Modelling the Full-Length Inactive PKC-δ Structure to Explore Regulatory Accessibility and Selective Targeting Opportunities
Abstract
1. Introduction
2. Results
2.1. Consensus Structural Model of the Kinase
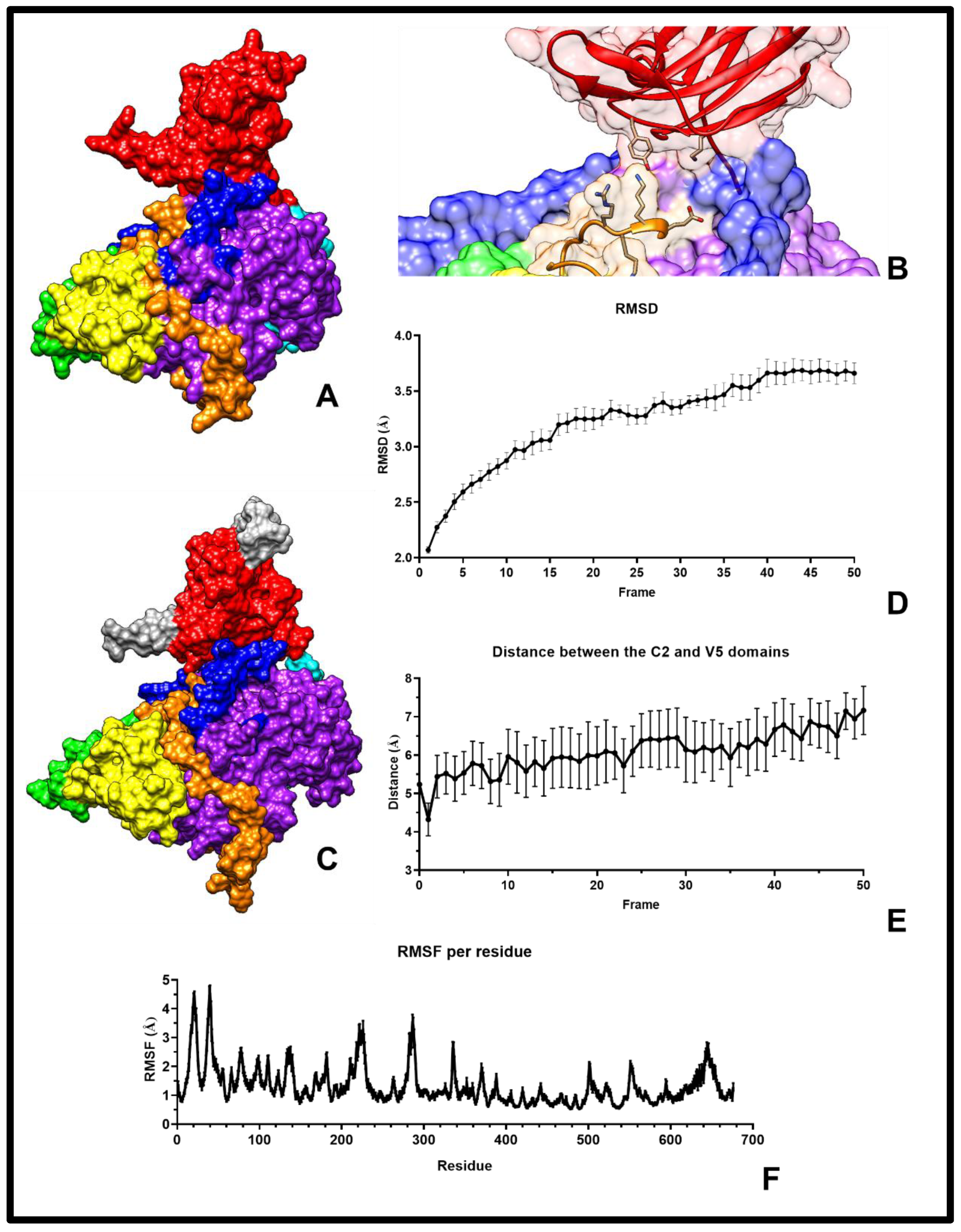
2.2. Model Refinement and Validation
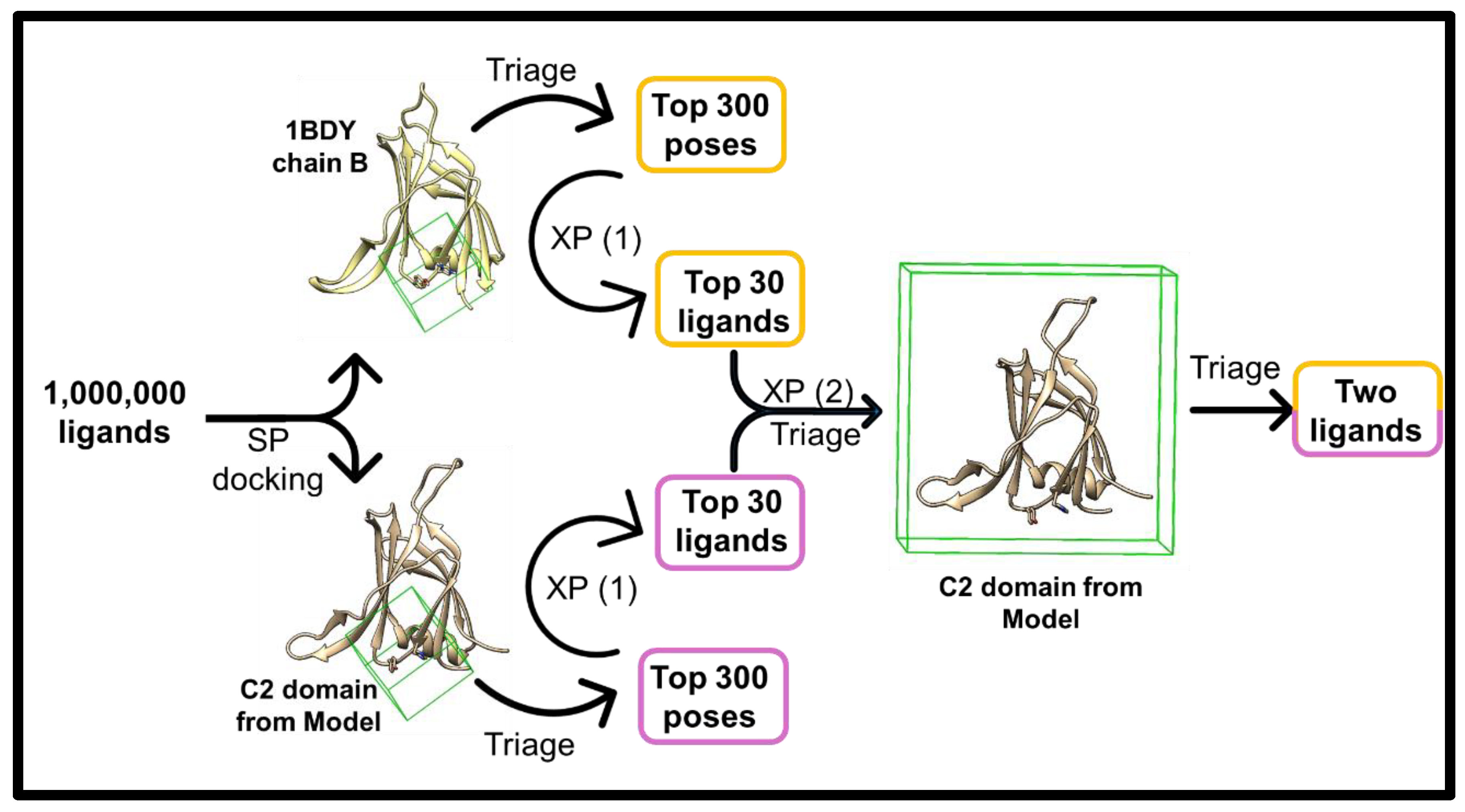
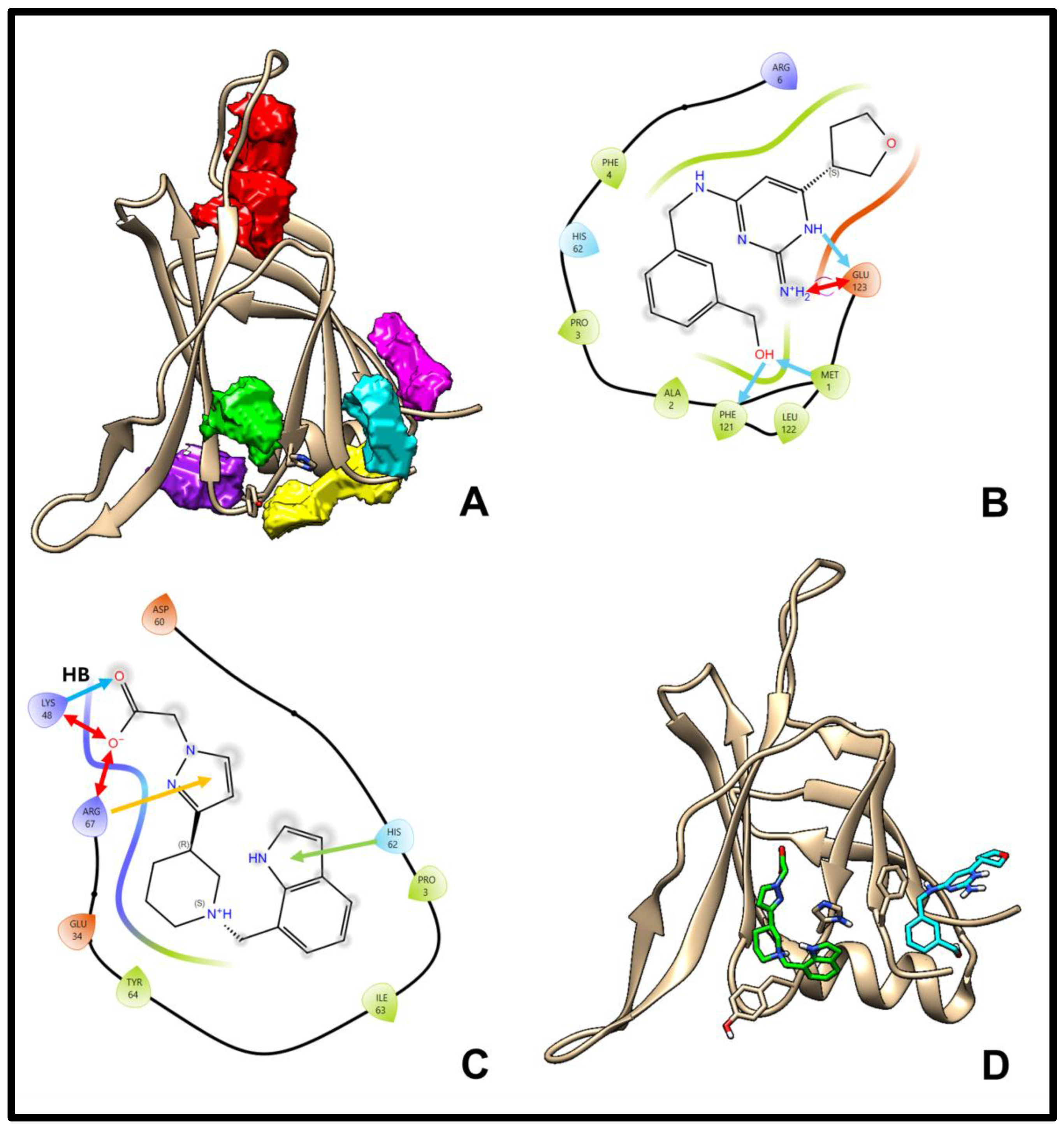
2.3. Ligand Screening and Pocket Specificity in the C2 Domain
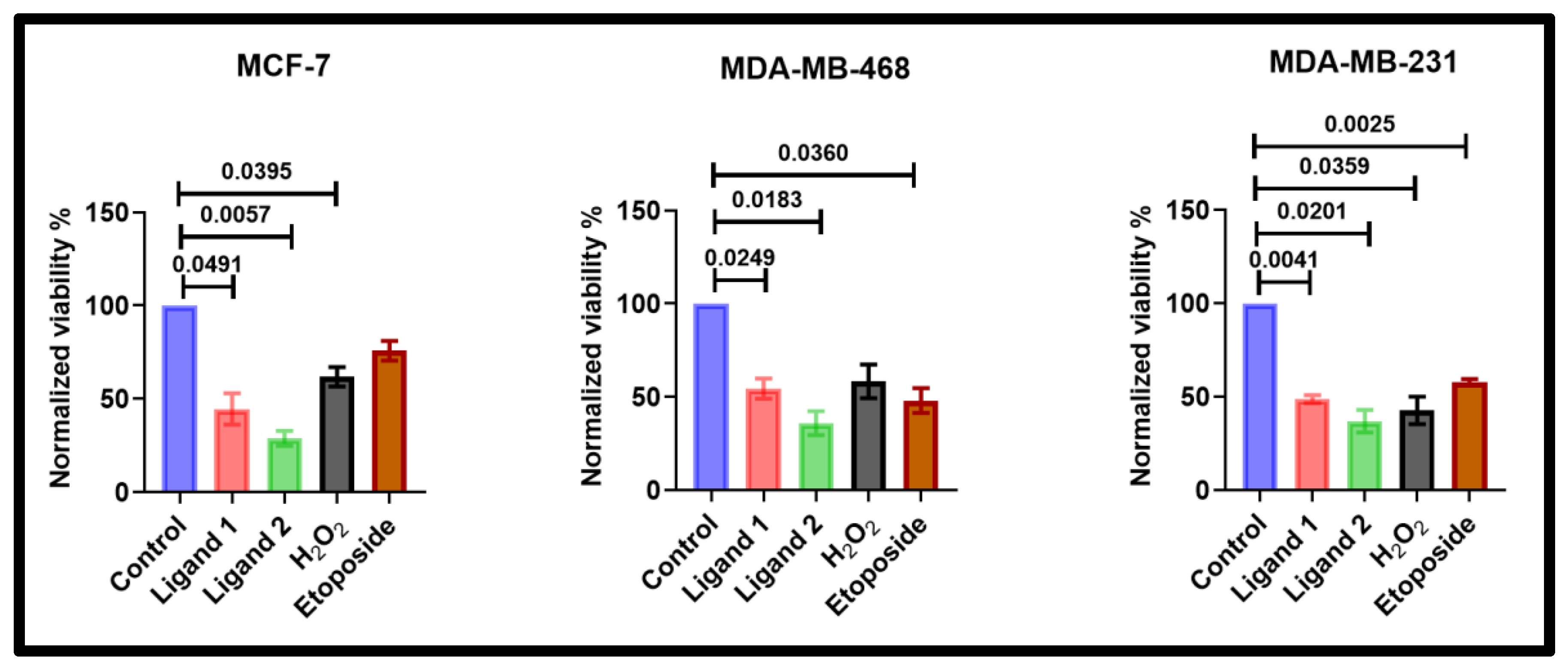
2.4. Ligands Reduce Wild-Type Breast Cancer Cell Lines Viability
2.5. Comparative Effects of Ligands on Viability in δC2- and Vector-Transfected MCF-7 Cells
3. Discussion
4. Methods
4.1. Protein Sequence and Template Selection
4.2. Comparative Modelling and Refinement
4.3. Molecular Dynamics Simulations
4.4. Pocket Detection and Ligand Screening
4.5. Cell Culture and Viability Assays
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antal, C.E.; Hudson, A.M.; Kang, E.; Zanca, C.; Wirth, C.; Stephenson, N.L.; Trotter, E.W.; Gallegos, L.L.; Miller, C.J.; Furnari, F.B.; et al. Cancer-Associated Protein Kinase C Mutations Reveal Kinase’s Role as Tumor Suppressor. Cell 2015, 160, 489–502. [Google Scholar] [CrossRef]
- Huse, M.; Kuriyan, J. The Conformational Plasticity of Protein Kinases. Cell 2002, 109, 275–282. [Google Scholar] [CrossRef]
- Newton, A.C. Protein Kinase C: Structural and Spatial Regulation by Phosphorylation, Cofactors, and Macromolecular Interactions. Chem. Rev. 2001, 101, 2353–2364. [Google Scholar] [CrossRef] [PubMed]
- Pappa, H.; Murray-Rust, J.; Dekker, L.V.; Parker, P.J.; McDonald, N.Q. Crystal Structure of the C2 Domain from Protein Kinase C-Delta. Structure 1998, 6, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Sondermann, H.; Kuriyan, J. C2 Can Do It, Too. Cell 2005, 121, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Leonard, T.A.; Różycki, B.; Saidi, L.F.; Hummer, G.; Hurley, J.H. Crystal Structure and Allosteric Activation of Protein Kinase C Βii. Cell 2011, 144, 55–66. [Google Scholar] [CrossRef]
- Hurley, J.H. Membrane Binding Domains. Biochim. Biophys. Acta 2006, 1761, 805–811. [Google Scholar] [CrossRef]
- Abe, H.; Miyamoto, K.; Tochio, N.; Saito, K.; Sasagawa, A.; Koshiba, S.; Inoue, M.; Kigawa, T.; Yokoyama, S. Solution Structure of the First Phorbol Esters/Diacylglycerol Binding Domain of Human Protein Kinase C, Delta. Protein Data Bank, 2008; to be published. [Google Scholar]
- Blass, M.; Kronfeld, I.; Kazimirsky, G.; Blumberg, P.M.; Brodie, C. Tyrosine Phosphorylation of Protein Kinase Cδ Is Essential for Its Apoptotic Effect in Response to Etoposide. Mol. Cell. Biol. 2002, 22, 182–195. [Google Scholar] [CrossRef]
- Lu, W.; Lee, H.K.; Xiang, C.; Finniss, S.; Brodie, C. The Phosphorylation of Tyrosine 332 Is Necessary for the Caspase 3-Dependent Cleavage of Pkcδ and the Regulation of Cell Apoptosis. Cell. Signal. 2007, 19, 2165–2173. [Google Scholar] [CrossRef]
- Newton, A.C. Lipid Activation of Protein Kinases. J. Lipid Res. 2009, 50, S266–S271. [Google Scholar] [CrossRef] [PubMed]
- Aminpour, M.; Montemagno, C.; Tuszynski, J.A. An Overview of Molecular Modeling for Drug Discovery with Specific Illustrative Examples of Applications. Molecules 2019, 24, 1693. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.; Wallner, B.; Lindahl, E.; Elofsson, A. Using Multiple Templates to Improve Quality of Homology Models in Automated Homology Modeling. Protein Sci. 2008, 17, 990–1002. [Google Scholar] [CrossRef]
- Makowske, M.; Rosen, O.M. Complete Activation of Protein Kinase C by an Antipeptide Antibody Directed against the Pseudosubstrate Prototope. J. Biol. Chem. 1989, 264, 16155–16159. [Google Scholar] [CrossRef]
- Malavez, Y.; Gonzalez-Mejia, M.E.; Doseff, A.I. Prkcd (Protein Kinase C, Delta). Atlas Genet. Cytogenet. Oncol. Haematol. 2008, 13, 28–42. [Google Scholar] [CrossRef]
- Orr, J.W.; Newton, A.C. Intrapeptide Regulation of Protein Kinase C. J. Biol. Chem. 1994, 269, 8383–8387. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Perry, J.K.; Shaw, D.E.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. Dogsitescorer: A Web Server for Automatic Binding Site Prediction, Analysis and Druggability Assessment. Bioinformatics 2012, 28, 2074–2075. Available online: https://proteins.plus/ (accessed on 25 September 2025). [CrossRef]
- Lu, W.; Finnis, S.; Xiang, C.; Lee, H.K.; Markowitz, Y.; Okhrimenko, H.; Brodie, C. Tyrosine 311 Is Phosphorylated by C-Abl and Promotes the Apoptotic Effect of Pkcδ in Glioma Cells. Biochem. Biophys. Res. Commun. 2006, 352, 431–436. [Google Scholar] [CrossRef]
- Khader, R.; Dekker, L.V. The C2 Domain of Pkc-Δ as a Dominant-Negative Modulator of Breast Cancer Cell Survival and Chemosensitivity. ACS Omega, 2025; manuscript submitted. [Google Scholar]
- Zheng, J.; Trafny, E.A.; Knighton, D.R.; Xuong, N.; Taylor, S.S.; Ten Eyck, L.F.; Sowadski, J.M. 2.2 Å Refined Crystal Structure of the Catalytic Subunit of Camp-Dependent Protein Kinase Complexed with Mnatp and a Peptide Inhibitor. Acta Crystallogr. D Biol. Crystallogr. 1993, 49, 362–365. [Google Scholar] [CrossRef] [PubMed]
- DeVries-Seimon, T.A.; Ohm, A.M.; Humphries, M.J.; Reyland, M.E. Induction of Apoptosis Is Driven by Nuclear Retention of Pkcdelta. J. Biol. Chem. 2007, 282, 22307–22314. [Google Scholar] [CrossRef]
- Antal, C.E.; Callender, J.A.; Kornev, A.P.; Taylor, S.S.; Newton, A.C. Intramolecular C2 Domain-Mediated Autoinhibition of Protein Kinase C BII. Cell Rep. 2015, 12, 1252–1260. [Google Scholar] [CrossRef]
- Stensman, H.; Larsson, C. Identification of Acidic Amino Acid Residues in the Protein Kinase C Alpha V5 Domain That Contribute to Its Insensitivity to Diacylglycerol. J. Biol. Chem. 2007, 282, 28627–28638. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with Alphafold. Nature 2021, 596, 583–589. Available online: https://alphafold.ebi.ac.uk/entry/AF-Q05655-F1 (accessed on 25 September 2025). [CrossRef]
- Osada, S.; Mizuno, K.; Saido, T.C.; Suzuki, K.; Kuroki, T.; Ohno, S. A New Member of the Protein Kinase C Family, Npkc Theta, Predominantly Expressed in Skeletal Muscle. Mol. Cell. Biol. 1992, 12, 3930–3938. [Google Scholar] [CrossRef] [PubMed]
- Adwan, T.S.; Ohm, A.M.; Jones, D.N.M.; Humphries, M.J.; Reyland, M.E. Regulated Binding of Importin-A to Protein Kinase C in Response to Apoptotic Signals Facilitates Nuclear Imports. J. Biol. Chem. 2011, 286, 35716–35724. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Matsuzaki, H.; Takaishi, H.; Yamamoto, T.; Fukunaga, M.; Ono, Y.; Kikkawa, U. Opposing Effects of Protein Kinase C Delta and Protein Kinase B Alpha on H2O2-Induced Apoptosis in Cho Cells. Biochem. Biophys. Res. Commun. 1999, 3, 840–846. [Google Scholar] [CrossRef]
- Humphries, M.J.; Ohm, A.M.; Schaack, J.; Adwan, T.S.; Reyland, M.E. Tyrosine Phosphorylation Regulates Nuclear Translocation of Pkcδ. Oncogene 2008, 27, 3045–3053. [Google Scholar] [CrossRef]
- Wie, S.M.; Adwan, T.S.; DeGregori, J.; Anderson, S.M.; Reyland, M.E. Inhibiting Tyrosine Phosphorylation of Protein Kinase Cδ (Pkcδ) Protects the Salivary Gland from Radiation Damage. J. Biol. Chem. 2014, 289, 10900–10908. [Google Scholar] [CrossRef]
- Benes, C.H.; Wu, N.; Elia, A.E.H.; Dharia, T.; Cantley, L.C.; Soltoff, S.P. The C2 Domain of Pkcdelta Is a Phosphotyrosine Binding Domain. Cell 2005, 121, 271–280. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (Refseq) Database at Ncbi: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. Available online: https://www.ncbi.nlm.nih.gov/refseq/ (accessed on 30 March 2025). [CrossRef]
- Burley, S.K.; Bhatt, R.; Bhikadiya, C.; Bi, C.; Biester, S.; Biswas, P.; Bittrich, S.; Blaumann, S.; Brown, R.; Chao, H.; et al. Updated Resources for Exploring Experimentally-Determined Pdb Structures and Computed Structure Models at the Rcsb Protein Data Bank. Nucleic Acids Res. 2025, 53, D564–D574. Available online: https://www.rcsb.org/ (accessed on 1 October 2025). [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. Cobalt: Constraint-Based Alignment Tool for Multiple Protein Sequences. Bioinformatics 2007, 23, 1073–1079. Available online: https://www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi (accessed on 1 March 2025). [CrossRef]
- Shanmugasundararaj, S.; Das, J.; Sandberg, W.S.; Zhou, X.; Wand, D.; Messing, R.O.; Bruzik, K.S.; Stehle, T.; Miller, K.W. Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta. Biophys. J. 2012, 103, 2331–2340. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.B.; Chaudhary, D.; Olland, S.; Wolfrom, S.; Czerwinski, R.; Malakain, K.; Lin, L.; Stahl, M.L.; Joseph-McCarthy, D.; Benander, C.; et al. Catalytic Domain Crystal Structure of Protein Kinase C-Theta (Pkctheta). J. Biol. Chem. 2004, 279, 50401–50409. [Google Scholar] [CrossRef] [PubMed]
- Hobson, A.D.; Judge, R.A.; Aguirre, A.L.; Brown, B.S.; Cui, Y.; Ding, P.; Domingues, E.; DiGiammarino, E.; Egan, D.A.; Freiberg, G.M.; et al. Identification of Selective Dual Rock1 and Rock2 Inhibitors Using Structure-Based Drug Design. J. Med. Chem. 2018, 61, 11074–11100. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. Molprobity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. Available online: https://molprobity.biochem.duke.edu/ (accessed on 1 October 2025). [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. Pep-Fold3: Faster De Novo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. Available online: https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/ (accessed on 5 July 2025). [CrossRef] [PubMed]
- Xu, D.; Zhang, Y. Improving the Physical Realism and Structural Accuracy of Protein Models by a Two-Step Atomic-Level Energy Minimization. Biophys. J. 2011, 101, 2525–2534. Available online: https://www.aideepmed.com/ModRefiner/ (accessed on 30 July 2025). [CrossRef]
- Heo, L.; Park, H.; Seok, C. Galaxyrefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. Available online: https://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE (accessed on 30 July 2025). [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. Charmm-Gui: A Web-Based Graphical User Interface for Charmm. J. Comput. Chem. 2008, 29, 1859–1865. Available online: https://www.charmm-gui.org/ (accessed on 30 August 2025). [CrossRef] [PubMed]
- Eastman, P.; Friedrichs, M.S.; Chodera, J.D.; Radmer, R.J.; Bruns, C.M.; Ku, J.P.; Beauchamp, K.A.; Lane, T.J.; Wang, L.P.; Shukla, D.; et al. Openmm 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J. Chem. Theor. Comput. 2013, 9, 461–469. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. Zinc 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khader, R.; Dekker, L.V. Modelling the Full-Length Inactive PKC-δ Structure to Explore Regulatory Accessibility and Selective Targeting Opportunities. Pharmaceuticals 2025, 18, 1760. https://doi.org/10.3390/ph18111760
Khader R, Dekker LV. Modelling the Full-Length Inactive PKC-δ Structure to Explore Regulatory Accessibility and Selective Targeting Opportunities. Pharmaceuticals. 2025; 18(11):1760. https://doi.org/10.3390/ph18111760
Chicago/Turabian StyleKhader, Rasha, and Lodewijk V. Dekker. 2025. "Modelling the Full-Length Inactive PKC-δ Structure to Explore Regulatory Accessibility and Selective Targeting Opportunities" Pharmaceuticals 18, no. 11: 1760. https://doi.org/10.3390/ph18111760
APA StyleKhader, R., & Dekker, L. V. (2025). Modelling the Full-Length Inactive PKC-δ Structure to Explore Regulatory Accessibility and Selective Targeting Opportunities. Pharmaceuticals, 18(11), 1760. https://doi.org/10.3390/ph18111760






