DPYD Genotyping, Fluoropyrimidine Dosage and Toxicity: An Umbrella Review of Systematic Reviews
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
- P: Oncologic patients with DPYD gene variants and undergoing treatment with fluoropyrimidines.
- I: Registry of severe adverse events (grades 3–5) related to fluoropyrimidine treatment in patients with DPYD gene variants.
- C: Patients without DPYD gene variants and undergoing treatment with fluoropyrimidines or without comparator.
- O: Variables related to toxicity and treatment: severe adverse events, DPYD gene variants detected, fluoropyrimidine dosage, and treatment regimen.
- S: Systematic review with/without meta-analysis.
2.2. Information Sources and Search Strategy
2.3. Selection and Data Collection Process
2.4. Quality Assessment
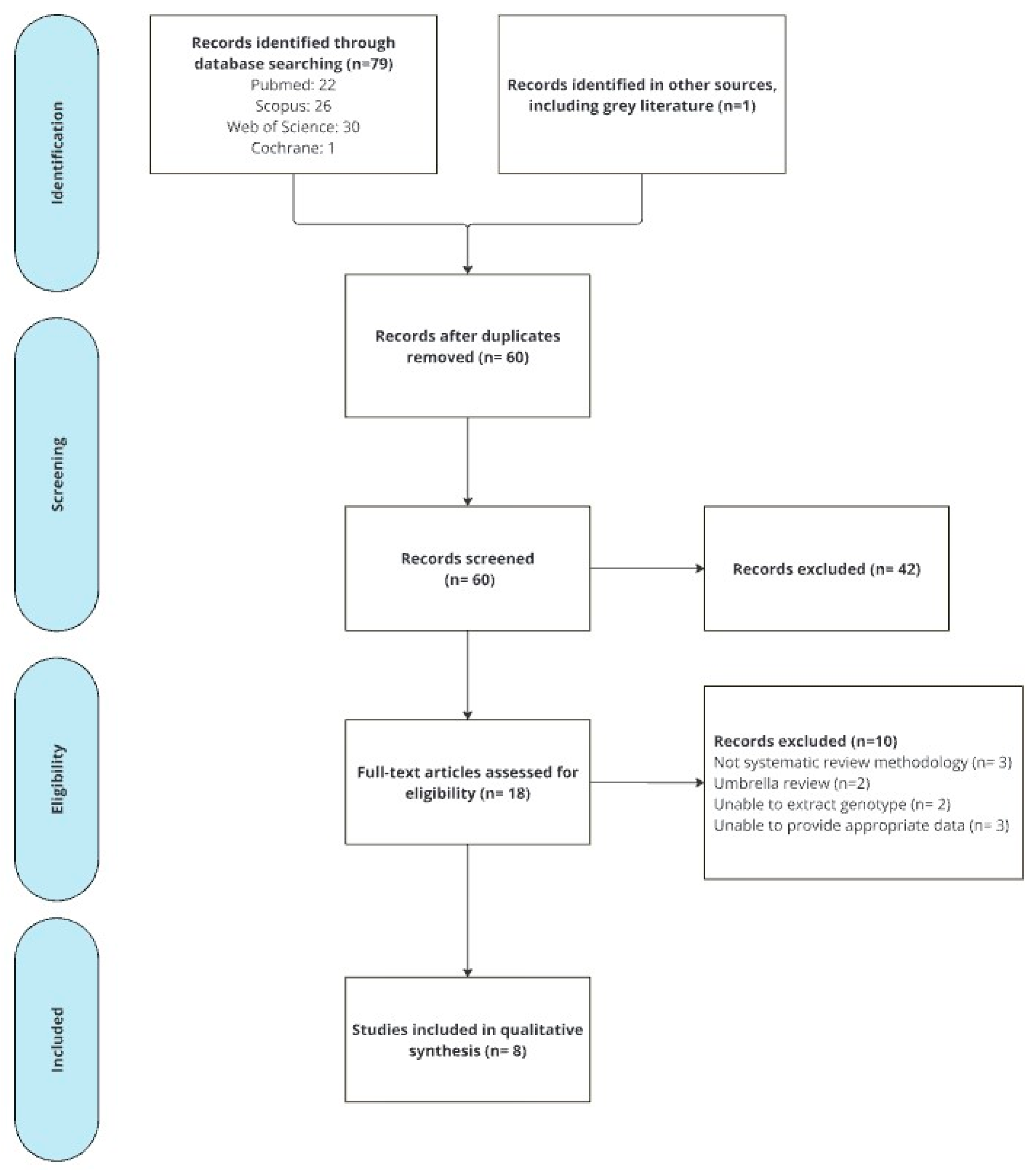
3. Results
3.1. Quality of the Systematic Reviews
3.2. Characteristics of Included Systematic Reviews
3.3. Fluoropyrimidine-Induced Toxicity DPYD
3.3.1. Germline Variations in the DPYD Gene and Fluoropyrimidine Toxicity (Table 3 and Table 4)
- Carriers of c.1905+1G>A (rs3918290) [also known as DPYD*2A] variant
- Carriers of c.1679T>G (rs55886062) [also known as DPYD*13] variant
- Carriers of c.2846A>T (rs67376798) variant
- Carriers of c.1236G>A (rs75017182) [also known as HapB3] variant
- Carriers of other rare variants vs. WT patients
- o
- Carriers of c.1601G>A (rs1801158) [DPYD*4] variant
- o
- Carriers of c.2194G>A (rs1801160) [DPYD*6] variant
- o
- Carriers of c.496A>G (rs2297595) variant
- o
- Carriers of c.1627A>G (rs1801159) [DPYD*5] variant
| Review | DPYD*2A | DPYD*13 | c.2846A>T | HapB3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | |
| Meulendijks et al. (2015) [15] | RR 2.9 (1.8–4.6) | p < 0.0001 | I2 = 73% p = 0.0013 | RR 4.4 (2.1–9.3) | p < 0.0001 | I2 = 85% p < 0.0001 | RR 3.0 (2.2–4.1) | p < 0.0001 | I2 = 80% p < 0.0001 | RR 1.6 (1.3–2.0) | p < 0.0001 | I2 = 23%, p = 0.26 |
| Terrazzino et al. (2013) [16] | OR 5.4 (2.8–10.5) | p < 0.001 | I2 = 13% p = 0.3 | - | OR 8.2 (2.7–25.3) | p < 0.001 | I2 = 47% p = 0.076 | - | ||||
| Rosmarin et al. (2014) [10] | Capecitabine: OR 3.0 (0.8–11.7) | p = 0.1 | I2 = 0% p > 0.05 | - | - | - | ||||||
| Infusional 5-FU: OR 6.7 (1.7–27.1) | p = 0.0075 | NR | ||||||||||
| Bolus 5-FU: OR 0.7 (0.5–1.0) | p = 0.062 | NR | ||||||||||
| Review | DPYD*4 | DPYD*6 | c.496A>G | DPYD*5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | Risk (95% CI) | p | Heterogeneity | |
| Meulendijks et al. (2015) [15] | RR 1.5 (0.9–2.7) | p = 0.15 | I2 = 91%, p < 0.0001 | |||||||||
| Rosmarin et al. (2014) [10] | - | - | Infusional 5-FU: OR 0.5 (0.1–3.1) | p = 0.48 | NR | Infusional 5-FU: OR 0.68 (0.3–1.8) | p = 0.43 | NR | ||||
| Bolus 5-FU: OR 1.3 (0.8–2.0) | p = 0.35 | NR | Bolus 5-FU: OR 0.7 (0.5–1.0) | p = 0.062 | NR | |||||||
| Kim et al. (2022) [17] | - | OR 1.7 (1.4–2.1) | p < 0.001 | I2 = 30% p = 0.21 | - | - | ||||||
3.3.2. Fluoropyrimidine Pharmacogenetics-Guided Dosing and Toxicity
3.4. Combined Genotyping and Phenotyping Approaches and Toxicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campbell, J.; Bateman, E.; Peters, M.; JM, B.; Keefe, D.; Stephenson, M. Fluoropyrimidine and platinum toxicity pharmacogenetics: An umbrella review of systematic reviews and meta-analyses Fluoropyrimidine. Pharmacogenomics 2016, 17, 435–451. [Google Scholar] [CrossRef] [PubMed]
- Agencia Española de Medicamentos y Productos Sanitarios (AEMPS). Technical Data Sheet: Capecitabina. 2016. Available online: https://cima.aemps.es/cima/dochtml/ft/76109/FichaTecnica_76109.html (accessed on 1 December 2024).
- Agencia Española de Medicamentos y Productos Sanitarios (AEMPS). Nota de Seguridad: Fluorouracilo, Capecitabina, Tegafur y Flucitosina en Pacientes con Déficit de Dihidropirimidina Deshidrogenasa. 2020. Available online: https://www.aemps.gob.es/informa/fluorouracilo-capecitabina-tegafur-y-flucitosina-en-pacientes-con-deficit-de-dihidropirimidina-deshidrogenasa/ (accessed on 1 December 2024).
- Amstutz, U.; Henricks, L.M.; Offer, S.M.; Barbarino, J.; Schellens, J.H.; Swen, J.J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef]
- García-Alfonso, P.; Saiz-Rodríguez, M.; Mondéjar, R.; Salazar, J.; Páez, D.; Borobia, A.M.; Safont, M.J.; García-García, I.; Colomer, R.; García-González, X.; et al. Consensus of experts from the Spanish Pharmacogenetics and Pharmacogenomics Society and the Spanish Society of Medical Oncology for the genotyping of DPYD in cancer patients who are candidates for treatment with fluoropyrimidines. Clin. Transl. Oncol. 2021, 24, 483–494. [Google Scholar] [CrossRef]
- Agencia Española de Medicamentos y Productos Sanitarios (AEMPS). Technical Data Sheet: Fluorouracil Accord 50 mg/mL for Injection or Infusion EFG. 2010. Available online: https://cima.aemps.es/cima/dochtml/ft/71868/FichaTecnica_71868.html (accessed on 1 December 2024).
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.O.; Pérez, O.M.; Mauriz, R.R.; Casamartina, E.F.; Martínez, S.F.; Estela, A.C. DPYD genotyping and 5-fluoropyrimidine toxicity: An overview of systematic reviews protocol. Farm. Hosp. 2023, 48, T79–T82. [Google Scholar] [CrossRef]
- Shea, B.J.; Reeves, B.C.; Wells, G.; Thuku, M.; Hamel, C.; Moran, J.; Moher, D.; Tugwell, P.; Welch, V.; Kristjansson, E.; et al. AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ 2017, 358, j4008. [Google Scholar] [CrossRef]
- Rosmarin, D.; Palles, C.; Church, D.; Domingo, E.; Jones, A.; Johnstone, E.; Wang, H.; Love, S.; Julier, P.; Scudder, C.; et al. Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: Investigation in the QUASAR2 study, systematic review, and meta-analysis. J. Clin. Oncol. 2014, 32, 1031–1039. [Google Scholar] [CrossRef]
- Conti, V.; De Bellis, E.; Manzo, V.; Sabbatino, F.; Iannello, F.; Piaz, F.D.; Izzo, V.; Charlier, B.; Stefanelli, B.; Torsiello, M.; et al. A genotyping/phenotyping approach with careful clinical monitoring to manage the fluoropyrimidines-based therapy: Clinical cases and systematic review of the literature. J. Pers. Med. 2020, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Ontario Health (Quality). DPYD Genotyping in Patients Who Have Planned Cancer Treatment With Fluoropyrimidines: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2021, 21, 1–186. [Google Scholar]
- Glewis, S.; Alexander, M.; Khabib, M.N.H.; Brennan, A.; Lazarakis, S.; Martin, J.; Tie, J.; Lingaratnam, S.; Michael, M. A systematic review and meta-analysis of toxicity and treatment outcomes with pharmacogenetic-guided dosing compared to standard of care BSA-based fluoropyrimidine dosing. Br. J. Cancer 2022, 127, 126–136. [Google Scholar] [CrossRef]
- Paulsen, N.H.; Vojdeman, F.; Andersen, S.E.; Bergmann, T.K.; Ewertz, M.; Plomgaard, P.; Hansen, M.R.; Esbech, P.S.; Pfeiffer, P.; Qvortrup, C.; et al. DPYD genotyping and dihydropyrimidine dehydrogenase (DPD) phenotyping in clinical oncology. A clinically focused minireview. Basic Clin. Pharmacol. Toxicol. 2022, 131, 325–346. [Google Scholar] [CrossRef]
- Meulendijks, D.; Henricks, L.M.; Sonke, G.S.; Deenen, M.J.; Froehlich, T.K.; Amstutz, U.; Largiadèr, C.R.; Jennings, B.A.; Marinaki, A.M.; Sanderson, J.D.; et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: A systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015, 16, 1639–1650. [Google Scholar] [CrossRef]
- Terrazzino, S.; Cargnin, S.; Del Re, M.; Danesi, R.; Canonico, P.; Genazzani, A. DPYD IVS14+1G>A and 2846A>T genotyping for the prediction of severe fluoropyrimidine-related toxicity: A meta-analysis. Pharmacogenomics 2013, 14, 1255–1272. [Google Scholar] [CrossRef]
- Kim, W.; Cho, Y.A.; Kim, D.C.; Lee, K.E. Elevated Risk of Fluoropyrimidine-Associated Toxicity in European Patients with DPYD Genetic Polymorphism: A Systematic Review and Meta-Analysis. J. Pers. Med. 2022, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health and Human Services. Common Terminology Criteria for Adverse Events (CTCAE) v.5.0. 2017. Available online: https://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm (accessed on 1 December 2024).
- Wigle, T.J.; Povitz, B.L.; Medwid, S.; Teft, W.A.; Legan, R.M.; Lenehan, J.; Nevison, S.; Panuganty, V.; Keller, D.; Mailloux, J.; et al. Impact of pretreatment dihydropyrimidine dehydrogenase genotype-guided fluoropyrimidine dosing on chemotherapy associated adverse events. Clin. Transl. Sci. 2021, 14, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Lunenburg, C.A.; Henricks, L.M.; Dreussi, E.; Peters, F.P.; Fiocco, M.; Meulendijks, D.; Toffoli, G.; Guchelaar, H.-J.; Swen, J.J.; Cecchin, E.; et al. Standard fluoropyrimidine dosages in chemoradiation therapy result in an increased risk of severe toxicity in DPYD variant allele carriers. Eur. J. Cancer 2018, 104, 210–218. [Google Scholar] [CrossRef]
- Henricks, L.; Lunenburg, C.A.T.C.; de Man, F.; Meulendijks, D.; Frederix, G.W.J.; Kienhuis, E.; Creemers, G.-J.; Baars, A.; Dezentjé, V.O.; Imholz, A.L.T.; et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: A prospective safety analysis. Lancet Oncol. 2018, 19, 1459–1467. [Google Scholar] [CrossRef] [PubMed]
- de With, M.; Knikman, J.; de Man, F.M.; Lunenburg, C.A.T.C.; Henricks, L.M.; van Kuilenburg, A.B.P.; Maring, J.G.; van Staveren, M.C.; de Vries, N.; Rosing, H.; et al. Dihydropyrimidine Dehydrogenase Phenotyping Using Pretreatment Uracil: A Note of Caution Based on a Large Prospective Clinical Study. Clin. Pharmacol. Ther. 2022, 112, 62–68. [Google Scholar] [CrossRef]
- Etienne-Grimaldi, M.-C.; Boyer, J.-C.; Beroud, C.; Mbatchi, L.; van Kuilenburg, A.; Bobin-Dubigeon, C.; Thomas, F.; Chatelut, E.; Merlin, J.-L.; Pinguet, F.; et al. New advances in DPYD genotype and risk of severe toxicity under capecitabine. PLoS ONE 2017, 12, e0175998. [Google Scholar] [CrossRef]
- Capitain, O.; Seegers, V.; Metges, J.-P.; Faroux, R.; Stampfli, C.; Ferec, M.; Budnik, T.M.; Senellart, H.; Rossi, V.; Blouin, N.; et al. Comparison of 4 Screening Methods for Detecting Fluoropyrimidine Toxicity Risk: Identification of the Most Effective, Cost-Efficient Method to Save Lives. Dose-Response 2020, 18, 1559325820951367. [Google Scholar] [CrossRef]
- Van Kuilenburg, A.B.; Haasjes, J.; Richel, D.J.; Zoetekouw, L.; Van Lenthe, H.; De Abreu, R.A.; Maring, J.G.; Vreken, P.; Van Gennip, A.H. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: Identification of new mutations in the DPD gene. Clin. Cancer Res. 2000, 6, 4705–4712. [Google Scholar] [PubMed]
- Boisdron-Celle, M.; Metges, J.P.; Capitain, O.; Adenis, A.; Raoul, J.L.; Lecomte, T.; Lam, Y.H.; Faroux, R.; Masliah, C.; Poirier, A.L.; et al. A multicenter phase II study of personalized FOLFIRI-cetuximab for safe dose intensification. Semin. Oncol. 2017, 44, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Lunenburg, C.A.T.C.; van der Wouden, C.H.; Nijenhuis, M.; Rhenen, M.H.C.-V.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2019, 28, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Quaranta, S.; Thomas, F. Pharmacogenetics of anti-cancer drugs: State of the art and implementation—Recommendations of the French National Network of Pharmacogenetics. Therapies 2017, 72, 205–215. [Google Scholar] [CrossRef]
- Sociedad Española de Farmacogénetica y Farmacogenómica (SEFF). Estrategia de Implementación de la Sociedad Española de Farmacogenética y Farmacogenómica: Recomendaciones de los Grupos de Trabajo para el gen DPYD y la Prescripción de Fluoropirimidinas (Fluorouracilo, Capecitabina y Tegafur). 2024. Available online: https://www.seff.es/fluoropirimidinas/# (accessed on 1 December 2024).
| General Variables |
|---|
| Author and year of publication |
| Aim of systematic review |
| Number of primary studies |
| Design of primary studies |
| Number of participants/Caucasians |
| Tumor type |
| Funding statement |
| Competing interest statement |
| Specific Variables |
| Severe adverse events (overall toxicity, gastrointestinal toxicity, hematological toxicity) |
| DPYD gene variants detected |
| Fluoropyrimidine guided dosing |
| Chemotherapeutic regimen |
| Author/Year | Aim | Primary Studies (n) | Primary Studies Design | Participants (n)/Caucasians (%) | Tumor Type | DPYD Genotype | Chemotherapeutic Regimens | Toxicity Criteria |
|---|---|---|---|---|---|---|---|---|
| Meulendijks et al. (2015) [15] | To assess the clinical relevance of DPYD*13, HapB3, and DPYD*4 as predictors of severe FIT. | 8 | Cohort studies and RCTs | 7365/85–100% | Colorectal, Gastric/gastroesophageal, hepatobiliary and pancreatic, breast and others. | DPYD*13: 5 studies (5616 patients) HapB3: 6 studies (4261 patients) DPYD*2A: 7 studies (5737 patients) c.2846A>T: 8 studies (7318 patients). DPYD*4: 5 studies (3900 patients) | Capecitabine: 2 studies 5-FU: 2 studies capecitabine and 5-FU regimens: 4 studies | NCI–CTC |
| Terrazzino et al. (2013) [16] | To quantify the impact of the DPYD*2A and 2846A>T variants on the risk of FIT, to determine sensitivity, and specificity testing for DPYD variants. | 15 | Prospective and retrospective studies | 4573/NR (mostly Caucasians) | Colorectal: predominant. Others: GI, head and neck and breast cancers. | DPYD*2A: 13 studies (3499 patients) c.2846A>T: 7 studies (2308 patients). | Capecitabine: 2 studies Tefagur-uracil: 1 study. In the remaining studies: 5-FU or capecitabine. | NCI–CTC: 13 studies WHO criteria: 2 studies |
| Kim et al. (2022) [17] | To investigate the association between DPYD*6 and FIT. | 6 | RCTs and cohort studies. | 6119/100% | Colorectal, breast, biliary, pancreatic, orofacial, esophageal, and gastric cancers. | DPYD*6 | Fluoropyrimidine-based regimens: 4 studies FOLFOX4: 1 study Capecitabine: 1 study | NCI–CTC |
| Conti et al. (2020) [11] | To analyze the variability of responses to fluoropyrimidine-based chemotherapy by DPYD genotyping combined with phenotyping methods and/or clinical monitoring. | 22 | Observational and RCTs. | 18,018/NR | NR | DPYD*13, HapB3, DPYD*2A, c.2846A>T and DPYD*6 | 5-FU or capecitabine. | NCI–CTC |
| Rosmarin et al. (2014) [10] | To investigate the associations between fluoropyrimidine- polymorphisms and FIT. | 16 | RCTs and cohort studies. | 4855/100% | NR | DPYD*9A, c.496A>G, HapB3, DPYD*4, DPYD*5, DPYD*2A, DPYD*6, and c.2846A>T | Bolus and infusional 5-FU or capecitabine. | NCI–CTC |
| Glewis et al. (2022) [13] | To evaluate treatment outcomes between PGD versus non-PGD and within PGD | 17 | Cohort studies and case-control study | 11,515/NR (mostly Caucasians) | Lower GI); upper GI; breast cancer; head and neck cancers); and gynecological cancers | Studies with majority testing for The bold formatting is not necessary, so we will remove it. 4o (15 studies) | 5-FU: 14 studies. Capecitabine: 11 studies | NCI–CTC |
| Ontario Health (2021) [12] | To evaluate the risk of severe FIT in carriers of the DPYD variants compared to patients with wild-type DPYD. | 29 | Observational studies, | 18,490/67–100% | Colorectal: predominant Other: Breast, GI, esophageal, and head and neck. | Four DPYD variants: 4 studies DPYD*2A: 20 studies c.2846A>T: 16 studies DPYD*13: 13 studies | 5-FU: 11 studies. Capecitabine: 4 studies. In the remaining studies: 12–91% of patients with 5-FU. | NCI–CTC |
| Paulsen et al. (2022) [14] | To present the current evidence for DPD testing in routine oncological practice. | 12 | Both prospective and retrospective studies | 10,696/NR | NR | HapB3 (322 patients) DPYD*2A (172 patients) D949V (18 patients) DPYD*13 (18 patients) | 5-FU, capecitabine or tegafur. | NCI–CTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otero-Torres, S.; Rodríguez-Mauriz, R.; Fort-Casamartina, E.; Clopés-Estela, A.; Soler-Rotllant, F.; Fontanals-Martínez, S.; Montero-Pérez, O. DPYD Genotyping, Fluoropyrimidine Dosage and Toxicity: An Umbrella Review of Systematic Reviews. Pharmaceuticals 2025, 18, 727. https://doi.org/10.3390/ph18050727
Otero-Torres S, Rodríguez-Mauriz R, Fort-Casamartina E, Clopés-Estela A, Soler-Rotllant F, Fontanals-Martínez S, Montero-Pérez O. DPYD Genotyping, Fluoropyrimidine Dosage and Toxicity: An Umbrella Review of Systematic Reviews. Pharmaceuticals. 2025; 18(5):727. https://doi.org/10.3390/ph18050727
Chicago/Turabian StyleOtero-Torres, Sara, Rosa Rodríguez-Mauriz, Eduard Fort-Casamartina, Ana Clopés-Estela, Francesc Soler-Rotllant, Sandra Fontanals-Martínez, and Olalla Montero-Pérez. 2025. "DPYD Genotyping, Fluoropyrimidine Dosage and Toxicity: An Umbrella Review of Systematic Reviews" Pharmaceuticals 18, no. 5: 727. https://doi.org/10.3390/ph18050727
APA StyleOtero-Torres, S., Rodríguez-Mauriz, R., Fort-Casamartina, E., Clopés-Estela, A., Soler-Rotllant, F., Fontanals-Martínez, S., & Montero-Pérez, O. (2025). DPYD Genotyping, Fluoropyrimidine Dosage and Toxicity: An Umbrella Review of Systematic Reviews. Pharmaceuticals, 18(5), 727. https://doi.org/10.3390/ph18050727







