In Silico Mining of the Streptome Database for Hunting Putative Candidates to Allosterically Inhibit the Dengue Virus (Serotype 2) RdRp
Abstract
1. Introduction
2. Results and Discussion
2.1. Docking Protocol Assessment
2.2. Virtual Screening of the Streptome Database
2.3. Molecular Dynamics Simulations (MDS)
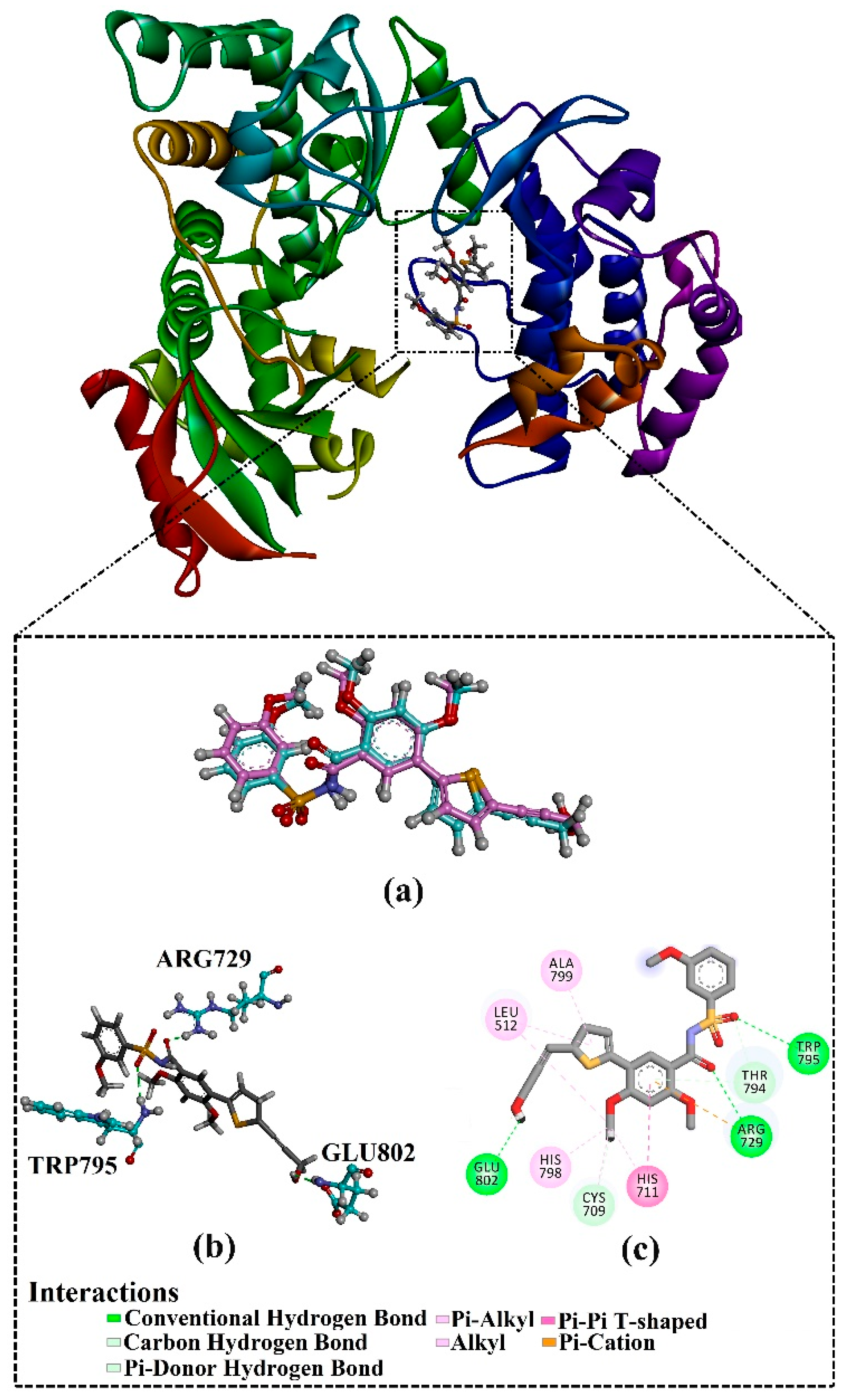
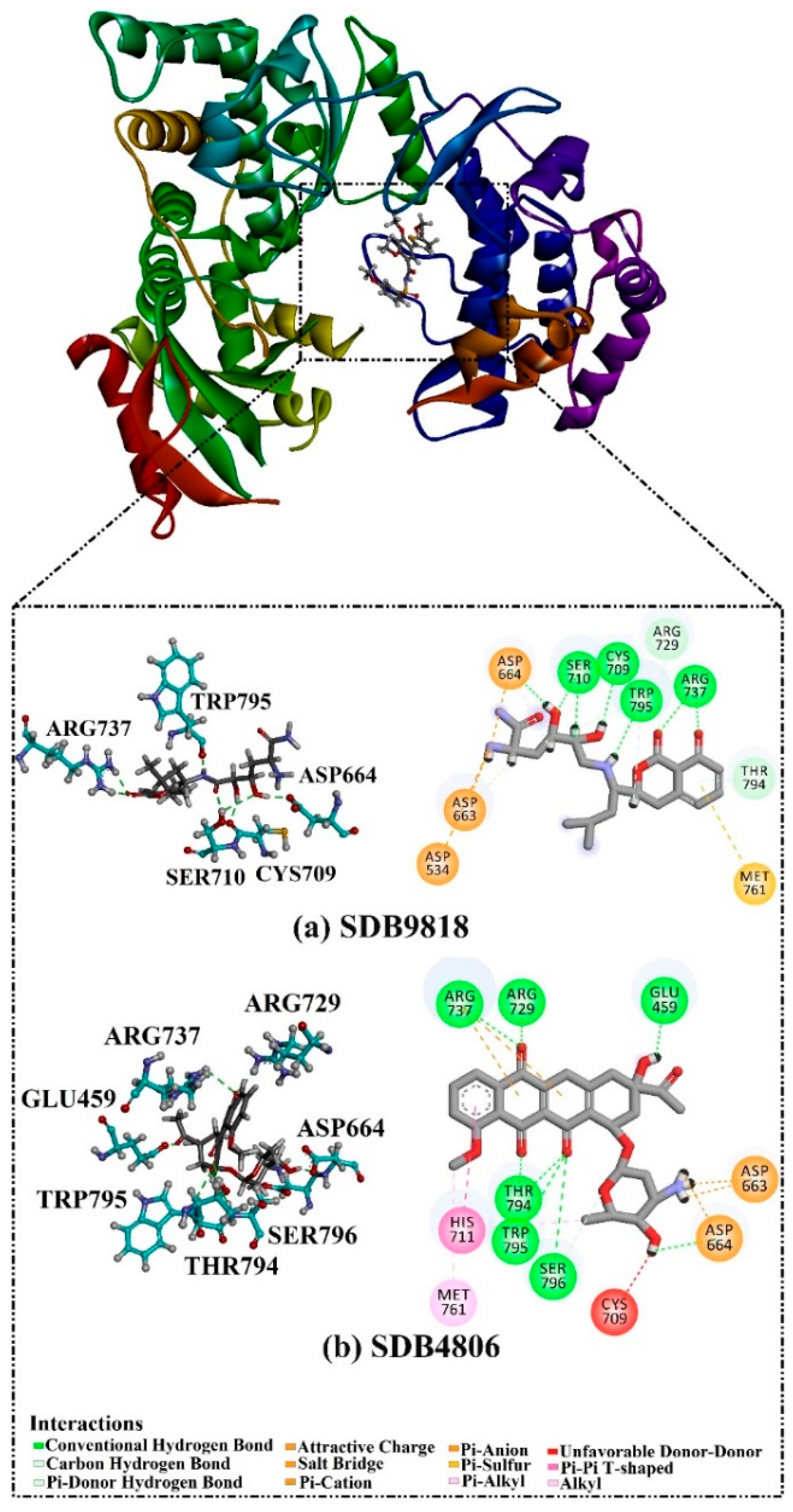
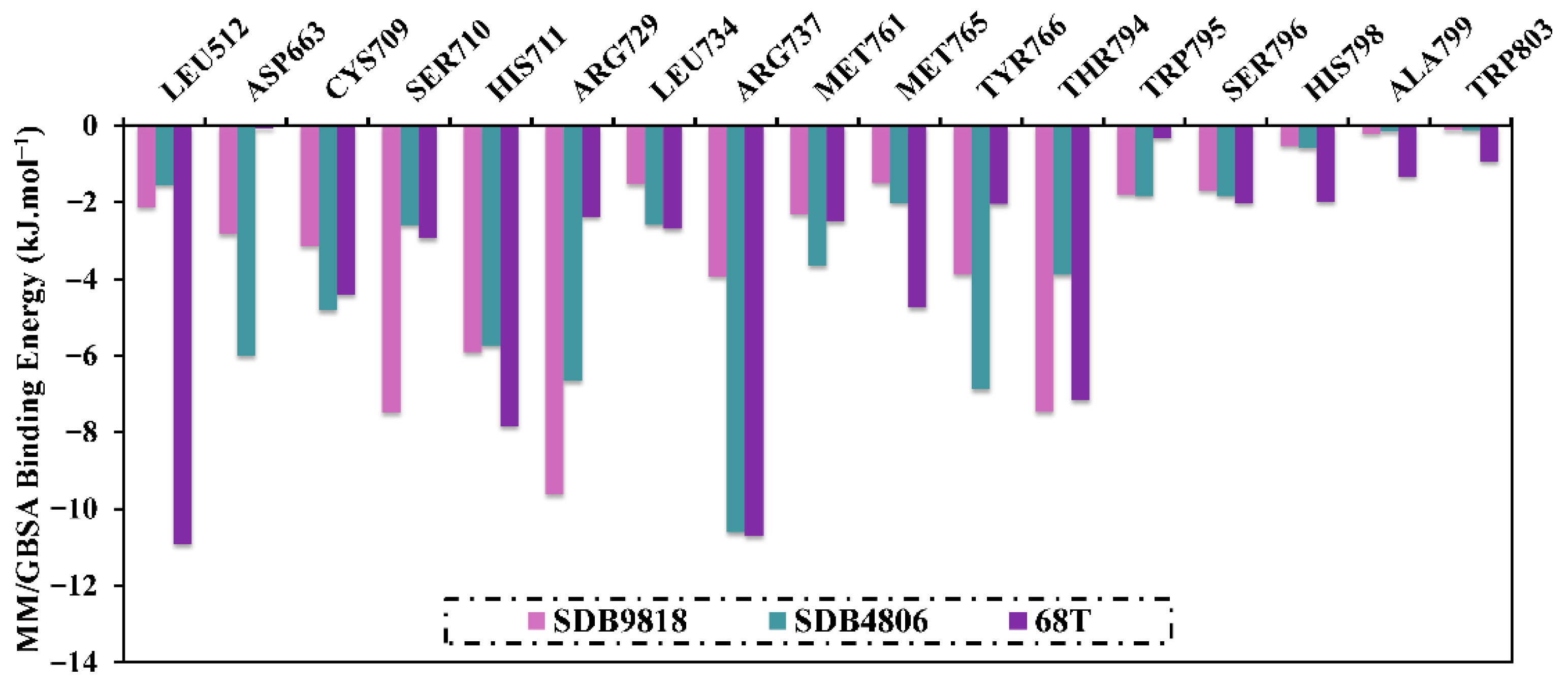
2.4. Post-MD Analyses
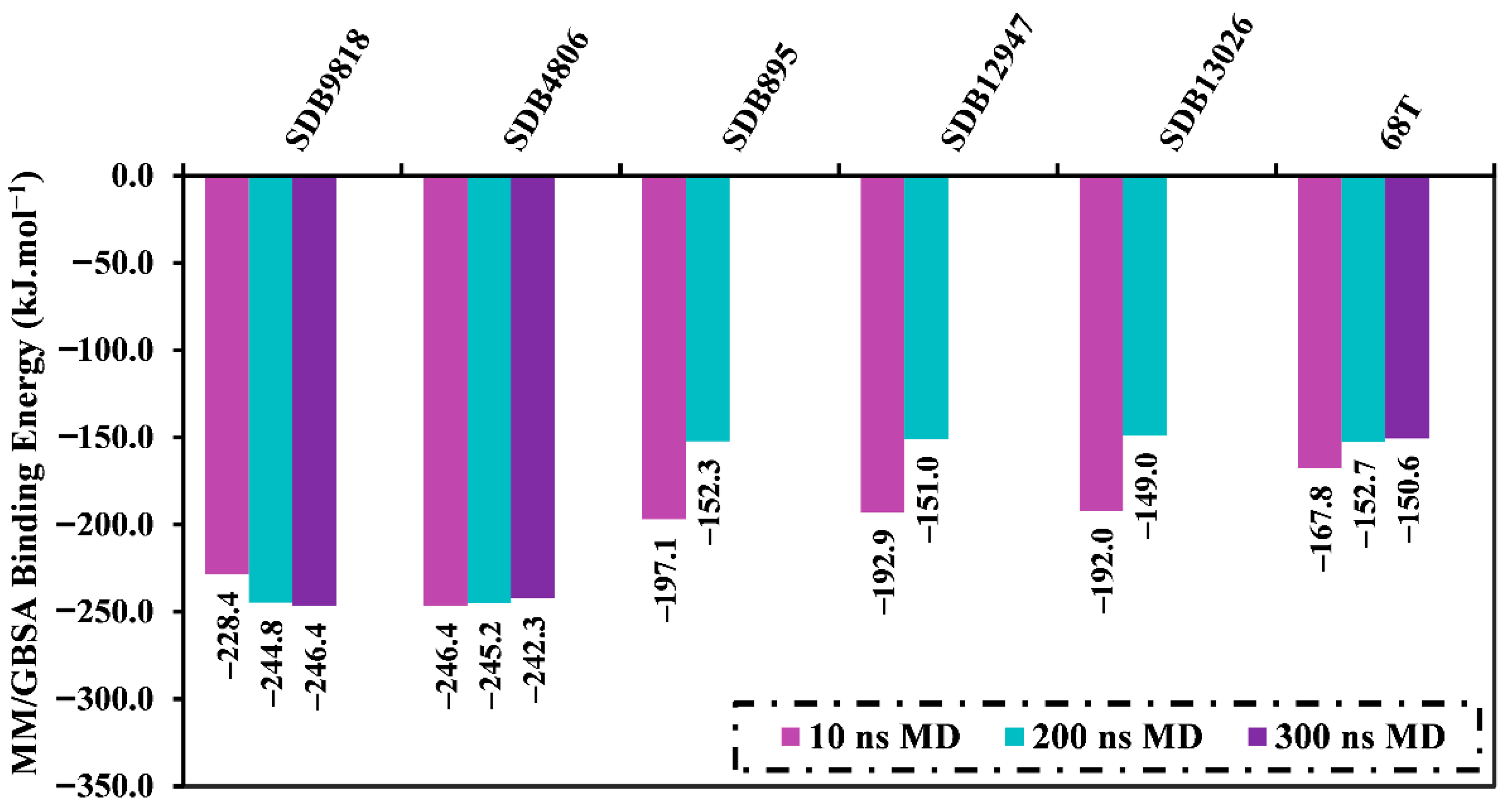
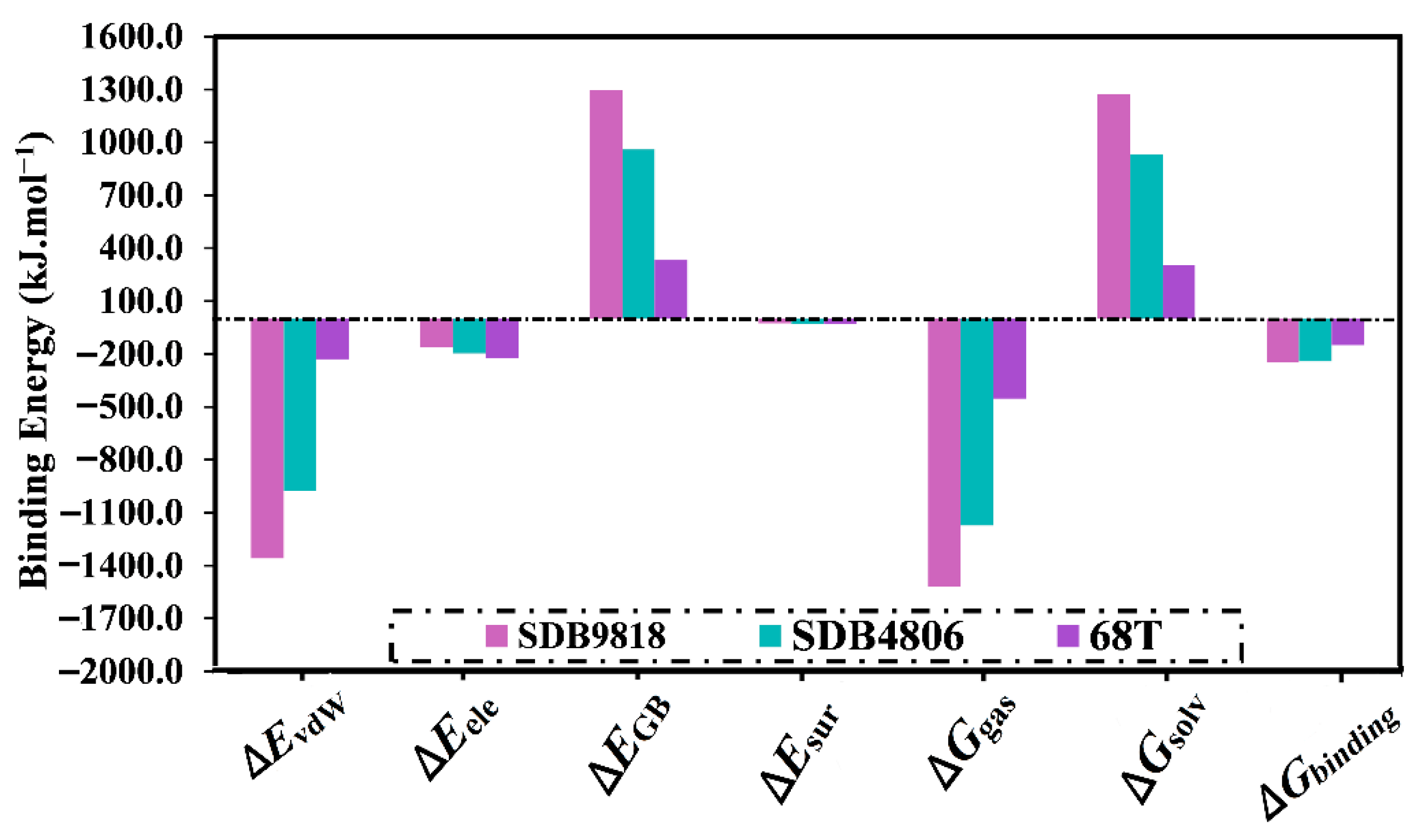
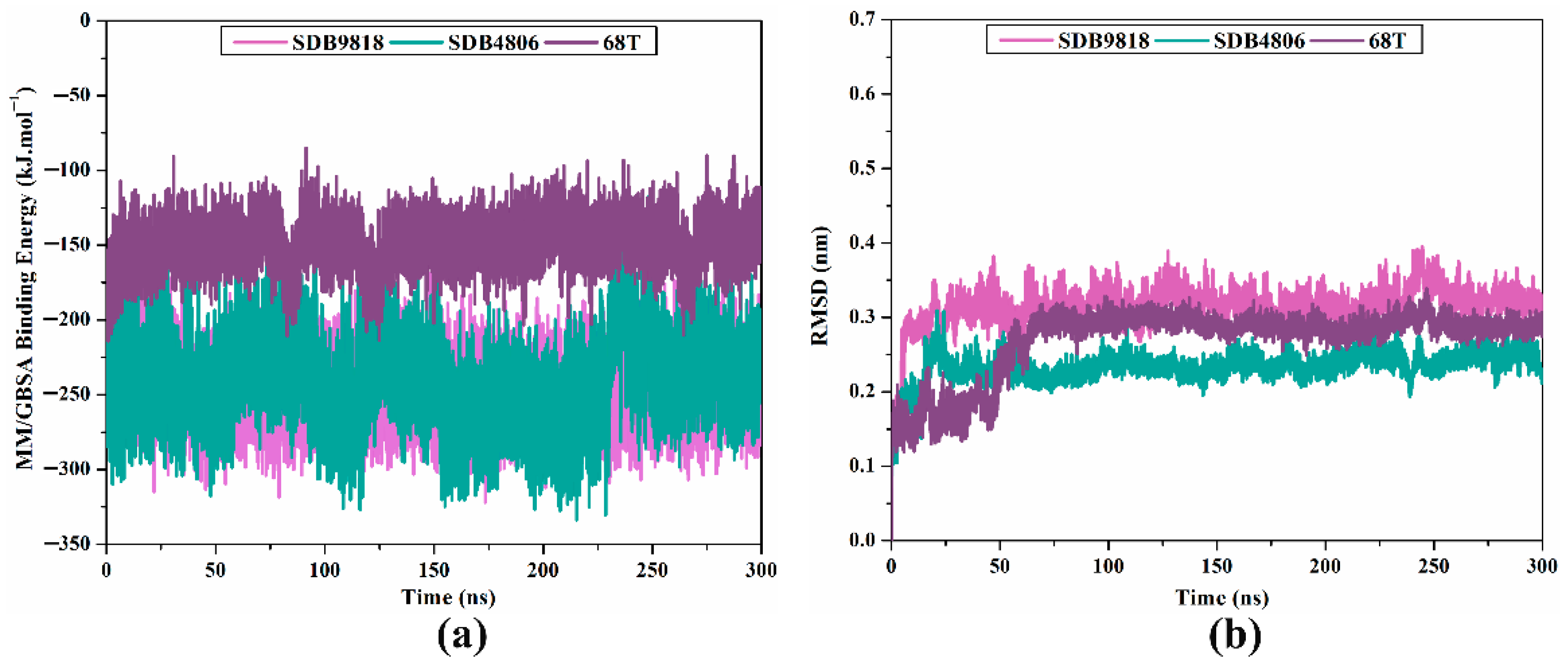
2.4.1. Binding Energy per Trajectory
2.4.2. RMSD Analysis
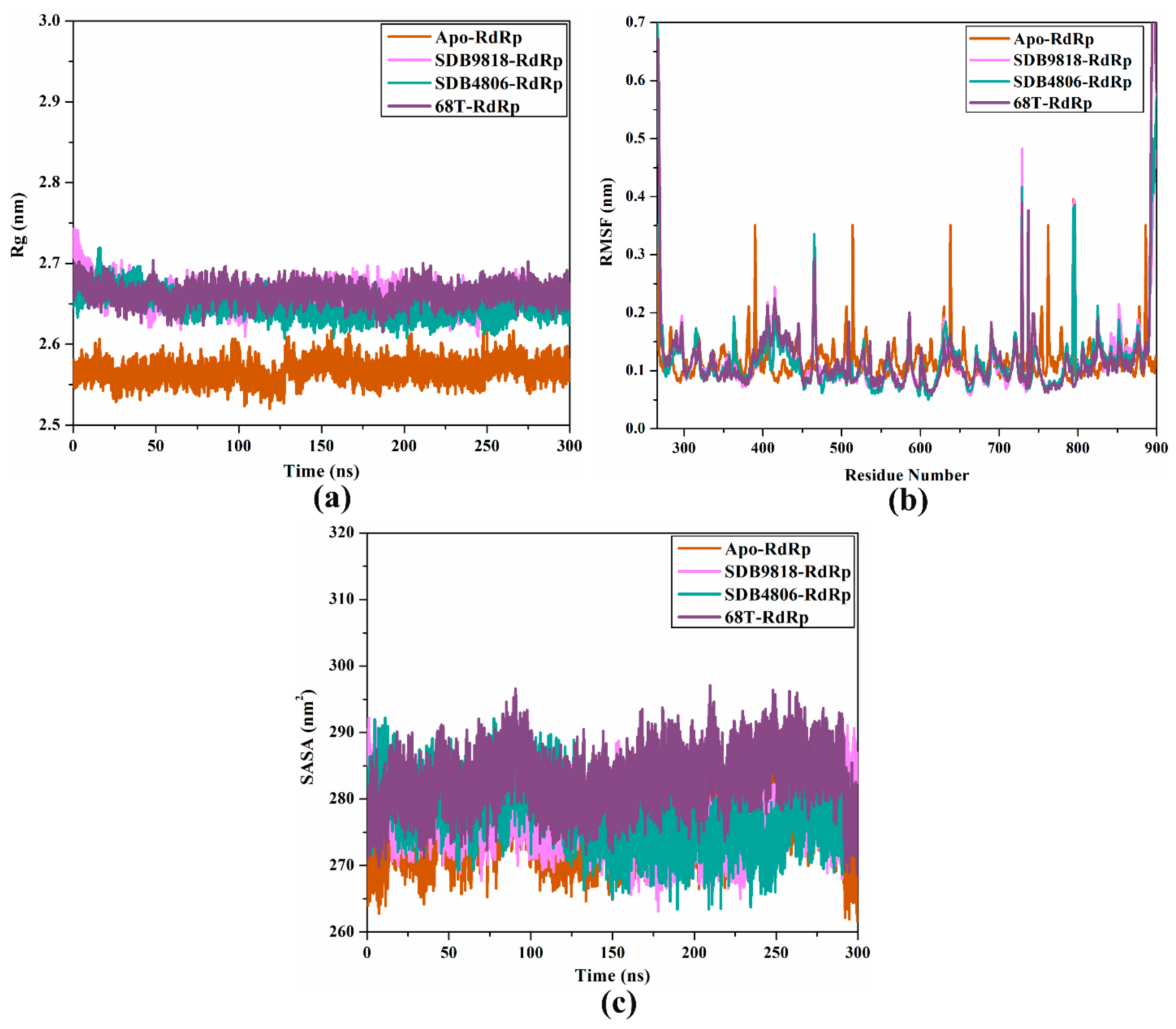
2.4.3. Rg Analysis
2.4.4. RMSF Analysis
2.4.5. SASA Analysis
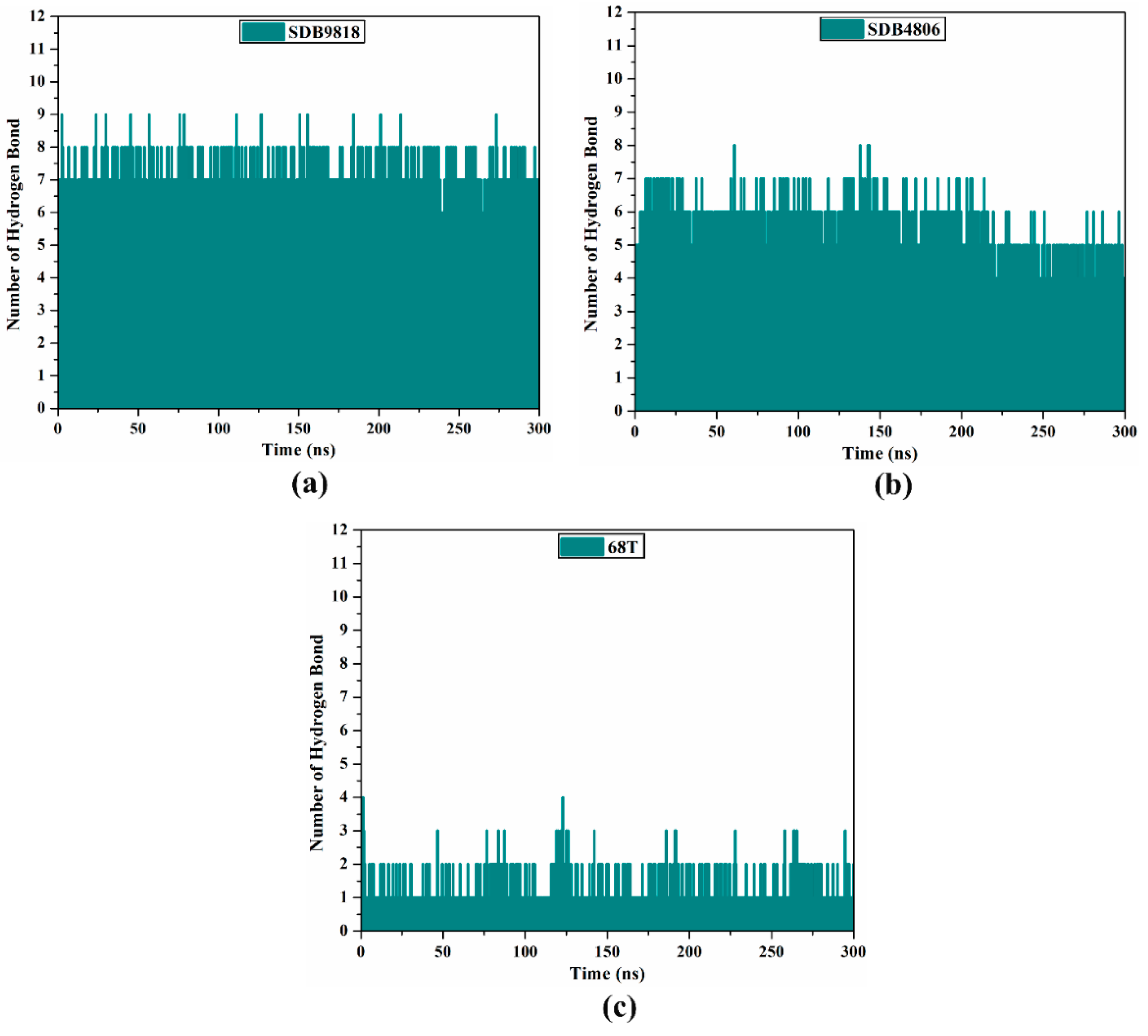
2.4.6. H-Bond Analysis
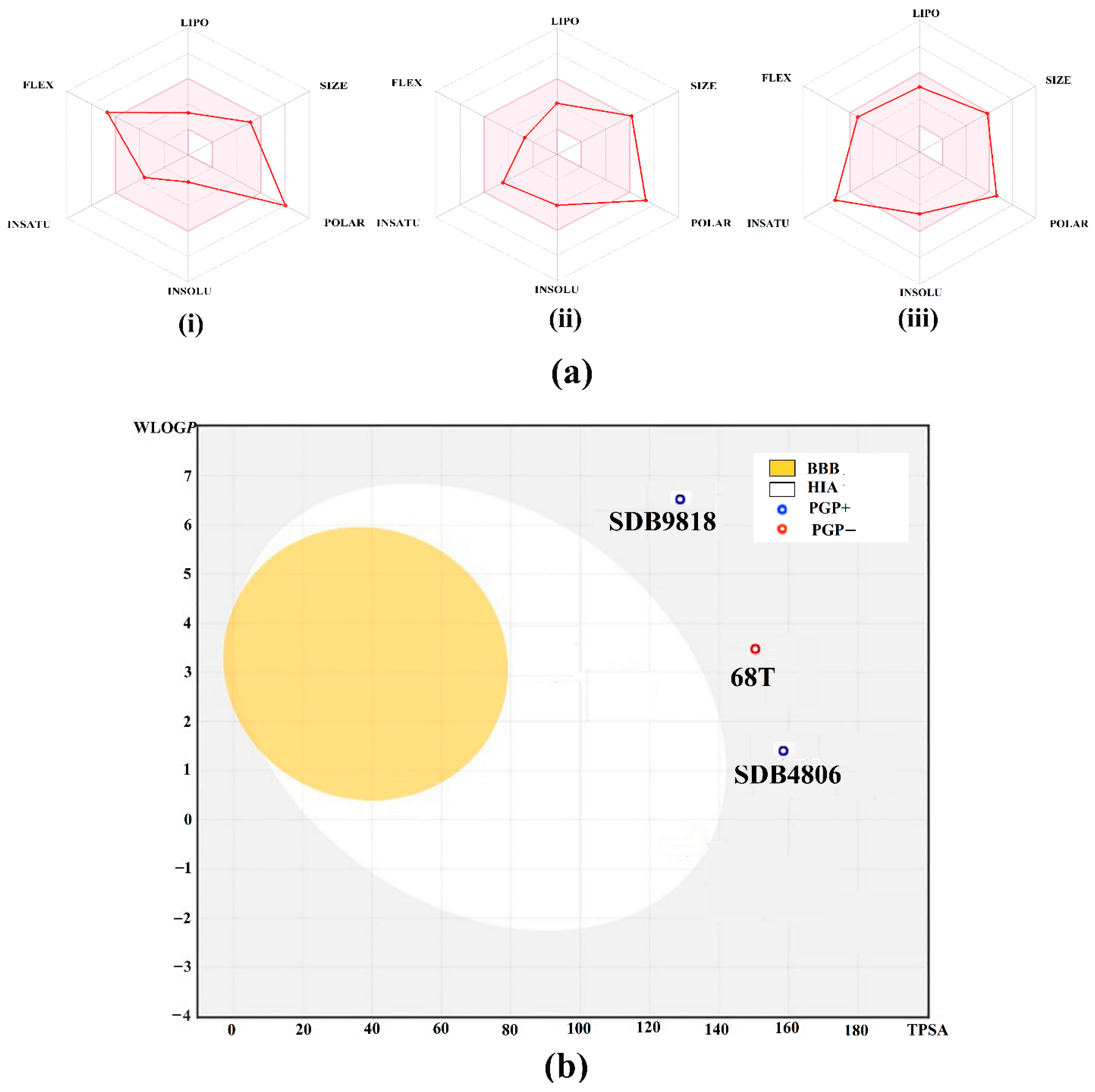
2.5. Physicochemical Characteristics
2.6. QM Computations
3. Computational Methodology
3.1. RdRp Preparation
3.2. Streptome Database Preparation
3.3. Docking Computation
3.4. MD Simulations (MDS)
3.5. Binding Energy Computations
3.6. Physicochemical Features
3.7. Quantum Mechanical (QM) Computations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; de Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; et al. Renaming of the genus Flavivirus to Orthoflavivirus and extension of binomial species names within the family Flaviviridae. Arch. Virol. 2023, 168, 224. [Google Scholar] [CrossRef]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martinez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef]
- Guzman, M.G.; Vazquez, S. The complexity of antibody-dependent enhancement of dengue virus infection. Viruses 2010, 2, 2649–2662. [Google Scholar] [CrossRef]
- Ferreira-de-Lima, V.H.; Lima-Camara, T.N. Natural vertical transmission of dengue virus in Aedes aegypti and Aedes albopictus: A systematic review. Parasites Vectors 2018, 11, 77. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Beatty, M.E.; Stone, A.; Fitzsimons, D.W.; Hanna, J.N.; Lam, S.K.; Vong, S.; Guzman, M.G.; Mendez-Galvan, J.F.; Halstead, S.B.; Letson, G.W.; et al. Best practices in dengue surveillance: A report from the Asia-Pacific and Americas Dengue Prevention Boards. PLoS Negl. Trop. Dis. 2010, 4, e890. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, A.; Pilankatta, R.; Teramoto, T.; Sajith, A.M.; Nwulia, E.; Kulkarni, A.; Padmanabhan, R. Inhibition of dengue virus by curcuminoids. Antivir. Res. 2019, 162, 71–78. [Google Scholar] [CrossRef]
- Normile, D. Tropical medicine. Surprising new dengue virus throws a spanner in disease control efforts. Science 2013, 342, 415. [Google Scholar] [CrossRef] [PubMed]
- Khetarpal, N.; Khanna, I. Dengue Fever: Causes, Complications, and Vaccine Strategies. J. Immunol. Res. 2016, 2016, 6803098. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, D.W.; Green, S.; Kalayanarooj, S.; Innis, B.L.; Nimmannitya, S.; Suntayakorn, S.; Endy, T.P.; Raengsakulrach, B.; Rothman, A.L.; Ennis, F.A.; et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000, 181, 2–9. [Google Scholar] [CrossRef]
- Murugesan, A.; Manoharan, M. Dengue Virus. In Emerging and Reemerging Viral Pathogens; Ennaji, M.M., Ed.; Academic Press: London, UK, 2020; pp. 281–359. [Google Scholar]
- Wei, Y.; Qin, C.; Jiang, T.; Li, X.; Zhao, H.; Liu, Z.; Deng, Y.; Liu, R.; Chen, S.; Yu, M.; et al. Translational regulation by the 3’ untranslated region of the dengue type 2 virus genome. Am. J. Trop. Med. Hyg. 2009, 81, 817–824. [Google Scholar] [CrossRef]
- Carocci, M.; Yang, P.L. Lactimidomycin is a broad-spectrum inhibitor of dengue and other RNA viruses. Antivir. Res. 2016, 128, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhou, Z.; Wen, Z.; Liu, Y.; Zeng, C.; Xiao, D.; Ou, M.; Han, Y.; Huang, S.; Liu, D.; et al. Global Epidemiology of Dengue Outbreaks in 1990–2015: A Systematic Review and Meta-Analysis. Front. Cell Infect. Microbiol. 2017, 7, 317. [Google Scholar] [CrossRef] [PubMed]
- Soe, A.M.; Ngwe Tun, M.M.; Nabeshima, T.; Myat, T.W.; Htun, M.M.; Lin, H.; Hom, N.S.; Inoue, S.; Nwe, K.M.; Aye, L.P.P.; et al. Emergence of a Novel Dengue Virus 3 (DENV-3) Genotype-I Coincident with Increased DENV-3 Cases in Yangon, Myanmar between 2017 and 2019. Viruses 2021, 13, 1152. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, G.; Visoso-Carvajal, G.; Garcia-Cordero, J.; Leon-Juarez, M.; Chavez-Munguia, B.; Lopez, T.; Nava, P.; Villegas-Sepulveda, N.; Cedillo-Barron, L. Dengue Virus Serotype 2 and Its Non-Structural Proteins 2A and 2B Activate NLRP3 Inflammasome. Front. Immunol. 2020, 11, 352. [Google Scholar] [CrossRef]
- da Fonseca, N.J., Jr.; Lima Afonso, M.Q.; Pedersolli, N.G.; de Oliveira, L.C.; Andrade, D.S.; Bleicher, L. Sequence, structure and function relationships in flaviviruses as assessed by evolutive aspects of its conserved non-structural protein domains. Biochem. Biophys. Res. Commun. 2017, 492, 565–571. [Google Scholar] [CrossRef]
- Verhaegen, M.; Vermeire, K. The endoplasmic reticulum (ER): A crucial cellular hub in flavivirus infection and potential target site for antiviral interventions. npj Viruses 2024, 2, 24. [Google Scholar] [CrossRef]
- Dey, D.; Poudyal, S.; Rehman, A.; Hasan, S.S. Structural and biochemical insights into flavivirus proteins. Virus Res. 2021, 296, 198343. [Google Scholar] [CrossRef]
- Cortese, M.; Mulder, K.; Chatel-Chaix, L.; Scaturro, P.; Cerikan, B.; Plaszczyca, A.; Haselmann, U.; Bartenschlager, M.; Neufeldt, C.J.; Bartenschlager, R. Determinants in Nonstructural Protein 4A of Dengue Virus Required for RNA Replication and Replication Organelle Biogenesis. J. Virol. 2021, 95, e0131021. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins. Membranes 2022, 12, 231. [Google Scholar] [CrossRef]
- Acosta, E.G.; Kumar, A.; Bartenschlager, R. Chapter One—Revisiting Dengue Virus–Host Cell Interaction: New Insights into Molecular and Cellular Virology. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Eds.; Academic Press: London, UK, 2014; Volume 88, pp. 1–109. [Google Scholar]
- Selisko, B.; Wang, C.; Harris, E.; Canard, B. Regulation of Flavivirus RNA synthesis and replication. Curr. Opin. Virol. 2014, 9, 74–83. [Google Scholar] [CrossRef]
- Bollati, M.; Milani, M.; Mastrangelo, E.; Ricagno, S.; Tedeschi, G.; Nonnis, S.; Decroly, E.; Selisko, B.; de Lamballerie, X.; Coutard, B.; et al. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: Implications for RNA-capping mechanisms in Flavivirus. J. Mol. Biol. 2009, 385, 140–152. [Google Scholar] [CrossRef]
- Holmes, E.C.; Twiddy, S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef]
- Lim, S.P.; Noble, C.G.; Shi, P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015, 119, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 1993, 74, 733–740. [Google Scholar] [CrossRef]
- Xu, H.T.; Colby-Germinario, S.P.; Hassounah, S.; Quashie, P.K.; Han, Y.; Oliveira, M.; Stranix, B.R.; Wainberg, M.A. Identification of a Pyridoxine-Derived Small-Molecule Inhibitor Targeting Dengue Virus RNA-Dependent RNA Polymerase. Antimicrob. Agents Chemother. 2016, 60, 600–608. [Google Scholar] [CrossRef]
- Cannalire, R.; Tarantino, D.; Astolfi, A.; Barreca, M.L.; Sabatini, S.; Massari, S.; Tabarrini, O.; Milani, M.; Querat, G.; Mastrangelo, E.; et al. Functionalized 2,1-benzothiazine 2,2-dioxides as new inhibitors of Dengue NS5 RNA-dependent RNA polymerase. Eur. J. Med. Chem. 2018, 143, 1667–1676. [Google Scholar] [CrossRef]
- Noble, C.G.; Lim, S.P.; Chen, Y.L.; Liew, C.W.; Yap, L.; Lescar, J.; Shi, P.Y. Conformational flexibility of the Dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013, 87, 5291–5295. [Google Scholar] [CrossRef]
- Manvar, D.; Kucukguzel, I.; Erensoy, G.; Tatar, E.; Deryabasogullari, G.; Reddy, H.; Talele, T.T.; Cevik, O.; Kaushik-Basu, N. Discovery of conjugated thiazolidinone-thiadiazole scaffold as anti-dengue virus polymerase inhibitors. Biochem. Biophys. Res. Commun. 2016, 469, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Anusuya, S.; Velmurugan, D.; Gromiha, M.M. Identification of dengue viral RNA-dependent RNA polymerase inhibitor using computational fragment-based approaches and molecular dynamics study. J. Biomol. Struct. Dyn. 2016, 34, 1512–1532. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Espiritu, M.V.; Abraham, J.; Thorpe, I.F. Computational predictions suggest that structural similarity in viral polymerases may lead to comparable allosteric binding sites. Virus Res. 2016, 222, 80–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crotty, S.; Maag, D.; Arnold, J.J.; Zhong, W.; Lau, J.Y.; Hong, Z.; Andino, R.; Cameron, C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000, 6, 1375–1379. [Google Scholar] [CrossRef]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; Van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef]
- Furuta, Y.; Takahashi, K.; Shiraki, K.; Sakamoto, K.; Smee, D.F.; Barnard, D.L.; Gowen, B.B.; Julander, J.G.; Morrey, J.D. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antivir. Res. 2009, 82, 95–102. [Google Scholar] [CrossRef]
- Brown, A.J.; Won, J.J.; Graham, R.L.; Dinnon, K.H., 3rd; Sims, A.C.; Feng, J.Y.; Cihlar, T.; Denison, M.R.; Baric, R.S.; Sheahan, T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019, 169, 104541. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Stessl, M.; Amartey, J.; Noe, C.R. Off-target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem 2010, 5, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.J.; Chang, W.; Furman, P.A.; Mosley, R.T.; Ross, B.S. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J. Med. Chem. 2012, 55, 2481–2531. [Google Scholar] [CrossRef]
- Denel-Bobrowska, M.; Olejniczak, A.B. Non-nucleoside structured compounds with antiviral activity-past 10 years (2010-2020). Eur. J. Med. Chem. 2022, 231, 114136. [Google Scholar] [CrossRef]
- Lim, S.P.; Noble, C.G.; Seh, C.C.; Soh, T.S.; El Sahili, A.; Chan, G.K.; Lescar, J.; Arora, R.; Benson, T.; Nilar, S.; et al. Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling. PLoS Pathog. 2016, 12, e1005737. [Google Scholar] [CrossRef]
- Yao, X.; Guo, S.; Wu, W.; Wang, J.; Wu, S.; He, S.; Wan, Y.; Nandakumar, K.S.; Chen, X.; Sun, N.; et al. Q63, a novel DENV2 RdRp non-nucleoside inhibitor, inhibited DENV2 replication and infection. J. Pharmacol. Sci. 2018, 138, 247–256. [Google Scholar] [CrossRef]
- Kimura, T.; Suga, T.; Kameoka, M.; Ueno, M.; Inahashi, Y.; Matsuo, H.; Iwatsuki, M.; Shigemura, K.; Shiomi, K.; Takahashi, Y.; et al. New tetrahydroquinoline and indoline compounds containing a hydroxy cyclopentenone, virantmycin B and C, produced by Streptomyces sp. AM-2504. J. Antibiot. 2019, 72, 169–173. [Google Scholar] [CrossRef]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y.; et al. Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef]
- Liu, M.; Ren, M.; Zhang, Y.; Wan, Z.; Wang, Y.; Wu, Z.; Wang, K.; Fang, W.; Yang, X. Antiviral Activity of Benzoheterocyclic Compounds from Soil-Derived Streptomyces jiujiangensis NBERC-24992. Molecules 2023, 28, 878. [Google Scholar] [CrossRef]
- Low, J.S.; Wu, K.X.; Chen, K.C.; Ng, M.M.; Chu, J.J. Narasin, a novel antiviral compound that blocks dengue virus protein expression. Antivir. Ther. 2011, 16, 1203–1218. [Google Scholar] [CrossRef]
- Lee, M.F.; Wu, Y.S.; Poh, C.L. Molecular Mechanisms of Antiviral Agents against Dengue Virus. Viruses 2023, 15, 705. [Google Scholar] [CrossRef]
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef]
- da Silva Costa, J.; da Silva Lopes Costa, K.; Cruz, J.V.; da Silva Ramos, R.; Silva, L.B.; Do Socorro Barros Brasil, D.; de Paula da Silva, C.H.T.; Dos Santos, C.B.R.; da Cruz Macedo, W.J. Virtual Screening and Statistical Analysis in the Design of New Caffeine Analogues Molecules with Potential Epithelial Anticancer Activity. Curr. Pharm. Des. 2018, 24, 576–594. [Google Scholar] [CrossRef]
- Gowthaman, U.; Jayakanthan, M.; Sundar, D. Molecular docking studies of dithionitrobenzoic acid and its related compounds to protein disulfide isomerase: Computational screening of inhibitors to HIV-1 entry. BMC Bioinform. 2008, 9 (Suppl. 12), S14. [Google Scholar] [CrossRef] [PubMed]
- McInnes, C. Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 2007, 11, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Rajan, B.M.; Kannabiran, K. Extraction and Identification of Antibacterial Secondary Metabolites from Marine Streptomyces sp. VITBRK2. Int. J. Mol. Cell. Med. 2014, 3, 130–137. [Google Scholar]
- Wagner, C.; Eckardt, K.; Tresselt, D.; Ihn, W.; Schumann, G.; Fleck, W.F. Leukaemomycin-geblockte Mutanten des Streptomyces griseus und ihre Pigmente III. 11-Desoxydaunomycinonderivate aus der Mutante ZIMET 43699/G44. J. Basic Microbiol. 2007, 25, 687–693. [Google Scholar] [CrossRef]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Benson, N.C.; Daggett, V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 8722–8731. [Google Scholar] [CrossRef] [PubMed]
- Bellissent-Funel, M.C.; Hassanali, A.; Havenith, M.; Henchman, R.; Pohl, P.; Sterpone, F.; van der Spoel, D.; Xu, Y.; Garcia, A.E. Water Determines the Structure and Dynamics of Proteins. Chem. Rev. 2016, 116, 7673–7697. [Google Scholar] [CrossRef]
- Caminero Gomes Soares, A.; Marques Sousa, G.H.; Calil, R.L.; Goulart Trossini, G.H. Absorption matters: A closer look at popular oral bioavailability rules for drug approvals. Mol. Inform. 2023, 42, e202300115. [Google Scholar] [CrossRef]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An updated compendium of streptomycetes natural products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef]
- Heller, S.R.; McNaught, A.; Pletnev, I.; Stein, S.; Tchekhovskoi, D. InChI, the IUPAC International Chemical Identifier. J. Cheminform. 2015, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- OpenEye Scientific Software. OMEGA 4.1.1.0; OpenEye Scientific Software: Santa Fe, NM, USA, 2021. [Google Scholar]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- Halgren, T.A. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 1999, 20, 720–729. [Google Scholar] [CrossRef]
- OpenEye Scientific Software. SZYBKI 2.4.0.0; OpenEye Scientific Software: Santa Fe, NM, USA, 2021. [Google Scholar]
- OpenEye Scientific Software. QUACPAC 2.1.3.0; OpenEye Scientific Software: Santa Fe, NM, USA, 2021. [Google Scholar]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Ibrahim, M.A.A.; Hassan, A.M.A.; Mekhemer, G.A.H.; Sidhom, P.A.; El-Tayeb, M.A.; Abdelbacki, A.M.M.; Khan, S.; Soliman, M.E.S.; Abdelrahman, A.H.M. Exploring marine natural products for identifying putative candidates as EBNA1 inhibitors: An insight from molecular docking, molecular dynamics, and DFT computations. Biochem. Biophys. Res. Commun. 2024, 735, 150856. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Hassan, A.M.A.; Mohamed, E.A.R.; Mekhemer, G.A.H.; Sidhom, P.A.; El-Tayeb, M.A.; Khan, S.; Shoeib, T.; Soliman, M.E.S.; Abdelrahman, A.H.M. Repurposing of drug candidates against Epstein-Barr virus: Virtual screening, docking computations, molecular dynamics, and quantum mechanical study. PLoS ONE 2024, 19, e0312100. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdelrahman, A.H.M.; Mohamed, D.E.M.; Abdeljawaad, K.A.A.; Naeem, M.A.; Gabr, G.A.; Shawky, A.M.; Soliman, M.E.S.; Sidhom, P.A.; Pare, P.W.; et al. Chetomin, a SARS-CoV-2 3C-like protease (3CLpro) inhibitor: In silico screening, enzyme docking, molecular dynamics and pharmacokinetics analysis. Viruses 2023, 15, 250. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision E01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Bayly, C.I.; Cieplak, P.; Cornell, W.D.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer, Version 2019; Dassault Systèmes: San Diego, CA, USA, 2019. [Google Scholar]
- Massova, I.; Kollman, P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. 2000, 18, 113–135. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 2004, 55, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: Strategies and applications in drug design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Sun, H.; Duan, L.; Chen, F.; Liu, H.; Wang, Z.; Pan, P.; Zhu, F.; Zhang, J.Z.H.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys Chem Chem Phys. 2018, 20, 14450–14460. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Ibrahim, M.A.A. Molecular mechanical perspective on halogen bonding. J. Mol. Model. 2012, 18, 4625–4638. [Google Scholar] [CrossRef] [PubMed]



| Compound Name/ID | 2D Chemical Structure | Docking Score (kJ.mol−1) | Intermolecular H-Bond | |
|---|---|---|---|---|
| Standard | Expensive | |||
| 68T | 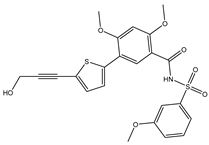 | −35.6 | −35.6 | ARG729 (3.11 Å), TRP795 (3.40 Å), GLU802 (2.67 Å) |
| SDB9818 | 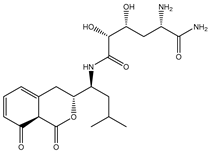 | −46.9 | −46.9 | ARG737 (2.04; 2.62; 1.83 Å), SER796 (3.01 Å), CYS709 (1.83 Å), ASP664 (1.74 Å), TRP795 (1.80 Å), SER710 (1.70 Å) |
| SDB4806 | 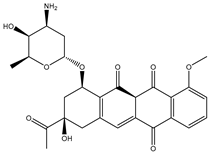 | −42.7 | −45.6 | GLU459 (1.63 Å), ASP664 (1.84 Å), ARG729 (2.16 Å), ARG737 (2.84 Å), THR794 (1.97; 2.44 Å), TRP795 (2.54 Å), SER796 (2.83 Å) |
| SDB895 | 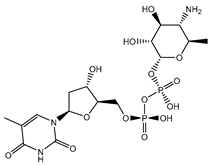 | –41.8 | –45.2 | LYS461 (2.90 Å), ASP664 (1.81 Å), ARG737 (3.72 Å), SER796 (2.22; 2.33; 2.08 Å), |
| SDB12947 | 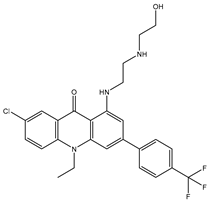 | –35.6 | –45.2 | ASP664 (1.88 Å), HIS798 (3.18 Å), SER796 (3.02; 3.18 Å), CYS709 (2.19 Å) |
| SDB13026 | 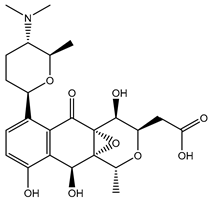 | –46.9 | –45.2 | LYS461 (2.09; 2.12; 2.22 Å), ASP664 (2.45 Å), ARG729 (2.33; 2.35 Å), TYR766 (2.83 Å), ARG737 (1.73 Å), |
| SDB9891 |  | –42.3 | –45.2 | LYS461 (1.90 Å), SER796 (1.72; 2.25; 3.15 Å), TYR766 (2.02; 2.15 Å), HIS798 (3.08 Å), CYS709 (1.83 Å) |
| SDB10285 | 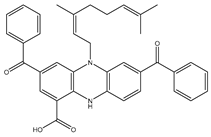 | –41.8 | –42.3 | LYS461 (2.24 Å), ARG472 (2.95 Å), ARG737 (2.53 Å), GLU802 (1.86; 2.36 Å) |
| SDB993 | 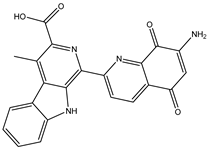 | –41.8 | –42.3 | SER796 (2.65 Å), ARG737 (2.01 Å) |
| SDB1014 | 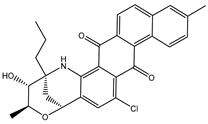 | –41.4 | –40.2 | LYS461 (2.60 Å), SER796 (2.84 Å) |
| SDB827 | 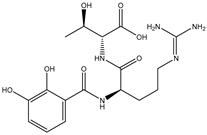 | –41.4 | –39.3 | ASP664 (2.13; 2.23 Å), ARG729 (3.07 Å), ARG737 (1.99, 2.57 Å), TYR766 (2.09, 2.01 Å), THR794 (2.41 Å), SER796 (2.12, 2.19 Å) |
| Compound Name/ID | LogP | MW (g/mol) | HBD | HBA |
|---|---|---|---|---|
| 68T | 4.23 | 487.55 | 2 | 7 |
| SDB9818 | 0.40 | 423.46 | 6 | 8 |
| SDB4806 | 2.24 | 511.52 | 4 | 10 |
| Compound Name/ID | EHOMO | ELUMO | EFL | Egap |
|---|---|---|---|---|
| 68T | −6.88 | −0.83 | −3.85 | 6.05 |
| SDB9818 | −7.99 | −1.34 | −4.67 | 6.65 |
| SDB4806 | −8.06 | −2.33 | −5.19 | 5.74 |
| Compound Name/ID | IP (eV) | EA (eV) | η (eV) | S (eV−1) |
|---|---|---|---|---|
| 68T | 6.88 | 0.83 | 3.03 | 0.33 |
| SDB9818 | 7.99 | 1.34 | 3.32 | 0.30 |
| SDB4806 | 8.06 | 2.33 | 2.87 | 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelrahman, A.H.M.; Mekhemer, G.A.H.; Sidhom, P.A.; Abalkhail, T.; Khan, S.; Ibrahim, M.A.A. In Silico Mining of the Streptome Database for Hunting Putative Candidates to Allosterically Inhibit the Dengue Virus (Serotype 2) RdRp. Pharmaceuticals 2025, 18, 1135. https://doi.org/10.3390/ph18081135
Abdelrahman AHM, Mekhemer GAH, Sidhom PA, Abalkhail T, Khan S, Ibrahim MAA. In Silico Mining of the Streptome Database for Hunting Putative Candidates to Allosterically Inhibit the Dengue Virus (Serotype 2) RdRp. Pharmaceuticals. 2025; 18(8):1135. https://doi.org/10.3390/ph18081135
Chicago/Turabian StyleAbdelrahman, Alaa H. M., Gamal A. H. Mekhemer, Peter A. Sidhom, Tarad Abalkhail, Shahzeb Khan, and Mahmoud A. A. Ibrahim. 2025. "In Silico Mining of the Streptome Database for Hunting Putative Candidates to Allosterically Inhibit the Dengue Virus (Serotype 2) RdRp" Pharmaceuticals 18, no. 8: 1135. https://doi.org/10.3390/ph18081135
APA StyleAbdelrahman, A. H. M., Mekhemer, G. A. H., Sidhom, P. A., Abalkhail, T., Khan, S., & Ibrahim, M. A. A. (2025). In Silico Mining of the Streptome Database for Hunting Putative Candidates to Allosterically Inhibit the Dengue Virus (Serotype 2) RdRp. Pharmaceuticals, 18(8), 1135. https://doi.org/10.3390/ph18081135








