Google Scholar] [CrossRef]
- Farc, O.; Cristea, V. An overview of the tumor microenvironment, from cells to complex networks (Review). Exp. Ther. Med. 2020, 21, 96. [Google Scholar] [CrossRef] [PubMed]
- Mafi, S.; Mansoori, B.; Taeb, S.; Sadeghi, H.; Abbasi, R.; Cho, W.C.; Rostamzadeh, D. mTOR-Mediated Regulation of Immune Responses in Cancer and Tumor Microenvironment. Front. Immunol. 2022, 12, 774103. [Google Scholar] [CrossRef] [PubMed]
- Naser, R.; Fakhoury, I.; El-Fouani, A.; Abi-Habib, R.; El-Sibai, M. Role of the tumor microenvironment in cancer hallmarks and targeted therapy (Review). Int. J. Oncol. 2023, 62, 23. [Google Scholar] [CrossRef] [PubMed]
- Domen, A.; Deben, C.; Verswyvel, J.; Flieswasser, T.; Prenen, H.; Peeters, M.; Lardon, F.; Wouters, A. Cellular senescence in cancer: Clinical detection and prognostic implications. J. Exp. Clin. Cancer Res. 2022, 41, 360. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Paez-Ribes, M.; González-Gualda, E.; Doherty, G.J.; Muñoz-Espín, D. Targeting senescent cells in translational medicine. EMBO Mol. Med. 2019, 11, e10234. [Google Scholar] [CrossRef]
- Majumder, P.K.; Grisanzio, C.; O’COnnell, F.; Barry, M.; Brito, J.M.; Xu, Q.; Guney, I.; Berger, R.; Herman, P.; Bikoff, R.; et al. A Prostatic Intraepithelial Neoplasia-Dependent p27Kip1 Checkpoint Induces Senescence and Inhibits Cell Proliferation and Cancer Progression. Cancer Cell 2008, 14, 146–155. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Vredeveld, L.C.W.; Douma, S.; van Doorn, R.; Desmet, C.J.; Aarden, L.A.; Mooi, W.J.; Peeper, D.S. Oncogene-Induced Senescence Relayed by an Interleukin-Dependent Inflammatory Network. Cell 2008, 133, 1019–1031. [Google Scholar] [CrossRef]
- Michaloglou, C.; Vredeveld, L.C.W.; Soengas, M.S.; Denoyelle, C.; Kuilman, T.; Van Der Horst, C.M.A.M.; Majoor, D.M.; Shay, J.W.; Mooi, W.J.; Peeper, D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005, 436, 720–724. [Google Scholar] [CrossRef]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Aquino-Acevedo, A.N.; Orengo-Orengo, J.A.; Cruz-Robles, M.E.; Saavedra, H.I. Mitotic kinases are emerging therapeutic targets against metastatic breast cancer. Cell Div. 2024, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.L.; Lan, L.; Zou, L. DNA repair defects in cancer and therapeutic opportunities. Genes. Dev. 2022, 34, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.P.L.; Foltz, D.R.; Cleveland, D.W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA 2004, 101, 8699–8704. [Google Scholar] [CrossRef]
- Borah, N.A.; Reddy, M.M. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021, 26, 1981. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef]
- di Rorà, A.G.L.; Cerchione, C.; Martinelli, G.; Simonetti, G. A WEE1 family business: Regulation of mitosis, cancer progression, and therapeutic target. J. Hematol. Oncol. 2020, 13, 126. [Google Scholar] [CrossRef]
- Ngoi, N.Y.L.; Pilié, P.G.; McGrail, D.J.; Zimmermann, M.; Schlacher, K.; Yap, T.A. Targeting ATR in patients with cancer. Nat. Rev. Clin. Oncol. 2024, 21, 278–293. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef]
- Chen, J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Seoane, M.; Costoya, J.A.; Arce, V.M. Uncoupling Oncogene-Induced Senescence (OIS) and DNA Damage Response (DDR) triggered by DNA hyper-replication: Lessons from primary mouse embryo astrocytes (MEA). Sci. Rep. 2017, 7, 12991. [Google Scholar] [CrossRef]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef]
- Jones, R.M.; Mortusewicz, O.; Afzal, I.; Lorvellec, M.; García, P.; Helleday, T.; Petermann, E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 2013, 32, 3744–3753. [Google Scholar] [CrossRef] [PubMed]
- Ganem, N.J.; Pellman, D. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 2012, 199, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689.e673. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Vogel, C.; Schneider, V.; Ertych, N.; Kienitz, A.; Yu, H.; Bastians, H. Pharmacologic abrogation of the mitotic spindle checkpoint by an indolocarbazole discovered by cellular screening efficiently kills cancer cells. Cancer Res. 2009, 69, 3874–3883. [Google Scholar] [CrossRef]
- Kwiatkowski, N.; Jelluma, N.; Filippakopoulos, P.; Soundararajan, M.; Manak, M.S.; Kwon, M.; Choi, H.G.; Sim, T.; Deveraux, Q.L.; Rottmann, S.; et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 2010, 6, 359–368. [Google Scholar] [CrossRef]
- Siri, S.O.; Martino, J.; Gottifredi, V. Structural Chromosome Instability: Types, Origins, Consequences, and Therapeutic Opportunities. Cancers 2021, 13, 3056. [Google Scholar] [CrossRef]
- Funk, L.C.; Zasadil, L.M.; Weaver, B.A. Living in CIN: Mitotic Infidelity and Its Consequences for Tumor Promotion and Suppression. Dev. Cell 2016, 39, 638–652. [Google Scholar] [CrossRef]
- Sansregret, L.; Swanton, C. The role of aneuploidy in cancer evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a028373. [Google Scholar] [CrossRef]
- Zasadil, L.M.; Britigan, E.M.C.; Ryan, S.D.; Kaur, C.; Guckenberger, D.J.; Beebe, D.J.; Moser, A.R.; Weaver, B.A. High rates of chromosome missegregation suppress tumor progression but do not inhibit tumor initiation. Mol. Biol. Cell 2016, 27, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Roylance, R.; Endesfelder, D.; Gorman, P.; Burrell, R.A.; Sander, J.; Tomlinson, I.; Hanby, A.M.; Speirs, V.; Richardson, A.L.; Birkbak, N.J.; et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Zhang, J.; Huang, G.; Yan, J.; Xu, C.; Dou, Z.; Sun, C.; Zhang, H. The crosstalk between HIFs and mitochondrial dysfunctions in cancer development. Cell Death Dis. 2021, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Yao, L.; Yuan, M.; Wang, Z.; Zhang, Q.; Jiang, Y.; Li, L. Mitochondrial dysfunction: A promising therapeutic target for liver diseases. Genes Dis. 2024, 11, 101115. [Google Scholar] [CrossRef]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef]
- Ramanathan, R.; Ali, A.H.; Ibdah, J.A. Mitochondrial Dysfunction Plays Central Role in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 7280. [Google Scholar] [CrossRef]
- Lee, H.Y.; Nga, H.T.; Tian, J.; Yi, H.S. Mitochondrial metabolic signatures in hepatocellular carcinoma. Cells 2021, 10, 1901. [Google Scholar] [CrossRef]
- Palmer, C.S.; Anderson, A.J.; Stojanovski, D. Mitochondrial protein import dysfunction: Mitochondrial disease, neurodegenerative disease and cancer. FEBS Lett. 2021, 595, 1107–1131. [Google Scholar] [CrossRef]
- Keerthiga, R.; Pei, D.-S.; Fu, A. Mitochondrial dysfunction, UPRmt signaling, and targeted therapy in metastasis tumor. Cell Biosci. 2021, 11, 186. [Google Scholar] [CrossRef]
- Chen, X.; Hao, B.; Li, D.; Reiter, R.J.; Bai, Y.; Abay, B.; Chen, G.; Lin, S.; Zheng, T.; Ren, Y.; et al. Melatonin inhibits lung cancer development by reversing the Warburg effect via stimulating the SIRT3/PDH axis. J. Pineal Res. 2021, 71, e12755. [Google Scholar] [CrossRef]
- Chiang, S.K.; Chen, S.E.; Chang, L.C. The role of HO-1 and its crosstalk with oxidative stress in cancer cell survival. Cells 2021, 10, 2401. [Google Scholar] [CrossRef]
- Middleton, P.; Vergis, N. Mitochondrial dysfunction and liver disease: Role, relevance, and potential for therapeutic modulation. Therap. Adv. Gastroenterol. 2021, 14, 17562848211031394. [Google Scholar] [CrossRef] [PubMed]
- Sperrhacke, M.; Fischer, J.; Wu, Z.; Klünder, S.; Sedlacek, R.; Schroeder, J.M.; Meyer-Hoffert, U.; Reiss, K. SPINK9 stimulates metalloprotease/EGFR-dependent keratinocyte migration via purinergic receptor activation. J. Investig. Dermatol. 2014, 134, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Ateeq, B.; Tomlins, S.A.; Laxman, B.; Asangani, I.A.; Cao, Q.; Cao, X.; Li, Y.; Wang, X.; Feng, F.Y.; Pienta, K.J.; et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci. Transl. Med. 2011, 3, 72ra17. [Google Scholar] [CrossRef] [PubMed]
- Ga, L. A Brief Review of SPINK1 Studies. Int. J. Biol. Life Sci. 2023, 4, 55–57. [Google Scholar] [CrossRef]
- Schmitt, M.; Greten, F.R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef]
- Pan, X.; Tan, J.; Yin, X.; Liu, Q.; Zheng, L.; Su, Z.; Zhou, Q.; Chen, N. The roles of mutated SPINK1 gene in prostate cancer cells. Mutagenesis 2022, 37, 238–247. [Google Scholar] [CrossRef]
- Räsänen, K.; Itkonen, O.; Koistinen, H.; Stenman, U.H. Emerging roles of SPINK1 in cancer. Clin. Chem. 2016, 62, 449–457. [Google Scholar] [CrossRef]
- Tiwari, R.; Manzar, N.; Bhatia, V.; Yadav, S.S.; Kumar, A.; Bhatia, N.; Goel, H.; Rajender, S.; Thulkar, S.; Singh, S.K.; et al. Androgen Deprivation Upregulates SPINK1 Expression and Potentiates Cellular Plasticity in Prostate Cancer. Cancers 2021, 13, 2795. [Google Scholar] [CrossRef]
- Weidle, U.H.; Epp, A.; Birzele, F.; Brinkmann, U. The functional role of prostate cancer metastasis-related Micro-RNAs. Cancer Genom. Proteom. 2019, 16, 1–19. [Google Scholar] [CrossRef]
- Soon, W.W.; Miller, L.D.; Black, M.A.; Dalmasso, C.; Chan, X.B.; Pang, B.; Ong, C.W.; Salto-Tellez, M.; Desai, K.V.; Liu, E.T. Combined genomic and phenotype screening reveals secretory factor SPINK1 as an invasion and survival factor associated with patient prognosis in breast cancer. EMBO Mol. Med. 2011, 3, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Wang, Q.; An, J.; Zhang, M.; Chen, J.; Li, X.; Xiao, L.; Wang, J.; Long, Q.; Liu, J.; et al. SPINKs in Tumors: Potential Therapeutic Targets. Front. Oncol. 2022, 12, 833741. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jiang, S.; Yuan, Z.; Jiang, J.; Yang, M.; Luo, J.; Ye, T. SPINK4 modulates inhibition of glycolysis against colorectal cancer progression. Biomol. Biomed. 2024, 24, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, J.; Chen, Z.; Liu, Z.; Sun, Y.; He, S.; Mi, Y.; Gao, Y.; Shen, D.; Lin, Q. SPINK5 inhibits esophageal squamous cell carcinoma metastasis via immune activity. J. Gene Med. 2024, 26, e3667. [Google Scholar] [CrossRef]
- Wei, L.; An, T.; An, Y.; He, Z.; Jia, T.; Li, B.; Lun, Y. Transcriptome analysis of the effect of a novel human serine protease inhibitor SPINK13 on gene expression in MHCC97-H cells. Transl. Cancer Res. 2021, 10, 4464–4477. [Google Scholar] [CrossRef]
- Shah, K.; Kazi, J.U. Phosphorylation-Dependent Regulation of WNT/Beta-Catenin Signaling. Front. Oncol. 2022, 12, 858782. [Google Scholar] [CrossRef]
- Peng, Y.; Xu, Y.; Zhang, X.; Deng, S.; Yuan, Y.; Luo, X.; Hossain, M.T.; Zhu, X.; Du, K.; Hu, F.; et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol. Cancer 2021, 20, 158. [Google Scholar] [CrossRef]
- Dolgova, N.; Wei, Z.; Spink, B.; Gui, L.; Hua, Q.; Truong, D.; Zhang, Z.; Zhang, Y. Low-Field Magnetic Stimulation Accelerates the Differentiation of Oligodendrocyte Precursor Cells via Non-canonical TGF-β Signaling Pathways. Mol. Neurobiol. 2021, 58, 855–866. [Google Scholar] [CrossRef]
- Teixeira, A.F.; Wu, S.; Luwor, R.; Zhu, H.J. A New Era of Integration between Multiomics and Spatio-Temporal Analysis for the Translation of EMT towards Clinical Applications in Cancer. Cells 2023, 12, 2740. [Google Scholar] [CrossRef]
- Gong, J.; Kim, D.M.; Freeman, M.R.; Kim, H.; Ellis, L.; Smith, B.; Theodorescu, D.; Posadas, E.; Figlin, R.; Bhowmick, N.; et al. Genetic and biological drivers of prostate cancer disparities in Black men. Nat. Rev. Urol. 2024, 21, 274–289. [Google Scholar] [CrossRef]
- Shetty, K.S.; Jose, A.; Bani, M.; Vinod, P.K. Network diffusion-based approach for survival prediction and identification of biomarkers using multi-omics data of papillary renal cell carcinoma. Mol. Genet. Genom. 2023, 298, 871–882. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Huang, W.; Rao, X.; Lai, Y. Pharmacological properties of indirubin and its derivatives. Biomed. Pharmacother. 2022, 151, 113112. [Google Scholar] [CrossRef] [PubMed]
- Lorenzin, F.; Demichelis, F. Past, Current, and Future Strategies to Target ERG Fusion-Positive Prostate Cancer. Cancers 2022, 14, 1118. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Choi, J.S.; Koo, B.M.; Kim, Y.J.; Song, J.Y.; Sung, M.; Chang, E.S.; Noh, K.W.; An, S.; Lee, M.S.; et al. TM4SF4 and LRRK2 are potential therapeutic targets in lung and breast cancers through outlier analysis. Cancer Res. Treat. 2021, 53, 9–24. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Q.; Ghareeb, W.M.; Zhang, Y.; Lu, X.; Huang, Y.; Huang, S.; Sun, Y.; Lin, J.; Liu, J.; et al. Downregulated SPINK4 is associated with poor survival in colorectal cancer. BMC Cancer 2019, 19, 1258. [Google Scholar] [CrossRef]
- Mangini, M.; Iaccino, E.; Mosca, M.G.; Mimmi, S.; D’Angelo, R.; Quinto, I.; Scala, G.; Mariggiò, S. Peptide-guided targeting of GPR55 for anti-cancer therapy. Oncotarget 2017, 8, 5179–5195. [Google Scholar] [CrossRef]
- Kazemizadeh, H.; Kashefizadeh, A. CRISPR-Cas9-mediated gene therapy in lung cancer. Clin. Transl. Oncol. 2023, 25, 1156–1166. [Google Scholar] [CrossRef]
- Karn, V.; Sandhya, S.; Hsu, W.; Parashar, D.; Singh, H.N.; Jha, N.K.; Gupta, S.; Dubey, N.K.; Kumar, S. CRISPR/Cas9 system in breast cancer therapy: Advancement, limitations and future scope. Cancer Cell Int. 2022, 22, 234. [Google Scholar] [CrossRef]
- Jain, P.; Jain, C.; Velcheti, V. Role of immune-checkpoint inhibitors in lung cancer. Ther. Adv. Respir. Dis. 2018, 12, 1753465817750075. [Google Scholar] [CrossRef]
- Ghidini, M.; Fusco, N.; Salati, M.; Khakoo, S.; Tomasello, G.; Petrelli, F.; Trapani, D.; Petrillo, A. The Emergence of Immune-checkpoint Inhibitors in Colorectal Cancer Therapy. Curr. Drug Targets 2021, 22, 1021–1033. [Google Scholar] [CrossRef]
- Ogundipe, O.D.; Olajubutu, O.; Adesina, S.K. Targeted drug conjugate systems for ovarian cancer chemotherapy. Biomed. Pharmacother. 2023, 165, 115151. [Google Scholar] [CrossRef]
- He, Y.; Xu, W.; Xiao, Y.T.; Huang, H.; Gu, D.; Ren, S. Targeting signaling pathways in prostate cancer: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2022, 7, 198. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Zhang, Y.; Cao, D.; He, H.; Cao, X.; Wang, Y.; Jia, Z.; Jiang, J. Dietary inflammatory index, and depression and mortality risk associations in U.S. adults, with a special focus on cancer survivors. Front. Nutr. 2022, 9, 1034323. [Google Scholar] [CrossRef]
- Edsjö, A.; Holmquist, L.; Geoerger, B.; Nowak, F.; Gomon, G.; Alix-Panabières, C.; Ploeger, C.; Lassen, U.; Le Tourneau, C.; Lehtiö, J.; et al. Precision cancer medicine: Concepts; current practice; future developments. J. Intern. Med. 2023, 294, 455–481. [Google Scholar] [CrossRef]
- Passaro, A.; Al Bakir, M.; Hamilton, E.G.; Diehn, M.; André, F.; Roy-Chowdhuri, S.; Mountzios, G.; Wistuba, I.I.; Swanton, C.; Peters, S. Cancer biomarkers: Emerging trends and clinical implications for personalized treatment. Cell 2024, 187, 1617–1635. [Google Scholar] [CrossRef]
- Mishra, A.K.; Ali, A.; Dutta, S.; Banday, S.; Malonia, S.K. Emerging Trends in Immunotherapy for Cancer. Diseases 2022, 10, 60. [Google Scholar] [CrossRef]
- Richard, G.; Princiotta, M.F.; Bridon, D.; Martin, W.D.; Steinberg, G.D.; De Groot, A.S. Neoantigen-based personalized cancer vaccines: The emergence of precision cancer immunotherapy. Expert. Rev. Vaccines 2022, 21, 173–184. [Google Scholar] [CrossRef]
- Lin, F.; Lin, E.Z.; Anekoji, M.; Ichim, T.E.; Hu, J.; Marincola, F.M.; Jones, L.D.; Kesari, S.; Ashili, S. Advancing personalized medicine in brain cancer: Exploring the role of mRNA vaccines. J. Transl. Med. 2023, 21, 830. [Google Scholar] [CrossRef]
- Morand, S.; Devanaboyina, M.; Staats, H.; Stanbery, L.; Nemunaitis, J. Ovarian cancer immunotherapy and personalized medicine. Int. J. Mol. Sci. 2021, 22, 6532. [Google Scholar] [CrossRef]
- Martino, E.; D’Onofrio, N.; Anastasio, C.; Abate, M.; Zappavigna, S.; Caraglia, M.; Balestrieri, M.L. MicroRNA-nanoparticles against cancer: Opportunities and challenges for personalized medicine. Mol. Ther. Nucleic Acids 2023, 32, 371–384. [Google Scholar] [CrossRef]
- Chehelgerdi, M.; Chehelgerdi, M. The use of RNA-based treatments in the field of cancer immunotherapy. Mol. Cancer 2023, 22, 106. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, J.; Tang, R.; Yang, J.; Wang, W.; Yu, X.; Shi, S. Novel research and future prospects of artificial intelligence in cancer diagnosis and treatment. J. Hematol. Oncol. 2023, 16, 114. [Google Scholar] [CrossRef]
- Faghfuri, E. Recent advances in personalized cancer immunotherapy with immune checkpoint inhibitors, T cells and vaccines. Pers. Med. 2024, 21, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Saeed, R.F.; Awan, U.A.; Saeed, S.; Mumtaz, S.; Akhtar, N.; Aslam, S. Targeted Therapy and Personalized Medicine. Cancer Treat. Res. 2023, 185, 177–205. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, G.; Karuppasamy, Y.; Krishnan, U.M. Emerging Trends in Nano-Driven Immunotherapy for Treatment of Cancer. Vaccines 2023, 11, 458. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wali, Z.; Neha; Shamsi, A.; Tasqeruddin, S.; Anwar, S. The SPINK Protein Family in Cancer: Emerging Roles in Tumor Progression, Therapeutic Resistance, and Precision Oncology. Pharmaceuticals 2025, 18, 1194. https://doi.org/10.3390/ph18081194
Wali Z, Neha, Shamsi A, Tasqeruddin S, Anwar S. The SPINK Protein Family in Cancer: Emerging Roles in Tumor Progression, Therapeutic Resistance, and Precision Oncology. Pharmaceuticals. 2025; 18(8):1194. https://doi.org/10.3390/ph18081194
Chicago/Turabian StyleWali, Zitin, Neha, Anas Shamsi, Syed Tasqeruddin, and Saleha Anwar. 2025. "The SPINK Protein Family in Cancer: Emerging Roles in Tumor Progression, Therapeutic Resistance, and Precision Oncology" Pharmaceuticals 18, no. 8: 1194. https://doi.org/10.3390/ph18081194
APA StyleWali, Z., Neha, Shamsi, A., Tasqeruddin, S., & Anwar, S. (2025). The SPINK Protein Family in Cancer: Emerging Roles in Tumor Progression, Therapeutic Resistance, and Precision Oncology. Pharmaceuticals, 18(8), 1194. https://doi.org/10.3390/ph18081194









