Exosomes in Ovarian Cancer: Towards Precision Oncology
Abstract
1. Ovarian Cancer
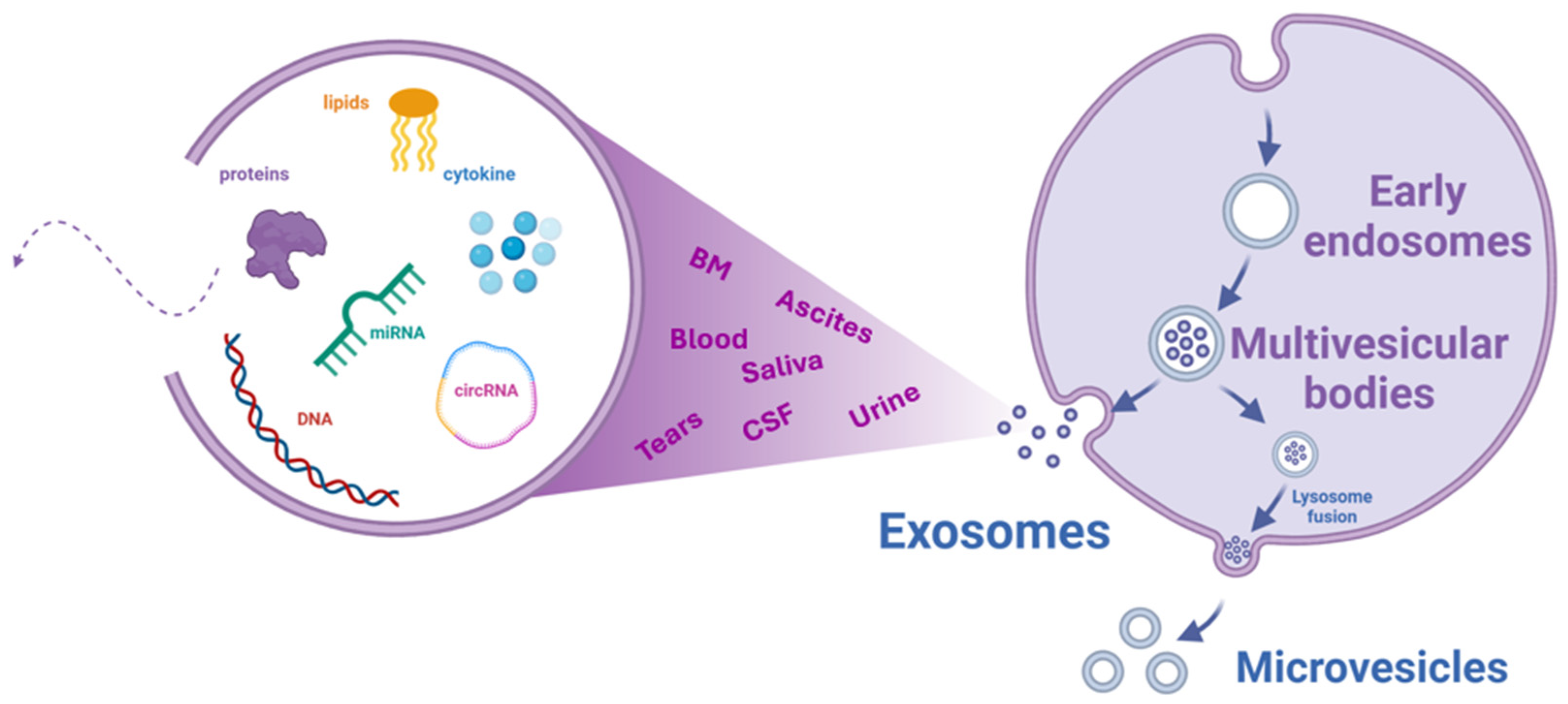
2. Exosomes
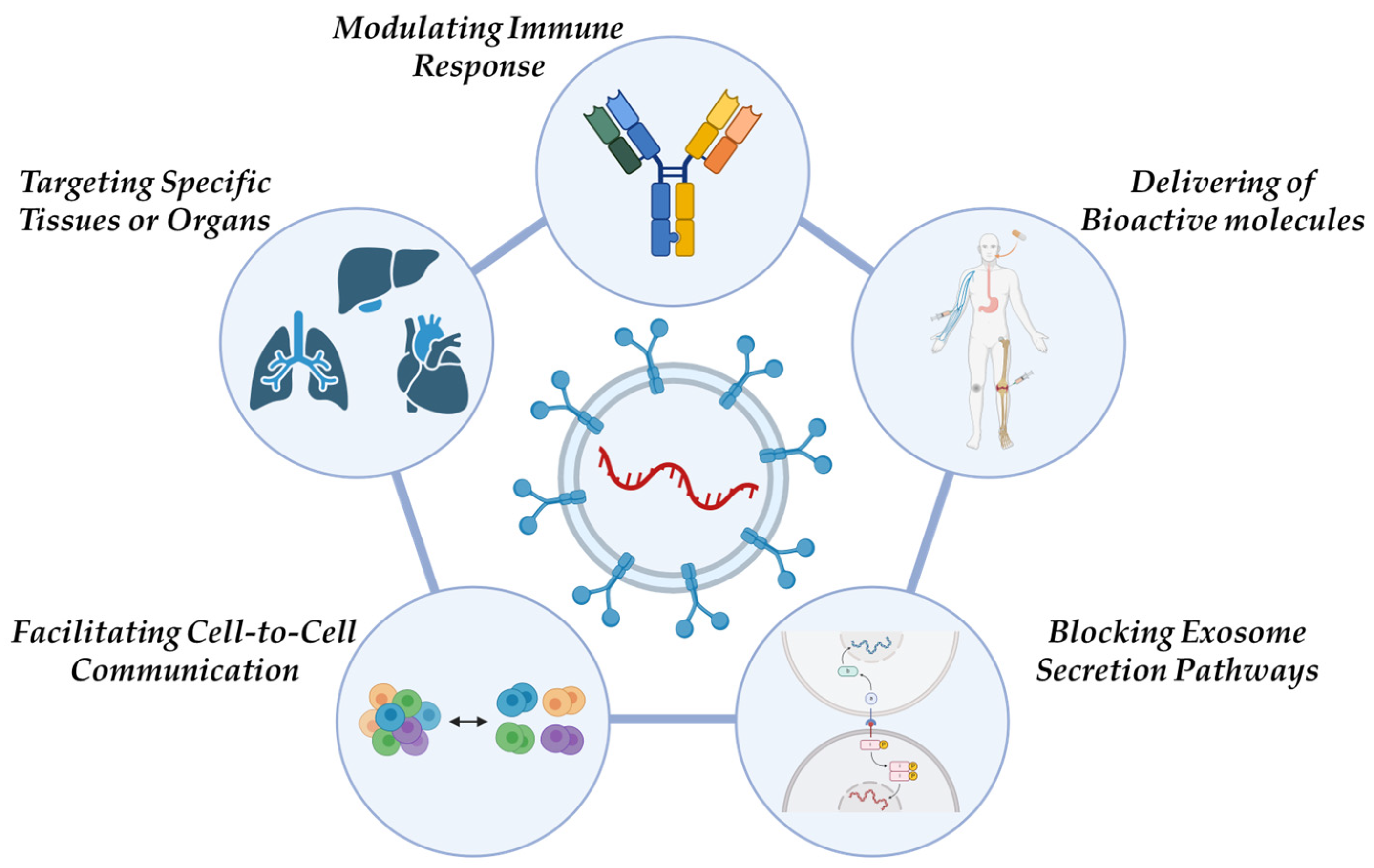
- Blocking Exosome Secretion Pathways: Cancer cells exploit exosome secretion to affect the tumor microenvironment, enhance tumor growth, drive invasion, and develop resistance to treatment. A potential therapeutic approach involves either blocking the mechanisms responsible for exosome dissemination within malignant cells or removing EVs from the blood circulatory system.
- Delivering Bioactive molecules: Exosomes, due to their lipid membrane composition, efficiently facilitate cellular uptake of bioactive molecules. This makes them ideal carriers for anticancer drugs, miRNAs, and siRNAs. Additionally, they can transport tumor antigens, nanobodies, apoptosis-inducing proteins, proteasomes, mutated or deficient anti-apoptotic proteins, tissue-specific peptides, transferrins, and lactoferrins to tumor cells.
- Targeting Specific Tissues or Organs: Due to their intrinsic cell tropism, exosomes can selectively target specific tissues or organs, offering a promising approach for precision medicine.
- Modulating Immune Responses: Exosomes play a role in immune system regulation, which has potential applications in the development of cancer vaccines aimed at slowing or preventing tumor progression.
- Facilitating Cell-to-Cell Communication: Exosomal miRNAs originating from tumor cells contribute to intercellular communication, influencing various cellular processes related to cancer progression.
2.1. Exosome Isolation and Purification Strategies
2.1.1. Sucrose Density Gradient Ultracentrifugation
2.1.2. Polymer-Based Precipitation
2.1.3. Ultrafiltration
2.1.4. Size-Exclusion Liquid Chromatography (SEC)
2.1.5. Immunomagnetic Beads and Nanoparticle Tracking Analysis (NTA)
2.1.6. Immunoaffinity System
2.1.7. Microfluidic Systems
- -
- Microfluidic immunoaffinity separation, which mimics traditional bead-based approaches by embedding antibodies within microfluidic channels to selectively capture exosomes. While highly specific, this method requires traditional RNA extraction, unlike bead-based systems that allow direct miRNA retrieval. Kabe et al. introduced a microfluidic system incorporating immunomagnetic beads for enhanced exosome isolation from plasma, demonstrating high diagnostic efficacy in ovarian cancer through multiplexed tumor marker detection [34];
- -
- Filtration using microfluidic chips with nanomembranes or nanowires offers a straightforward approach for exosome isolation, relying on size exclusion. This technique includes the use of porous polymer monoliths in a microfluidic system with DC electrophoresis to prevent clogging. Moreover, the development of the Exosome Total Isolation Chip (ExoTIC), a modular platform designed to isolate exosomes from urine, plasma, and cell culture media, achieves yields up to 1000-fold higher than ultracentrifugation within three hours. While it offers significant efficiency, the complexity of nanowire-based structures may present challenges in clinical implementation [35,36];
- -
- Deterministic Lateral Displacement (DLD) classifies particles based on size using micro-structured columns that guide smaller particles through predefined trajectories while redirecting larger ones. Nano-DLD arrays have successfully separated particles ranging from 20 to 110 nm with high precision. However, traditional DLD systems require high hydrodynamic resistance, necessitating pressures exceeding 200 kPa. Wu et al. optimized this method by incorporating electro-osmotic flow, significantly reducing the required pressure while maintaining continuous and precise separation [37];
- -
- Acoustic-based exosome isolation is a label-free approach that utilizes ultrasound standing waves to achieve high selectivity and biocompatibility. Acoustic forces act on particles based on their volume, allowing for precise size-based separation. Recent developments have integrated acoustics with microfluidics into a two-module system: the first step removes large blood components, while the second isolates exosomes, achieving 82.4% recovery and 98.4% purity. This method provides a streamlined, high-purity exosome isolation process directly from whole blood with minimal processing [38];
- -
- Electrokinetic approaches, such as di-electrophoresis (DEP) and electrophoresis (EP), leverage alternating current (AC) voltage to separate exosomes. DEP exploits differences in particle polarization in a non-uniform electric field, while EP directs charged particles based on their electrophoretic mobility. An alternating current electrokinetic microarray chip has been developed to isolate exosomes from plasma in 30 min by attracting them to high-field regions while larger particles migrate to low-field areas [39];
- -
- Viscoelastic microfluidic separation is a label-free technique that exploits differential elastic lift forces among particles of varying sizes. Due to their small size, exosomes experience minimal viscoelastic effects. Wang et al. developed a microfluidic system with two inlets and three outlets, where polyoxyethylene polymers are used to enhance the viscoelasticity of exosomes. In this system, larger particles exit through the central outlet, while exosomes are collected at the side outlets. This approach achieves high separation purity (>90%) and recovery (80%) without the need for complex procedures [40].
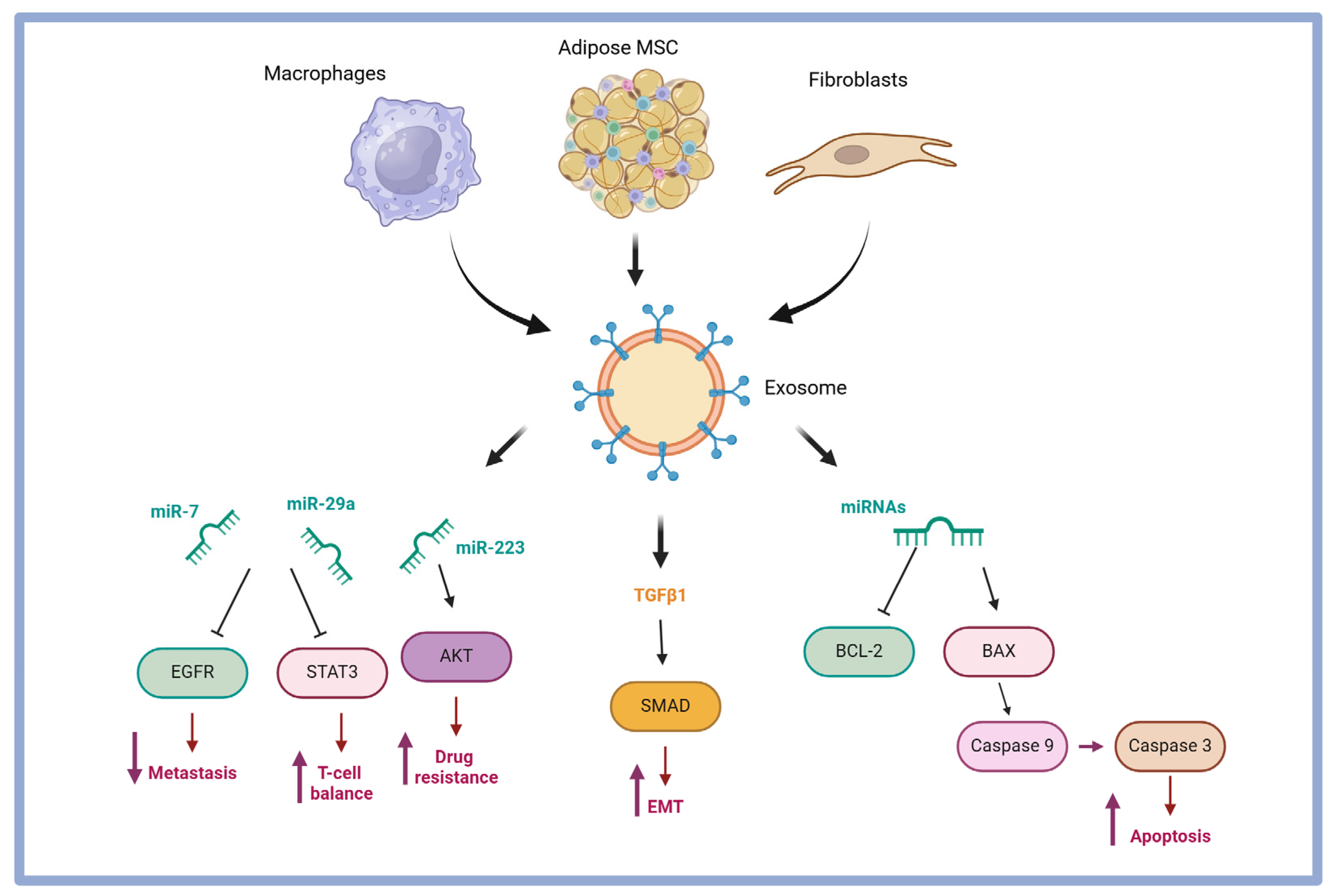
3. Exosomes Isolated from Ovarian Cancer: Content and Role in Tumor Malignancy
4. Ovarian Cancer-Specific Exosome Cargo
4.1. Lipidomic Content of OC-Exosomes
4.2. Proteomic Content of Exosomes
4.3. RNAs Transported by Exosomes
4.3.1. Long Non-Coding RNAs (lncRNAs)
4.3.2. microRNAs (miRNAs)
4.3.3. circularRNAs (circRNAs)
| CircRNA | Regulation in OC | Prognosis |
|---|---|---|
| circ-ITCH | down | poor OS [60] |
| circ-ABCB10 | up | poor OS [61] |
| circ-1656 | up | poor OS [62] |
| circHIPK3 | up | poor OS/poor disease-free survival (DFS) [63] |
| circLARP4 | down | poor OS/poor disease-free survival (DFS) [64] |
5. Exosomes as Drug Delivery Systems for Treating Ovarian Cancer
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ovarian Cancer Statistics at a Glance. Available online: https://ocrahope.org/for-patients/gynecologic-cancers/ovarian-cancer/ovarian-cancer-statistics/ (accessed on 6 February 2025).
- Can Ovarian Cancer Be Found Early? Available online: https://www.cancer.org/cancer/types/ovarian-cancer/detection-diagnosis-staging/detection.html (accessed on 6 February 2025).
- National Cancer Institute. Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Ovarian Cancer. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 6 February 2025).
- Cancer Facts & Figures 2018—Special Section: Ovarian Cancer. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-special-section-ovarian-cancer-2018.pdf (accessed on 30 November 2023).
- Scilimati, A.; Ferorelli, S.; Iaselli, M.; Miciaccia, M.; Pati, M.; Fortuna, C.G.; Aleem, A.M.; Marnett, L.J.; Perrone, M.G. Targeting COX-1 by Mofezolac-Based Fluorescent Probes for Ovarian Cancer Detection. Eur. J. Med. Chem. 2019, 179, 16–25. [Google Scholar] [CrossRef]
- Perrone, M.G.; Vitale, P.; Miciaccia, M.; Ferorelli, S.; Centonze, A.; Solidoro, R.; Munzone, C.; Bonaccorso, C.; Fortuna, C.G.; Kleinmanns, K.; et al. Fluorochrome Selection for Imaging Intraoperative Ovarian Cancer Probes. Pharmaceuticals 2022, 15, 668. [Google Scholar] [CrossRef] [PubMed]
- Solidoro, R.; Centonze, A.; Miciaccia, M.; Baldelli, O.M.; Armenise, D.; Ferorelli, S.; Perrone, M.G.; Scilimati, A. Fluorescent imaging probes for in vivo ovarian cancer targeted detection and surgery. Med. Res. Rev. 2024, 44, 1800–1866. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; De Grassi, A.; Ferorelli, S.; Perrone, G.; Zalfa, F.; Scilimati, A. Genome-Wide Identification and Validation of Gene Expression Biomarkers in the Diagnosis of Ovarian Serous Cystadenocarcinoma. Cancers 2022, 14, 3764. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; Luisi, O.; De Grassi, A.; Ferorelli, S.; Cormio, G.; Scilimati, A. Translational Theragnosis of Ovarian Cancer: Where Do We Stand? Curr. Med. Chem. 2020, 27, 5675–5715. [Google Scholar] [CrossRef]
- Zhang, R.; Siu, M.K.Y.; Ngan, H.Y.S.; Chan, K.K.L. Molecular Biomarkers for the Early Detection of Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 12041. [Google Scholar] [CrossRef]
- Barr, C.E.; Funston, G.; Jeevan, D.; Sundar, S.; Mounce, L.T.A.; Crosbie, E.J. The Performance of HE4 Alone and in Combination with CA125 for the Detection of Ovarian Cancer in an Enriched Primary Care Population. Cancers 2022, 14, 2124. [Google Scholar] [CrossRef]
- Stephens, A.N.; Hobbs, S.J.; Kang, S.-W.; Bilandzic, M.; Rainczuk, A.; Oehler, M.K.; Jobling, T.W.; Plebanski, M.; Allman, R. A Novel Predictive Multi-Marker Test for the Pre-Surgical Identification of Ovarian Cancer. Cancers 2023, 15, 5267. [Google Scholar] [CrossRef]
- Anastasi, E.; Farina, A.; Granato, T.; Colaiacovo, F.; Pucci, B.; Tartaglione, S.; Angeloni, A. Recent Insight about HE4 Role in Ovarian Cancer Oncogenesis. Int. J. Mol. Sci. 2023, 24, 10479. [Google Scholar] [CrossRef]
- Samborski, A.; Miller, M.C.; Blackman, A.; MacLaughlan-David, S.; Jackson, A.; Lambert-Messerlian, G.; Rowswell-Turner, R.; Moore, R.G. HE4 and CA125 Serum Biomarker Monitoring in Women with Epithelial Ovarian Cancer. Tumour Biol. 2022, 44, 205–213. [Google Scholar] [CrossRef]
- Dzhugashvili, E.; Tamkovich, S. Exosomal Cargo in Ovarian Cancer Dissemination. Curr. Issues Mol. Biol. 2023, 45, 9851–9867. [Google Scholar] [CrossRef] [PubMed]
- Kok, V.C.; Yu, C.C. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef]
- Hu, J.; Zhu, J.; Chai, J.; Zhao, Y.; Luan, J.; Wang, Y. Application of Exosomes as Nanocarriers in Cancer Therapy. J. Mater. Chem. B 2023, 11, 10595–10612. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.B.; Collie, S.P.; Parent, C.A. The Ins-and-Outs of Exosome Biogenesis, Secretion, and Internalization. Trends Cell Biol. 2024, 34, 90–108. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, S.; Zhang, K.; Qing, Y.; Xu, T. A Comprehensive Overview of Exosomes in Ovarian Cancer: Emerging Biomarkers and Therapeutic Strategies. J. Ovarian Res. 2017, 10, 73. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, J.; Li, X.; Wang, X.; Lin, Y.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 2018, 435, 80–91, Corrigendum in Cancer Lett. 2023, 568, 216292. [Google Scholar] [CrossRef]
- Vaksman, O.; Tropé, C.; Davidson, B.; Reich, R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 2014, 35, 2113–2120. [Google Scholar] [CrossRef]
- Cai, J.; Gong, L.; Li, G.; Guo, J.; Yi, X.; Wang, Z. Exosomes in ovarian cancer ascites promote epithelial-mesenchymal transition of ovarian cancer cells by delivery of miR-6780b-5p. Cell Death Dis. 2021, 12, 210. [Google Scholar] [CrossRef]
- Ma, X.; Peng, L.; Zhu, X.; Chu, T.; Yang, C.; Zhou, B.; Sun, X.; Gao, T.; Zhang, M.; Chen, P.; et al. Isolation, Identification, and Challenges of Extracellular Vesicles: Emerging Players in Clinical Applications. Apoptosis 2024, 30, 422–445. [Google Scholar] [CrossRef]
- Kluszczyńska, K.; Pęczek, Ł.; Różański, A.; Czernek, L.; Duechler, M. U6/miR-211 Expression Ratio as a Purity Parameter for HEK293 Cell-Derived Exosomes. Acta Biochim. Pol. 2022, 69, 409–415. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-Scale Production, Isolation, Drug Loading Efficiency, and Biodistribution and Uptake. J. Control. Release 2022, 347, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, Opportunity, and Perspective on Exosome Isolation—Efforts for Efficient Exosome-Based Theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-‘t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Tiwari, S.; Kumar, V.; Randhawa, S.; Verma, S.K. Preparation and Characterization of Extracellular Vesicles. Am. J. Reprod. Immunol. 2021, 85, e13367. [Google Scholar] [CrossRef]
- Giannopoulou, L.; Zavridou, M.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid biopsy in ovarian cancer: The potential of circulating miRNAs and exosomes. Transl. Res. 2019, 205, 77–91. [Google Scholar] [CrossRef]
- Yousif, G.; Qadri, S.; Parray, A.; Akhthar, N.; Shuaib, A.; Haik, Y. Exosomes Derived Neuronal Markers: Immunoaffinity Isolation and Characterization. Neuromolecular Med. 2022, 24, 339–351. [Google Scholar] [CrossRef]
- Lai, J.J.; Chau, Z.L.; Chen, S.Y.; Hill, J.J.; Korpany, K.V.; Liang, N.W.; Lin, L.H.; Lin, Y.H.; Liu, J.K.; Liu, Y.C.; et al. . Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222. [Google Scholar] [CrossRef]
- Kabe, Y.; Suematsu, M.; Sakamoto, S.; Hirai, M.; Koike, I.; Hishiki, T.; Matsuda, A.; Hasegawa, Y.; Tsujita, K.; Ono, M.; et al. Development of a highly sensitive device for counting the number of disease-specific exosomes in human sera. Clin. Chem. 2018, 64, 1463–1473. [Google Scholar] [CrossRef]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef] [PubMed]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids down to 20 nm. Nat. Nanotechnol. 2016, 11, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of Extracellular Vesicles from Blood Plasma Using Electrophoretic Migration through Porous Membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Z.; Tayebi, M.; Ai, Y. Submicron Particle Focusing and Exosome Sorting by Wavy Microchannel Structures within Viscoelastic Fluids. Anal. Chem. 2019, 91, 4577–4584. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Yang, Y.; Jacobs, B.; Chen, S.; Iwuchukwu, I.; Gaines, K.J.; Chen, Y.; Simman, R.; Lv, G.; et al. The Novel Methods for Analysis of Exosomes Released from Endothelial Cells and Endothelial Progenitor Cells. Stem Cells Int. 2016, 2016, 2639728. [Google Scholar] [CrossRef]
- Kholafazad Kordasht, H.; Hasanzadeh, M. Biomedical analysis of exosomes using biosensing methods: Recent progress. Anal. Methods 2020, 12, 2795–2811. [Google Scholar] [CrossRef]
- Regimbeau, M.; Abrey, J.; Vautrot, V.; Causse, S.; Gobbo, J.; Garrido, C. Heat shock proteins and exosomes in cancer theranostics. Semin. Cancer Biol. 2022, 86, 46–57. [Google Scholar] [CrossRef]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labelled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Tayebi, M.; Zhou, Y.; Tripathi, P.; Chandramohanadas, R.; Ai, Y. Exosome Purification and Analysis Using a Facile Microfluidic Hydrodynamic Trapping Device. Anal. Chem. 2020, 92, 10733–10742. [Google Scholar] [CrossRef]
- Le, M.N.; Fan, Z.H. Exosome Isolation Using Nanostructures and Microfluidic Devices. Biomed. Mater. 2021, 16, 022005. [Google Scholar] [CrossRef] [PubMed]
- Kotrbová, A.; Štěpka, K.; Maška, M.; Pálenik, J.J.; Ilkovics, L.; Klemová, D.; Kravec, M.; Hubatka, F.; Dave, Z.; Hampl, A.; et al. TEM ExosomeAnalyzer: A computer-assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. J. Extracell. Vesicles 2019, 8, 1560808. [Google Scholar] [CrossRef] [PubMed]
- Herzog, M.; Verdenik, I.; Černe, K.; Kobal, B. Extracellular Vesicle Characteristics in Local Fluid and Plasma Measured by Nanoparticle Tracking Analysis Can Help Differentiate High-Grade Serous Carcinoma from Benign Ovarian Pathology. Diagnostics 2024, 14, 2235. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal Lipid Composition and the Role of Ether Lipids and Phosphoinositides in Exosome Biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, K.; Qing, Y.; Li, D.; Cui, M.; Jin, P.; Xu, T. Proteomic and Lipidomic Analysis of Exosomes Derived from Ovarian Cancer Cells and Ovarian Surface Epithelial Cells. J. Ovarian Res. 2020, 13, 9. [Google Scholar] [CrossRef]
- Song, W.J.; Dong, Y.; Luo, C.; Chen, Y.Y. P38MAPK family isoform p38α and activating transcription factor 2 are associated with the malignant phenotypes and poor prognosis of patients with ovarian adenocarcinoma. Pathol. Res. Pract. 2017, 213, 1282–1288. [Google Scholar] [CrossRef]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial Ovarian Cancer-Secreted Exosomal miR-222-3p Induces Polarization of Tumor-Associated Macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef]
- Khayati, S.; Dehnavi, S.; Sadeghi, M.; Tavakol Afshari, J.; Esmaeili, S.A.; Mohammadi, M. The Potential Role of miRNA in Regulating Macrophage Polarization. Heliyon 2023, 9, e21615. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, X.; Ying, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wu, Y.; Wang, X. Suppression of Endothelial Cell Migration by Tumor-Associated Macrophage-Derived Exosomes Is Reversed by Epithelial Ovarian Cancer Exosomal lncRNA. Cancer Cell Int. 2017, 17, 62. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Mu, J.; Yang, D.; Gu, X.; Zhang, J. Exosomal miR-21-5p Contributes to Ovarian Cancer Progression by Regulating CDK6. Hum. Cell 2021, 34, 1185–1196. [Google Scholar] [CrossRef]
- Yoshida, K.; Yokoi, A.; Kato, T.; Ochiya, T.; Yamamoto, Y. The Clinical Impact of Intra- and Extracellular miRNAs in Ovarian Cancer. Cancer Sci. 2020, 111, 3435–3444. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.J.; Cheng, H.F. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 322–328. [Google Scholar]
- Lu, S.; Liu, W.; Shi, H.; Zhou, H. Exosomal miR-34b inhibits proliferation and the epithelial-mesenchymal transition by targeting Notch2 in ovarian cancer. Oncol. Lett. 2020, 20, 2721–2728. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Samsami, M. Emerging role of circular RNAs in the pathogenesis of ovarian cancer. Cancer Cell Int. 2022, 22, 172. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Xu, X.; Yang, Q.; Liang, L.; Qiao, S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020, 20, 336. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chen, Y.; Ye, X.; Xia, X. Circular RNA ABCB10 promotes cell proliferation and invasion, but inhibits apoptosis via regulating the microRNA-1271 mediated Capn4/Wnt/β-catenin signaling pathway in epithelial ovarian cancer. Mol. Med. Rep. 2021, 23, 387. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Liu, Y.; Wang, M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high-grade serous ovarian cancer. Biosci. Trends 2019, 13, 204–211. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Zhang, L.Y.; Wang, L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3713–3718. [Google Scholar]
- Zou, T.; Wang, P.L.; Gao, Y.; Liang, W.T. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7178–7182. [Google Scholar]
- Qin, T.; Chen, F.; Zhu, J.; Ding, Y.; Zhang, Q. Advances in Exosomal microRNAs and Proteins in Ovarian Cancer Diagnosis, Prognosis, and Treatment. Curr. Mol. Med. 2023, 23, 509–520. [Google Scholar] [CrossRef]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.E.; Leonard, J.N. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles 2016, 5, 31027. [Google Scholar] [CrossRef] [PubMed]
- System Biosciences. XPack™ Exosome Protein Engineering. Available online: https://systembio.com/shop/xpack-mscv-xp-mcs-ef1%CE%B1-puro-cloning-lentivector (accessed on 4 February 2025).
- Yim, N.; Ryu, S.W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.H.; et al. Exosome Engineering for Efficient Intracellular Delivery of Soluble Proteins Using Optically Reversible Protein–Protein Interaction Module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef] [PubMed]
- Perrone, M.G.; Malerba, P.; Uddin, M.J.; Vitale, P.; Panella, A.; Crews, B.C.; Daniel, C.K.; Ghebreselasie, K.; Nickels, M.; Tantawy, M.N.; et al. PET Radiotracer [18F]-P6 Selectively Targeting COX-1 as a Novel Biomarker in Ovarian Cancer: Preliminary Investigation. Eur. J. Med. Chem. 2014, 80, 562–568. [Google Scholar] [CrossRef]
- Brescia, V.; Lovero, R.; Fontana, A.; Di Serio, F.; Colella, M.; Carbone, V.; Giliberti, M.; Perrone, M.G.; Scilimati, A.; Palmirotta, R. Analytical Interference of Burosumab Therapy on Intact Fibroblast Growth Factor 23 (iFGF23) Measurements Using an Immunoassay: Preliminary Evaluation. J. Immunoass. Immunochem. 2025, 46, 89–105. [Google Scholar] [CrossRef]
- Dellino, M.; Cascardi, E.; Leoni, C.; Fortunato, F.; Fusco, A.; Tinelli, F.; Cazzato, G.; Scacco, S.; Gnoni, A.; Scilimati, A.; et al. Effects of Oral Supplementation with Myo-Inositol and D-Chiro-Inositol on Ovarian Functions in Female Long-Term Survivors of Lymphoma: Results from a Prospective Case–Control Analysis. J. Pers. Med. 2022, 12, 1536. [Google Scholar] [CrossRef]
- Vitale, P.; Perna, F.M.; Perrone, M.G.; Scilimati, A. Screening on the use of Kluyveromyces marxianus CBS 6556 growing cells as enantioselective biocatalysts for ketone reductions. Tetrahedron Asymmetry 2011, 22, 1985–1993. [Google Scholar] [CrossRef]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial Ovarian Cancer: Evolution of Management in the Era of Precision Medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef]

| Methods | Advantages | Disadvantages | |
|---|---|---|---|
| Sucrose density gradient ultracentrifugation | Efficient isolation of exosomes of different densities, most commonly used, easy approach | Labor and time-consuming, special equipment required (ultracentrifuge), low efficiency | |
| Polymeric precipitation | Small sample, simple one-step method | Formation of protein aggregates | |
| Ultrafiltration | High exosome purity, faster, easy to handle compared to ultracentrifugation | Deformation/breaking up of vesicles (splat factor), filtration rate may be affected by pore size and sample concentration | |
| Size-exclusion liquid chromatography (SEC) | High exosome purity, efficacy to remove debris and contaminants | Labor and time consuming, sample contamination with lipoproteins, formation of protein aggregates, membrane damage or disruption during isolation may impact exosome function and properties | |
| Immunomagnetic beads | Efficient isolation of exosomes, potential for downstream analysis | Low levels of the target protein | |
| Nanoparticle tracking analysis | Ideal for small particles | Accuracy depends on exosome content | |
| Immunoaffinity system | Fast, easy to use, no specialized equipment | Exosome separation with targeted protein only | |
| Microfluidic systems | Filtration | Small volumes of biological samples, reduced isolation time | Requires staff competent in the microfluidic platform |
| Deterministic Lateral Displacement (DLD) | |||
| Acoustic-wave-based device | |||
| Electrical-field-based device | |||
| Viscoelastic-flow based |
| miRNA | Function | Mechanism | Clinical Implications |
|---|---|---|---|
| miR-222-3p | Angiogenesis, lymphangiogenesis | Promotes blood and lymphatic vessel formation under hypoxic conditions | Enhances OC progression [52] |
| miR-940, miR-21-3p, miR-125b-5p, miR-181d-5p | Macrophage polarization | Drive macrophages toward a tumor-supportive M2 phenotype, enhancing cancer cell proliferation and migration | Create a tumor-promoting microenvironment [53] |
| miR-146b-5p | Endothelial cell migration | Modulates TRAF6, regulating inflammation, immune response, and apoptosis | Influences tumor progression by affecting cell migration [53] |
| miR-21 | Apoptosis regulation | Suppresses PDCD4, reducing programmed cell death | Associated with poor prognosis and tumor progression [55] |
| miR-100, miR-200b, miR-320 | Prognostic markers | Increased levels in patients with advanced OC (Stage III–IV) | Correlated with reduced survival and advanced disease stage [56] |
| miR-30a-5p | Early diagnosis | More abundant in early-stage OC (Stage I–II) compared to advanced stages (Stage III–IV) | Potential biomarker for early OC detection [56] |
| miR-1290 | Early diagnosis | Highly expressed in serum, especially in high-grade serous ovarian cancer (HGSOC) | Possible tool for early OC detection [57] |
| miR-34b | Early diagnosis | Inhibitory effect on cell proliferation and epithelial–mesenchymal transition (EMT) | Potential biomarker for early OC detection [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrone, M.G.; Filieri, S.; Azzariti, A.; Armenise, D.; Baldelli, O.M.; Liturri, A.; Sardanelli, A.M.; Ferorelli, S.; Miciaccia, M.; Scilimati, A. Exosomes in Ovarian Cancer: Towards Precision Oncology. Pharmaceuticals 2025, 18, 371. https://doi.org/10.3390/ph18030371
Perrone MG, Filieri S, Azzariti A, Armenise D, Baldelli OM, Liturri A, Sardanelli AM, Ferorelli S, Miciaccia M, Scilimati A. Exosomes in Ovarian Cancer: Towards Precision Oncology. Pharmaceuticals. 2025; 18(3):371. https://doi.org/10.3390/ph18030371
Chicago/Turabian StylePerrone, Maria Grazia, Silvana Filieri, Amalia Azzariti, Domenico Armenise, Olga Maria Baldelli, Anselma Liturri, Anna Maria Sardanelli, Savina Ferorelli, Morena Miciaccia, and Antonio Scilimati. 2025. "Exosomes in Ovarian Cancer: Towards Precision Oncology" Pharmaceuticals 18, no. 3: 371. https://doi.org/10.3390/ph18030371
APA StylePerrone, M. G., Filieri, S., Azzariti, A., Armenise, D., Baldelli, O. M., Liturri, A., Sardanelli, A. M., Ferorelli, S., Miciaccia, M., & Scilimati, A. (2025). Exosomes in Ovarian Cancer: Towards Precision Oncology. Pharmaceuticals, 18(3), 371. https://doi.org/10.3390/ph18030371











