5′-Guanidino Xylofuranosyl Nucleosides as Novel Types of 5′-Functionalized Nucleosides with Biological Potential
Abstract
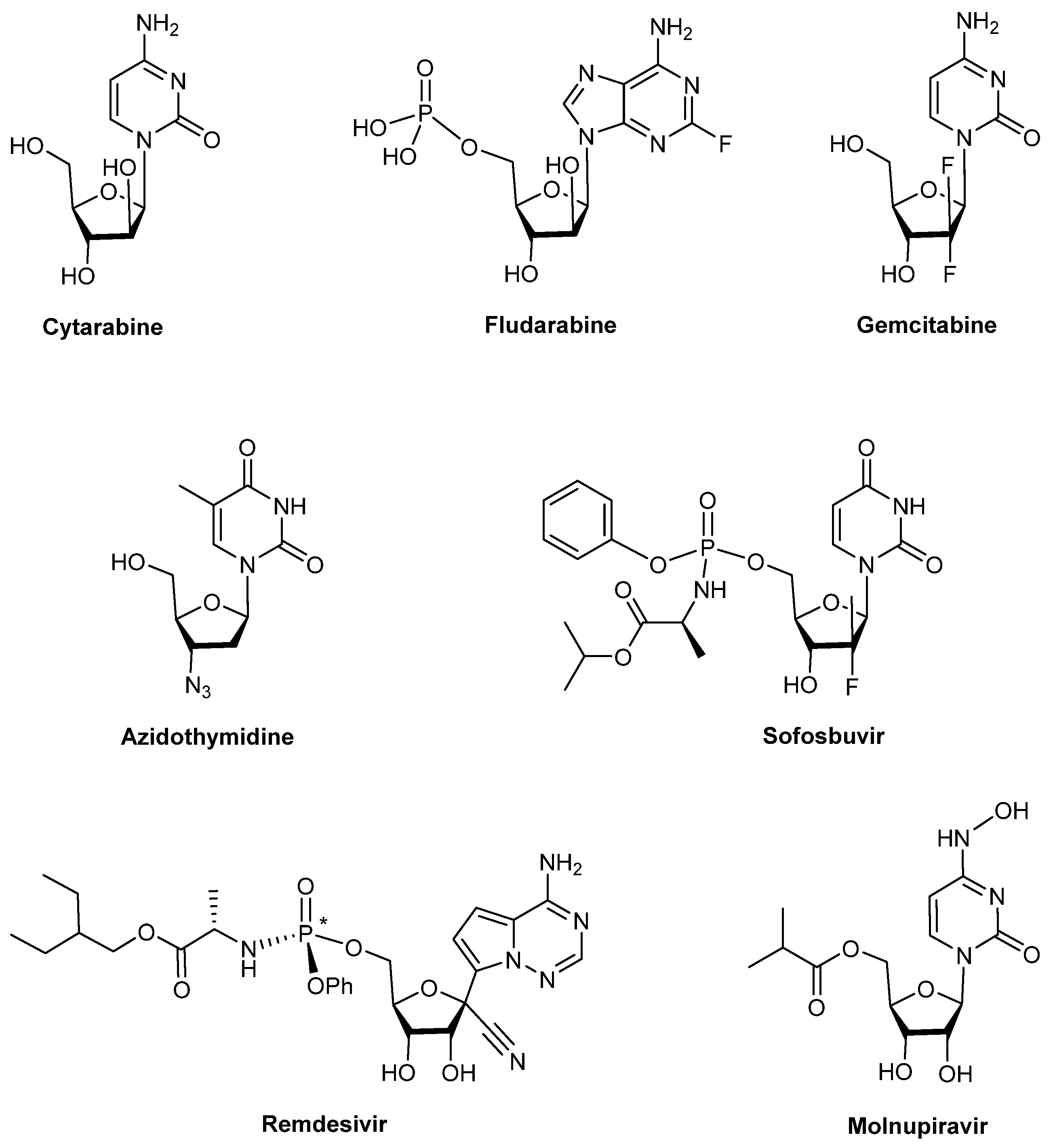
:1. Introduction
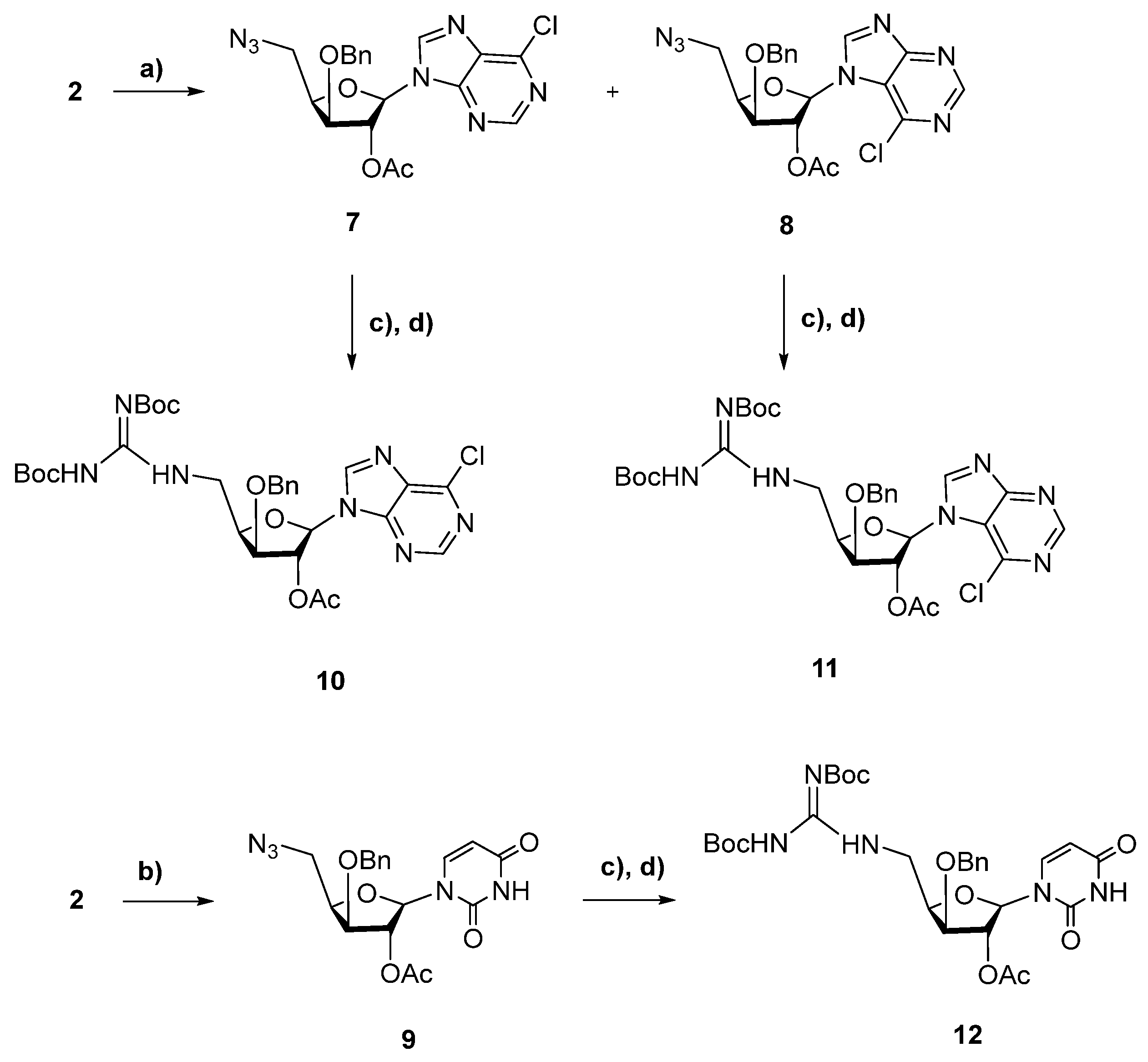
2. Results and Discussion
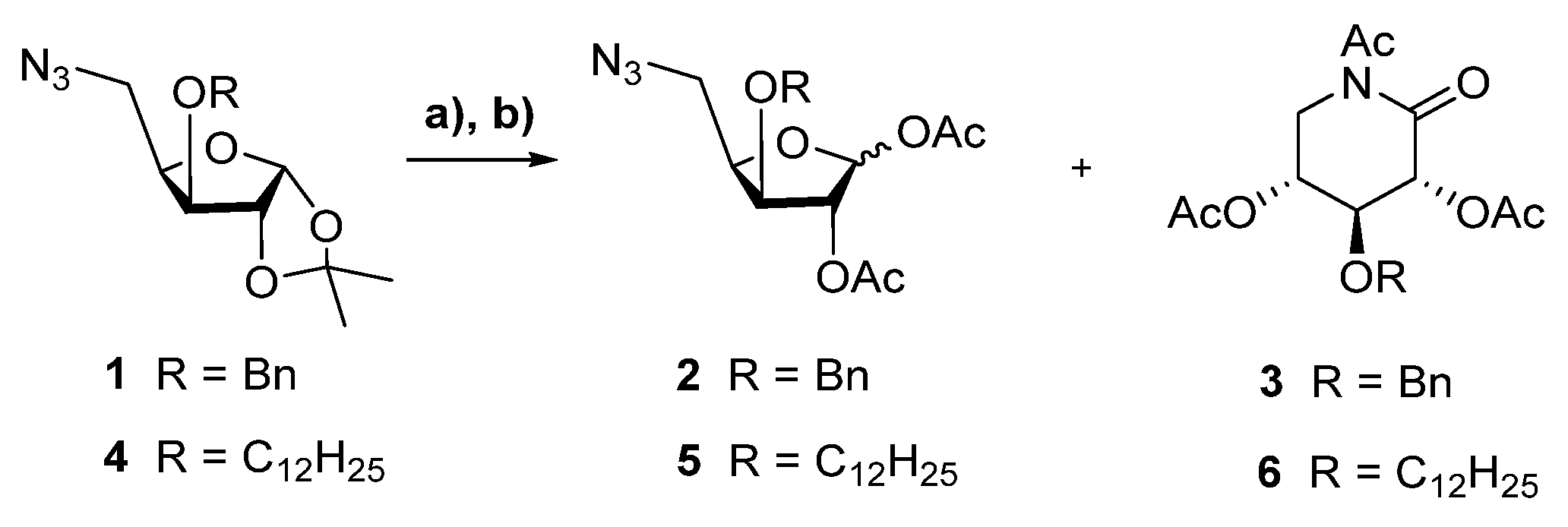
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Cholinesterase Inhibition Assays
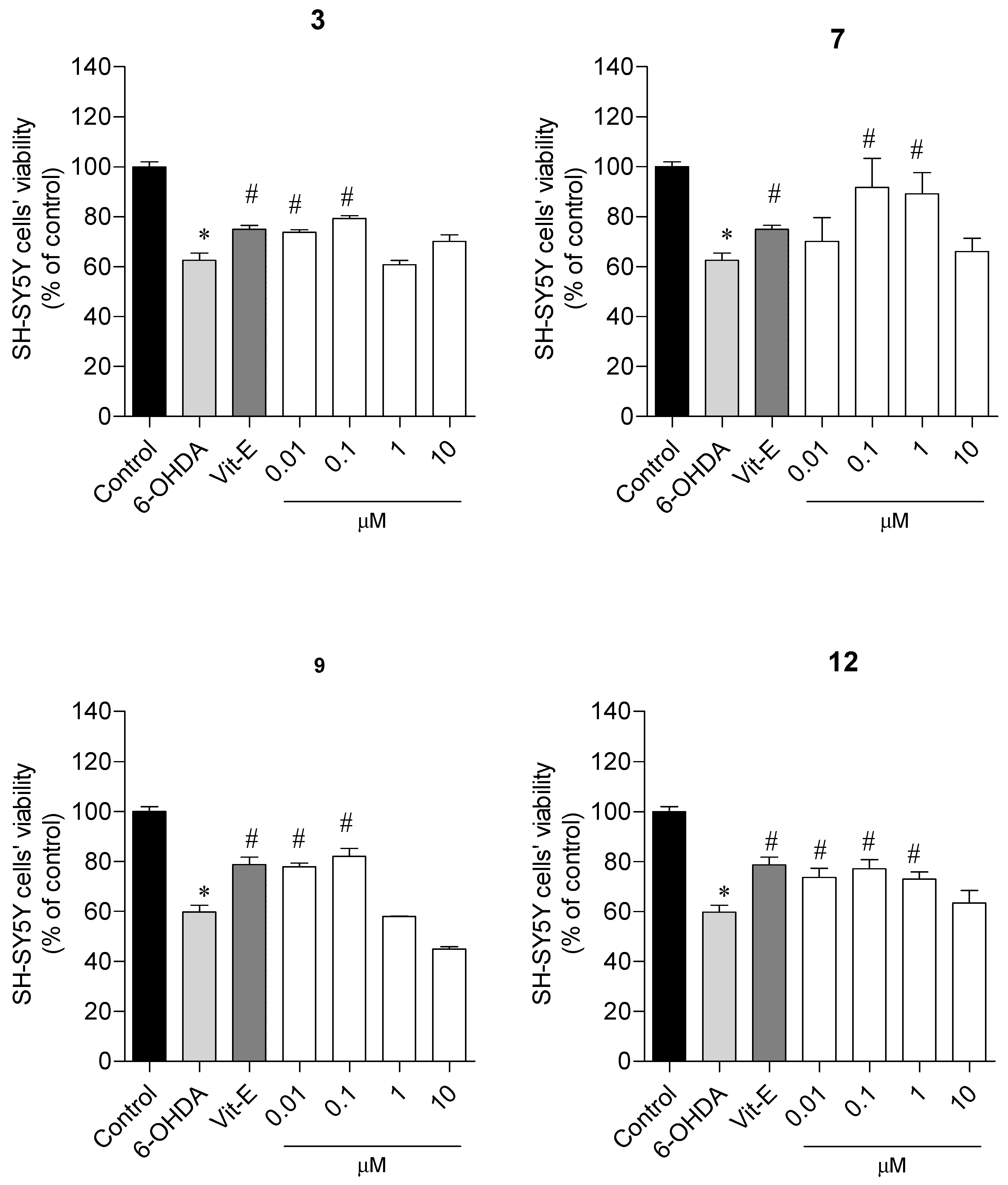
2.2.2. Neuroprotective Capacity on a Parkinson’s Disease Cellular Model
2.2.3. Cytotoxicity on Different Malignant Cell Lines
2.3. Computacional Studies
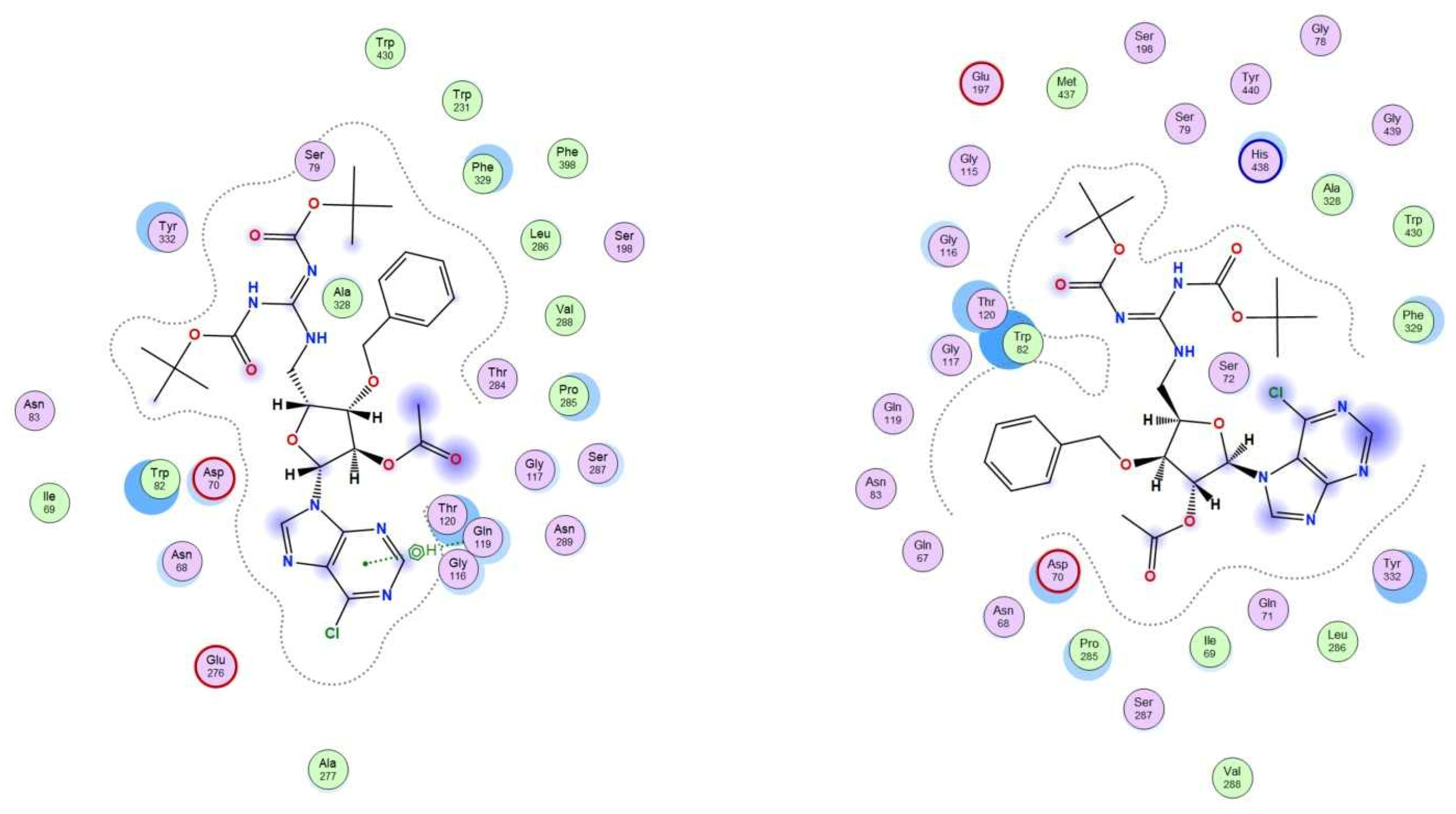
2.3.1. Molecular Docking Studies
2.3.2. In Silico log p Prediction
3. Conclusions
4. Experimental Section
4.1. Chemistry
4.1.1. General Methods
4.1.2. General Procedure for TFA-Mediated Acetonide Hydrolysis of 5-Azido 3-O-Protected-5-Deoxy-1,2-O-Isopropylidene-α-D-Xylofuranose and Further Acetylation
4.2. Biological Assays
4.2.1. Cholinesterase Inhibition Assays
Preparation of the Solutions
Kinetic Studies
4.2.2. Neuroprotective Capacity on a PD Cellular Model
Differentiation of Human SH-SY5Y Neuronal Cells
Cell Viability and Neuroprotective Effects on Differentiated SH-SY5Y Cells
4.2.3. Cytotoxicity Evaluation
Cell Culture Maintenance
Cytotoxicity Assay
4.3. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef]
- Shelton, J.; Lu, X.; Hollenbaugh, J.A.; Cho, J.H.; Amblard, F.; Schinazi, R.F. Metabolism, biochemical actions, and chemical synthesis of anticancer nucleosides, nucleotides, and base analogs. Chem. Rev. 2016, 116, 14379–14455. [Google Scholar] [CrossRef] [PubMed]
- Hruba, L.; Das, V.; Hajduch, M.; Dzubak, P. Nucleoside-based anticancer drugs: Mechanism of action and drug resistance. Biochem. Pharmacol. 2023, 215, 115741. [Google Scholar] [CrossRef] [PubMed]
- Shabestari, S.M.; Pourmadadi, M.; Abdouss, H.; Ghanbari, T.; Bazari, S.; Abdouss, M.; Rahdar, A.; Ferreira, L.F.R. Unlocking the potential of cytarabine: A comprehensive review from molecular insights to advanced nanoformulations and Co-delivery strategies for enhanced drug efficacy. J. Drug. Deliv. Sci. Technol. 2024, 102, 106346. [Google Scholar] [CrossRef]
- Ricci, F.; Tedeschi, A.; Morra, E.; Montillo, M. Fludarabine in the treatment of chronic lymphocytic leukemia: A review. Ther. Clin. Risk Manag. 2009, 5, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.; Goa, K.L. Gemcitabine. A review of its pharmacology and clinical potential in non-small cell lung cancer and pancreatic cancer. Drugs 1997, 54, 447–472. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, G.; Brisdelli, F.; Bozzi, A. AZT: An old drug with new perspectives. Curr. Clin. Pharmacol. 2008, 3, 20–37. [Google Scholar] [CrossRef]
- Keating, G.M. Sofosbuvir: A review of its use in patients with chronic hepatitis C. Drugs. 2014, 74, 1127–1146. [Google Scholar] [CrossRef]
- Godwin, P.O.; Polsonetti, B.; Caron, M.F.; Oppelt, T.F. Remdesivir for the Treatment of COVID-19: A Narrative Review. Infect Dis. Ther. 2024, 13, 1–19. [Google Scholar] [CrossRef]
- Mukherjee, T.; Mal, P.; Upadhyay, A.K.; Mohanty, S.; Nayak, N.; Singh, R.P.; Pattnaik, A.; Das, T.; Basak, S. Safety Profile of Molnupiravir with Significant Effect on COVID-19: A Review. Curr. Drug Ther. 2023, 18, 183–193. [Google Scholar] [CrossRef]
- Thomson, J.M.; Lamont, I.L. Nucleoside Analogues as Antibacterial Agents. Front. Microbiol. 2019, 10, 952. [Google Scholar] [CrossRef] [PubMed]
- Serpi, M.; Ferrari, V.; Pertusati, F. Nucleoside Derived Antibiotics to Fight Microbial Drug Resistance: New Utilities for an Established Class of Drugs? J. Med. Chem. 2016, 59, 10343–10382. [Google Scholar] [CrossRef]
- Bugg, T.D.H. Nucleoside Natural Product Antibiotics Targeting Microbial Cell Wall Biosynthesis. In Antibacterials, Topics in Medicinal Chemistry; Fisher, J., Mobashery, S., Miller, M., Eds.; Springer: Cham, Switzerland, 2017; Volume 26. [Google Scholar] [CrossRef]
- Motter, J.; Benckendorff, C.M.M.; Westarp, S.; Sunde-Brown, P.; Neubauer, P.; Kurreck, A.; Miller, G.J. Purine nucleoside antibiotics: Recent synthetic advances harnessing chemistry and biology. Nat. Prod. Rep. 2024, 41, 873–884. [Google Scholar] [CrossRef]
- Schwarz, S.; Csuk, R.; Rauter, A.P. Microwave-assisted synthesis of novel purine nucleosides as selective cholinesterase inhibitors. Org. Biomol. Chem. 2014, 12, 2446–2456. [Google Scholar] [CrossRef]
- Batista, D.; Schwarz, S.; Loesche, A.; Csuk, R.; Costa, P.J.; Oliveira, M.C.; Xavier, N.M. Synthesis of glucopyranos-6′-yl purine and pyrimidine isonucleosides as potential cholinesterase inhibitors. Access to pyrimidine-linked pseudodisaccharides through Mitsunobu reaction. Pure Appl. Chem. 2016, 88, 363–379. [Google Scholar] [CrossRef]
- Pereira, R.G.; Pereira, M.P.; Serra, S.G.; Loesche, A.; Csuk, R.; Silvestre, S.; Costa, P.J.; Oliveira, M.C.; Xavier, N.M. Furanosyl nucleoside analogs embodying triazole or theobromine units as potential lead molecules for Alzheimer’s disease. Eur. J. Org. Chem. 2018, 2018, 2667–2681. [Google Scholar] [CrossRef]
- Xavier, N.M.; de Sousa, E.C.; Pereira, M.P.; Loesche, A.; Serbian, I.; Csuk, R.; Oliveira, M.C. Synthesis and biological evaluation of structurally varied 5′-/6′-isonucleosides and theobromine-containing N-isonucleosidyl derivatives. Pharmaceuticals 2019, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Maria, C.; Rauter, A.P. Nucleoside analogues: N-glycosylation methodologies, synthesis of antiviral and antitumor drugs and potential against drug-resistant bacteria and Alzheimer’s disease. Carbohydr. Res. 2023, 532, 108889. [Google Scholar] [CrossRef] [PubMed]
- Cachatra, V.; Martins, A.; Oliveira, M.C.; Oliveira, M.C.; Gano, L.; Paulo, A.; López, Ó.; Fernández-Bolaños, J.G.; Contino, M.; Colabufo, N.A.; et al. Purine nucleosides as selective inhibitors of butyrylcholinesterase—A multidisciplinary study. Org. Biomol. Chem. 2025, 23, 3845–3859. [Google Scholar] [CrossRef]
- Tsesmetzis, N.; Paulin, C.B.J.; Rudd, S.G.; Herold, N. Nucleobase and Nucleoside Analogues: Resistance and Re-Sensitisation at the Level of Pharmacokinetics, Pharmacodynamics and Metabolism. Cancers 2018, 10, 240. [Google Scholar] [CrossRef]
- Xavier, N.M.; Goncalves-Pereira, R.; Jorda, R.; Řezníčková, E.; Kryštof, V.; Oliveira, M.C. Synthesis and antiproliferative evaluation of novel azido nucleosides and their phosphoramidate derivatives. Pure Appl. Chem. 2017, 89, 1267–1281. [Google Scholar] [CrossRef]
- Xavier, N.M.; Goncalves-Pereira, R.; Jorda, R.; Hendrychová, D.; Oliveira, M.C. Novel dodecyl-containing azido and glucuronamide-based nucleosides exhibiting anticancer potential. Pure Appl. Chem. 2019, 91, 1085–1105. [Google Scholar] [CrossRef]
- Marucci, G.; Buccioni, M.; Dal Ben, D.; Lambertucci, C.; Volpini, R.; Amenta, F. Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology 2021, 190, 108352. [Google Scholar] [CrossRef]
- Milanković, V.; Tasić, T.; Lazarević-Pašti, T. The role of acetylcholinesterase in cancer development and possible therapeutic applications. In Pathophysiological Aspects of Proteases in Cancer; Chakraborti, S., Das, S., Kim, C.-H., Eds.; Academic Press: Cambridge, MA, USA, 2025; pp. 283–307. [Google Scholar] [CrossRef]
- Pérez-Aguilar, B.; Marquardt, J.U.; Muñoz-Delgado, E.; López-Durán, R.M.; Gutiérrez-Ruiz, M.C.; Gomez-Quiroz, L.E.; Gómez-Olivares, J.L. Changes in the Acetylcholinesterase Enzymatic Activity in Tumor Development and Progression. Cancers 2023, 15, 4629. [Google Scholar] [CrossRef] [PubMed]
- Richbart, S.D.; Merritt, J.C.; Nolan, N.A.; Dasgupta, P. Acetylcholinesterase and human cancers. In Advances in Cancer Research; Tew, K.D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 152, pp. 1–66. [Google Scholar] [CrossRef]
- Xi, H.J.; Wu, R.P.; Liu, J.J.; Zhang, L.J.; Li, Z.S. Role of acetylcholinesterase in lung cancer. Thorac. Cancer 2015, 6, 390–398. [Google Scholar] [CrossRef]
- Fortuna, A.; Gonçalves-Pereira, R.; Costa, P.J.; Jorda, R.; Vojáčková, V.; Gonzalez, G.; Heise, N.V.; Csuk, R.; Oliveira, M.C.; Xavier, N.M. Synthesis and exploitation of the biological profile of novel guanidino xylofuranose derivatives. ChemMedChem 2022, 17, e202200180. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Shrestha, A.R.; Akisawa, T.; Piao, H.; Kizawa, H.; Ohmiya, Y.; Kurita, R. Development of gapmer antisense oligonucleotide with deoxyribonucleic guanidine (DNG) modifications. Nucleosides Nucleotides Nucleic Acids 2020, 39, 258–269. [Google Scholar] [CrossRef]
- Muttathukattil, A.N.; Srinivasan, S.; Halder, A.; Reddy, G. Role of Guanidinium-Carboxylate Ion Interaction in Enzyme Inhibition with Implications for Drug Design. J. Phys. Chem. B 2019, 123, 9302–9311. [Google Scholar] [CrossRef]
- Kim, S.-H.; Semenya, D.; Castagnolo, D. Antimicrobial drugs bearing guanidine moieties: A review. Eur. J. Med. Chem. 2021, 216, 113293. [Google Scholar] [CrossRef]
- Gomes, A.R.; Varela, C.L.; Pires, A.S.; Tavares-da-Silva, E.J.; Roleira, F.M.F. Synthetic and natural guanidine derivatives as antitumor and antimicrobial agents: A review. Bioorg. Chem. 2023, 138, 106600. [Google Scholar] [CrossRef]
- Lee, H.-L.; Aubé, J. Intramolecular and Intermolecular Schmidt Reactions of Alkyl Azides with Aldehydes. Tetrahedron 2007, 63, 9007–9015. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, R.; Gao, F.; Li, X. An efficient synthesis of δ-glyconolactams by intramolecular Schmidt–Boyer reaction under microwave radiation. Tetrahedron Lett. 2012, 53, 7147–7149. [Google Scholar] [CrossRef]
- Greig, N.H.; Utsuki, T.; Ingram, D.K.; Wang, Y.; Pepeu, G.; Scali, C.; Yu, Q.-S.; Mamczarz, J.; Holloway, H.W.; Giordano, T.; et al. Selective butyrylcholinesterase inhibition elevates brain acetylcholine, augments learning and lowers Alzheimer beta-amyloid peptide in rodent. Proc. Natl. Acad. Sci. USA 2005, 102, 17213–17218. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Richbart, S.D.; Merritt, J.C.; Brown, K.C.; Nolan, N.A.; Akers, A.T.; Lau, J.K.; Robateau, Z.R.; Miles, S.L.; Dasgupta, P. Acetylcholine signaling system in progression of lung cancers. Pharmacol. Ther. 2019, 194, 222–254. [Google Scholar] [CrossRef]
- Nie, M.; Chen, N.; Pang, H.; Jiang, T.; Jiang, W.; Tian, P.; Yao, L.; Chen, Y.; DeBerardinis Ralph, J.; Li, W.; et al. Targeting acetylcholine signaling modulates persistent drug tolerance in EGFR-mutant lung cancer and impedes tumor relapse. J. Clin. Investig. 2022, 132, e160152. [Google Scholar] [CrossRef]
- Obreshkova, D.; Atanasov, P.; Chaneva, M.; Ivanova, S.; Balkanski, S.; Peikova, L. Cytotoxic activity of Galantamine hydrobromide against HeLa cell line. Pharmacia 2023, 70, 619–623. [Google Scholar] [CrossRef]
- Ki, Y.S.; Park, E.Y.; Lee, H.-W.; Oh, M.S.; Cho, Y.-W.; Kwon, Y.K.; Moon, J.H.; Lee, K.-T. Donepezil, a Potent Acetylcholinesterase Inhibitor, Induces Caspase-Dependent Apoptosis in Human Promyelocytic Leukemia HL-60 Cells. Biol. Pharm. Bull. 2010, 33, 1054–1059. [Google Scholar] [CrossRef]
- Skrzypek, A.; Karpińska, M.; Juszczak, M.; Grabarska, A.; Wietrzyk, J.; Krajewska-Kułak, E.; Studziński, M.; Paszko, T.; Matysiak, J. Cholinesterases Inhibition, Anticancer and Antioxidant Activity of Novel Benzoxazole and Naphthoxazole Analogs. Molecules 2022, 27, 8511. [Google Scholar] [CrossRef]
- Holmgard, I.C.V.; González-Bakker, A.; Poeta, E.; Puerta, A.; Fernandes, M.X.; Monti, B.; Fernández-Bolaños, J.G.; Padrón, J.M.; López, O.; Lindbäck, E. Coumarin–azasugar–benzyl conjugates as non-neurotoxic dual inhibitors of butyrylcholinesterase and cancer cell growth. Org. Biomol. Chem. 2024, 22, 3425–3438. [Google Scholar] [CrossRef]
- Niazi, I.U.; Tariq, S.A.; Ali, A. Effect of doxorubicin and daunorubicin on the activity of acetylcholinesterase in acute lymphoblastic leukamia. J. Ayub. Med. Coll. Abbottabad 2011, 23, 45–47. [Google Scholar]
- Hyatt, J.L.; Tsurkan, L.; Morton, C.L.; Yoon, K.J.P.; Harel, M.; Brumshtein, B.; Silman, I.; Sussman, J.L.; Wadkins, R.M.; Potter, P.M. Inhibition of acetylcholinesterase by the anticancer prodrug CPT-11. Chem. Biol. Interact. 2005, 157, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, J.; Xiang, S.; Zhao, K.; Yu, J.; Zheng, J.; Xu, D.; Mak, S.; Hu, S.; Nirasha, S.; et al. Sunitinib, a Clinically Used Anticancer Drug, Is a Potent AChE Inhibitor and Attenuates Cognitive Impairments in Mice. ACS Chem. Neurosci. 2016, 7, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Razab, S.; Prabhu, K.; Ray, S.; Prakash, B. Serum butyrylcholinesterase and zinc in breast cancer. J. Can. Res. Ther. 2017, 13, 367–370. [Google Scholar] [CrossRef]
- Liu, J.; Tian, T.; Liu, X.; Cui, Z. BCHE as a Prognostic Biomarker in Endometrial Cancer and Its Correlation with Immunity. J. Immunol. Res. 2022, 2022, 6051092. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, G.Y.; Osman, A.A.; Mukhtar, A. Acetylcholinesterase enzyme among cancer patients a potential diagnostic and prognostic indicator a multicenter case–control study. Sci. Rep. 2024, 14, 5127. [Google Scholar] [CrossRef]
- Gu, Y.; Chow, M.J.; Kapoor, A.A.; Mei, W.; Jiang, Y.; Yan, J.; De Melo, J.; Seliman, M.; Yang, H.; Cutz, J.-C.; et al. Biphasic alteration of butyrylcholinesterase (BChE) during prostate cancer development. Transl. Oncol. 2018, 11, 1012–1022. [Google Scholar] [CrossRef]
- Guo, J.D.; Zhao, X.; Li, Y.; Li, G.R.; Liu, X.L. Damage to dopaminergic neurons by oxidative stress in Parkinson’s disease (Review). Int. J. Mol. Med. 2018, 41, 1817–1825. [Google Scholar] [CrossRef]
- Hernandez-Baltazar, D.; Zavala-Flores, L.M.; Villanueva-Olivo, A. The 6-hydroxydopamine model and parkinsonian pathophysiology: Novel findings in an older model. Neurología 2017, 32, 533–539. [Google Scholar] [CrossRef]
- Ferreira, P.S.; Nogueira, T.B.; Costa, V.M.; Branco, P.S.; Ferreira, L.M.; Fernandes, E.; Bastos, M.L.; Meisel, A.; Carvalho, F.; Capela, J.P. Neurotoxicity of “ecstasy” and its metabolites in human dopaminergic differentiated SH-SY5Y cells. Toxicol. Lett. 2013, 216, 159–170. [Google Scholar] [CrossRef]
- Lopez-Suarez, L.; Awabdh, S.A.; Coumoul, X.; Chauvet, C. The SH-SY5Y human neuroblastoma cell line, a relevant in vitro cell model for investigating neurotoxicology in human: Focus on organic pollutants. Neurotoxicology 2022, 92, 131–155. [Google Scholar] [CrossRef]
- Arnott, J.A.; Planey, S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Tetko, I.V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V.A.; Radchenko, E.V.; Zefirov, N.S.; Makarenko, A.S.; et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aid. Mol. Des. 2005, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- VCCLAB. Virtual Computational Chemistry Laboratory. 2005. Available online: https://vcclab.org (accessed on 10 March 2025).
- Ekpenyong, O.; Gao, X.; Ma, J.; Cooper, C.; Nguyen, L.; Olaleye, O.A.; Liang, D.; Xie, H. Pre-Clinical Pharmacokinetics, Tissue Distribution and Physicochemical Studies of CLBQ14, a Novel Methionine Aminopeptidase Inhibitor for the Treatment of Infectious Diseases. Drug Des. Devel. Ther. 2020, 14, 1263–1277. [Google Scholar] [CrossRef]
- Handbook of Preformulation: Chemical, Biological, and Botanical Drugs; Niazi, S.K., Ed.; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Pajouhesh, H.; Lenz, G.R. Medicinal chemical properties of successful central nervous system drugs. NeuroRx 2005, 2, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Björkroth, J.P.; Leo, A. Hydrophobicity and Central Nervous System Agents: On the Principle of Minimal Hydrophobicity in Drug Design. J. Pharm. Sci. 1987, 76, 663–687. [Google Scholar] [CrossRef]
- Silva, J.; Alves, C.; Pinteus, S.; Susano, P.; Simões, M.; Guedes, M.; Martins, A.; Rehfeldt, S.; Gaspar, H.; Goettert, M.; et al. Disclosing the potential of eleganolone for Parkinson’s disease therapeutics: Neuroprotective and anti-inflammatory activities. Pharmacol. Res. 2021, 168, 105589. [Google Scholar] [CrossRef]
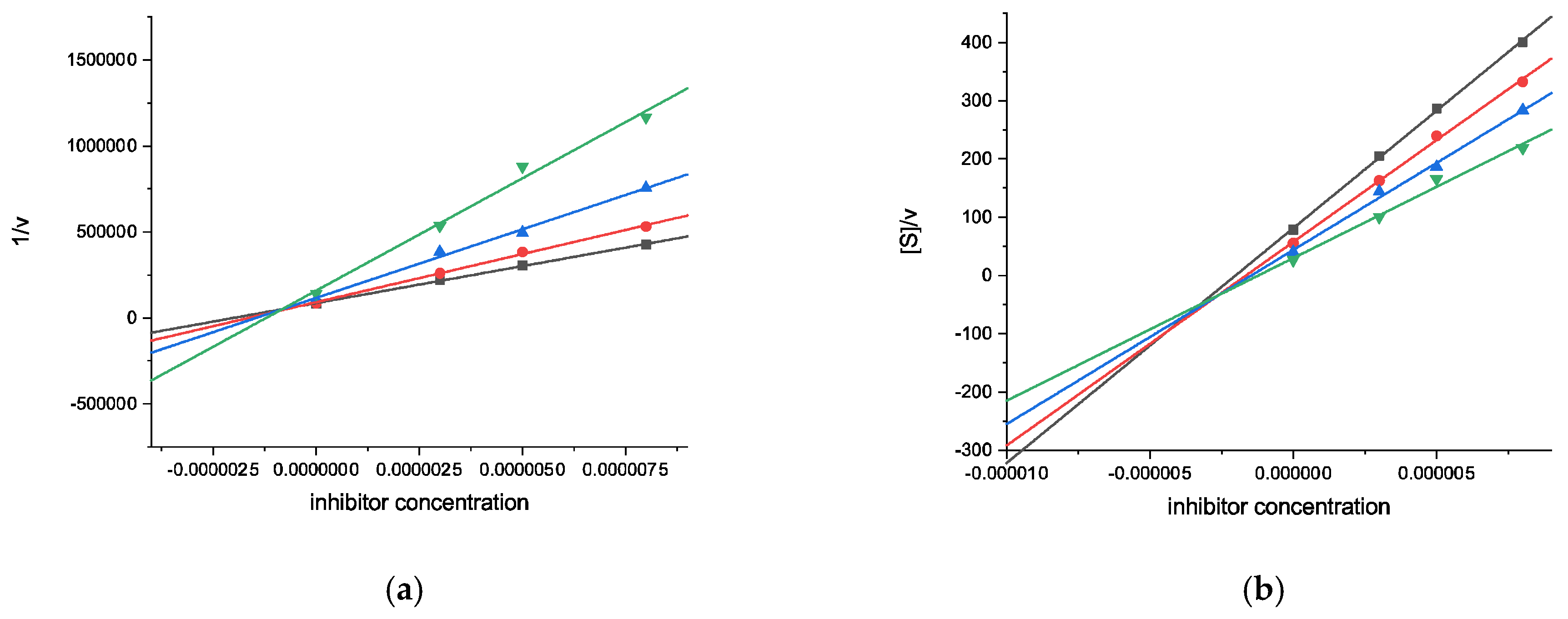
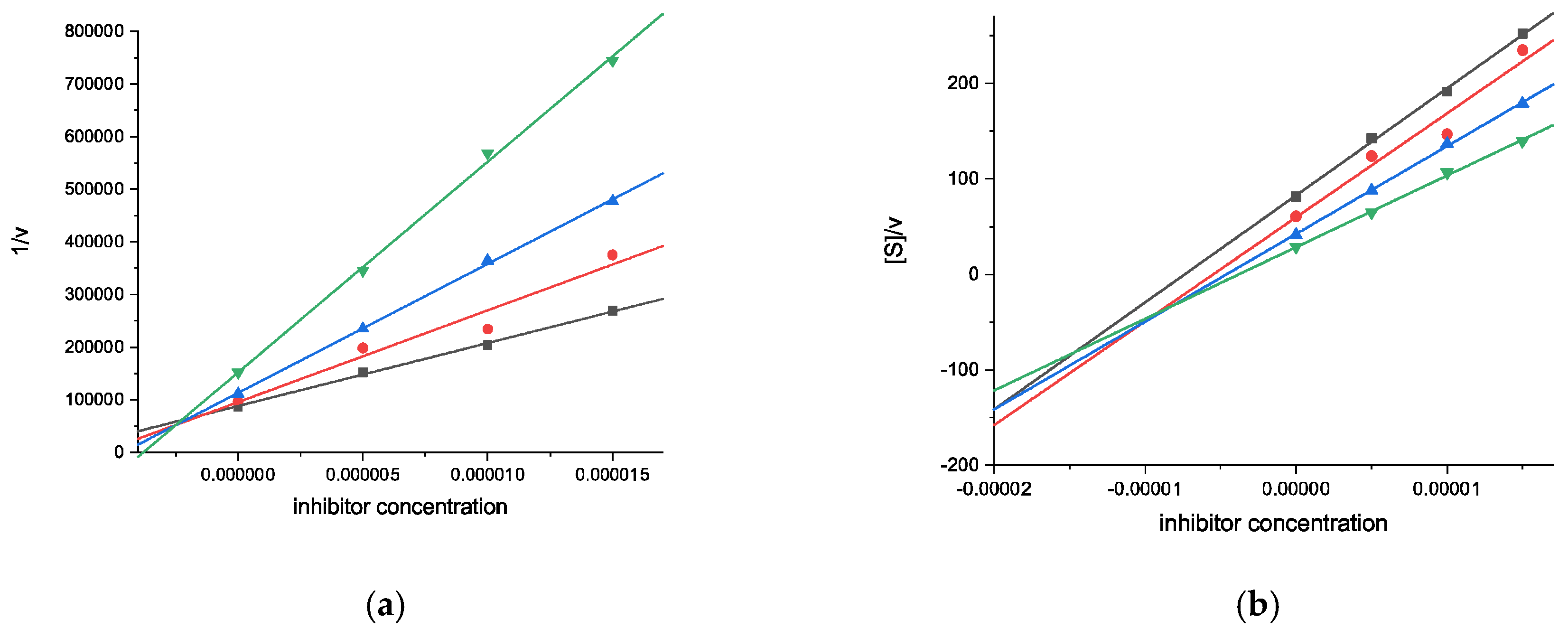

| Compound | AChE | BChE | ||
|---|---|---|---|---|
| Inhibition (%) a | Ki (μM) (Type of Inhibition) | Inhibition (%) a | Ki (μM) [Ki′ (μM)] (Type of Inhibition) | |
| 10 | 39.14 ± 0.9 | n.d. | 83.82 ± 1.3 | 0.89 ± 0.08 [2.96 ± 0.39] (mixed-type) |
| 11 | 34.64 ± 1.4 | n.d. | 65.23 ± 0.9 | 2.34 ± 0.20 [10.6 ± 1.4] (mixed-type) |
| Galantamine | 88.19 ± 0.5 | 0.54 ± 0.01 (competitive) | 52.00 ± 0.5 | 9.37 ± 0.67 (competitive) |
| Compound | IC50 (μM) | ||||
|---|---|---|---|---|---|
| HCT-15 | MCF-7 | DU-145 | SH-SY5Y | FL83B | |
| δ-Xylonolactam | |||||
| 3 | >100 | >100 | >100 | >100 | n.d. |
| 5′-Azido nucleosides | |||||
| 7 | 72.73 | >100 | >100 | >100 | 22.74 |
| 8 | >100 | >100 | >100 | >100 | n.d. |
| 9 | >100 | >100 | >100 | >100 | n.d. |
| 5′-Guanidino nucleosides | |||||
| 10 | >100 | >100 | 27.63 | >100 | 73.74 |
| 11 | 64.07 | 43.67 | 24.48 | 12.14 | 24.08 |
| 12 | 76.02 | >100 | >100 | >100 | >100 |
| 5-Fluorouracil | 155.5 | n.d. | n.d. | n.d. | >100 |
| Cisplatin | n.d. | n.d. | >100 | 13.92 | 25.71 |
| Tamoxifen | n.d. | 27.19 | n.d. | n.d. | 19.51 |
| Compound | c log p a |
|---|---|
| δ-Xylonolactam | |
| 3 | 1.33 |
| 5′-Azido nucleosides | |
| 7 | 2.83 |
| 8 | 2.84 |
| 9 | 2.18 |
| 5′-Guanidino nucleosides | |
| 10 | 4.50 |
| 11 | 4.51 |
| 12 | 2.83 |
| Reference Drugs | |
| 5-Fluorouracil | −0.58 |
| Cisplatin | 0.43 |
| Tamoxifen | 5.93 |
| Galantamine | 1.39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szilagyi, J.; Moreira, T.; Nunes, R.S.; Silva, J.; Alves, C.; Martins, A.; Alvariño, R.; Heise, N.V.; Csuk, R.; Xavier, N.M. 5′-Guanidino Xylofuranosyl Nucleosides as Novel Types of 5′-Functionalized Nucleosides with Biological Potential. Pharmaceuticals 2025, 18, 734. https://doi.org/10.3390/ph18050734
Szilagyi J, Moreira T, Nunes RS, Silva J, Alves C, Martins A, Alvariño R, Heise NV, Csuk R, Xavier NM. 5′-Guanidino Xylofuranosyl Nucleosides as Novel Types of 5′-Functionalized Nucleosides with Biological Potential. Pharmaceuticals. 2025; 18(5):734. https://doi.org/10.3390/ph18050734
Chicago/Turabian StyleSzilagyi, Jennifer, Tânia Moreira, Rafael Santana Nunes, Joana Silva, Celso Alves, Alice Martins, Rebeca Alvariño, Niels V. Heise, René Csuk, and Nuno M. Xavier. 2025. "5′-Guanidino Xylofuranosyl Nucleosides as Novel Types of 5′-Functionalized Nucleosides with Biological Potential" Pharmaceuticals 18, no. 5: 734. https://doi.org/10.3390/ph18050734
APA StyleSzilagyi, J., Moreira, T., Nunes, R. S., Silva, J., Alves, C., Martins, A., Alvariño, R., Heise, N. V., Csuk, R., & Xavier, N. M. (2025). 5′-Guanidino Xylofuranosyl Nucleosides as Novel Types of 5′-Functionalized Nucleosides with Biological Potential. Pharmaceuticals, 18(5), 734. https://doi.org/10.3390/ph18050734












