Unveiling the Therapeutic Mechanisms of Chinese Herbs in Heart Failure: Integrating Network Pharmacology, Molecular Docking, and Simulation Analysis
Abstract
1. Introduction
2. Result
2.1. Active Compounds in Herbs and Targets
2.2. Differential Expression Genes in Heart Failure
2.3. Construction of PPI Network for Common Targets
2.4. Evaluation of mRNA Levels and Target-Organ Analysis
2.5. GO Enrichment Analysis
2.6. Docking and Evaluation
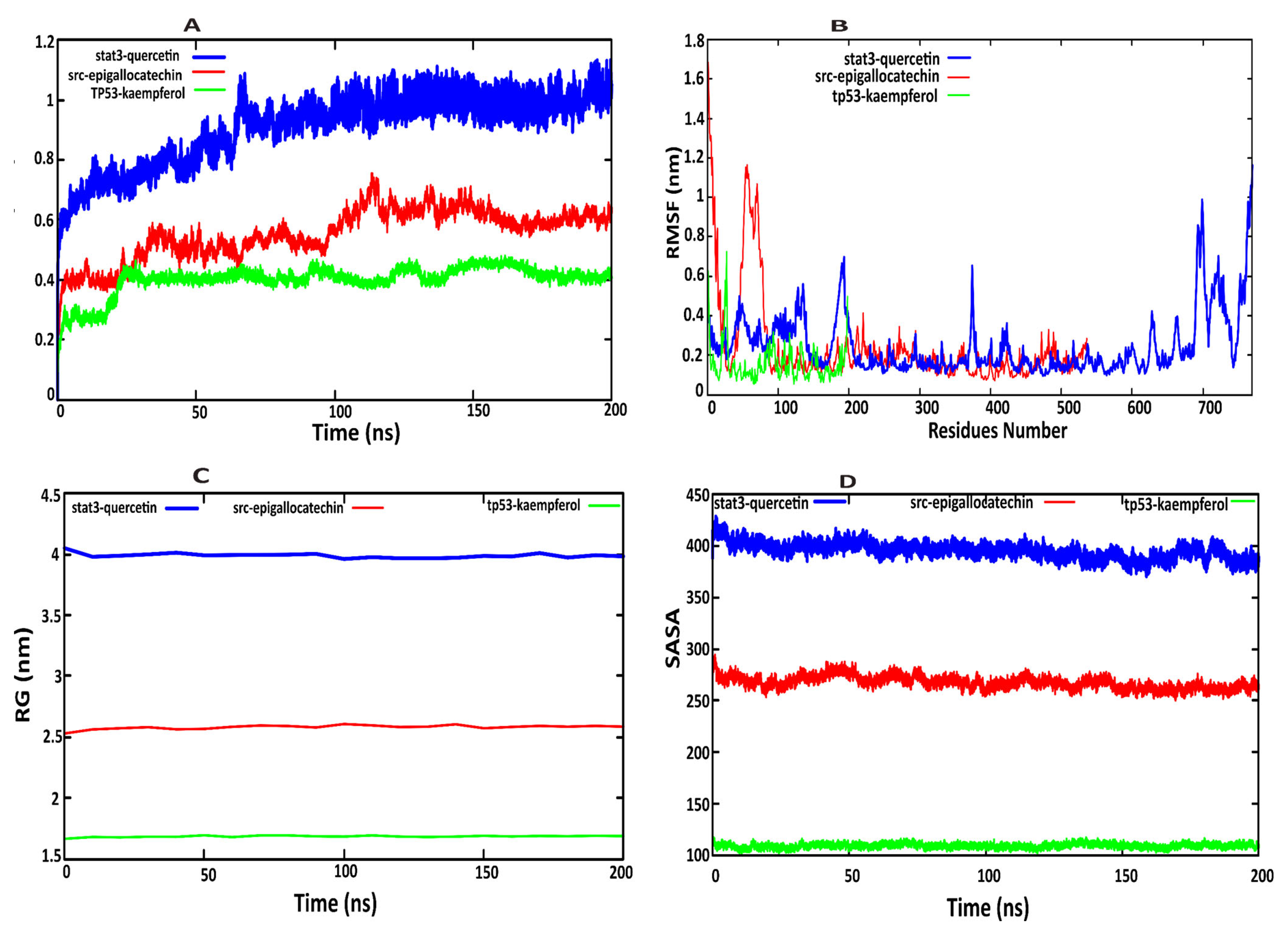
2.7. Molecular Dynamics Simulation Analysis
3. Discussion
4. Materials and Methods
4.1. Evaluation of Bioactive Compounds and Associated Molecular Targets
4.2. Integration of Target Gene Identification Methods
4.3. Protein–Protein Interaction (PPI) Network Construction
4.4. Biological Functional Enrichment Analysis
4.5. Molecular Docking
4.6. Molecular Dynamics (MD) Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sapna, F.; Raveena, F.; Chandio, M.; Bai, K.; Sayyar, M.; Varrassi, G.; Khatri, M.; Kumar, S.; Mohamad, T. Advancements in Heart Failure Management: A Comprehensive Narrative Review of Emerging Therapies. Cureus 2023, 15, e46486. [Google Scholar] [CrossRef]
- Bhandari, B.; Rodriguez, B.S.Q.; Masood, W. Ischemic Cardiomyopathy; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Huang, Y.; Xuan, L.; Xu, Q.; Wang, J.; Liu, J. Differences in T cell-associated serum markers between ischemic cardiomyopathy and dilated cardiomyopathy. J. Thorac. Dis. 2024, 16, 4655–4665. [Google Scholar] [CrossRef]
- Cipriani, A.; Bauce, B.; De Lazzari, M.; Rigato, I.; Bariani, R.; Meneghin, S.; Pilichou, K.; Motta, R.; Aliberti, C.; Thiene, G.; et al. Arrhythmogenic Right Ventricular Cardiomyopathy: Characterization of Left Ventricular Phenotype and Differential Diagnosis with Dilated Cardiomyopathy. J. Am. Heart Assoc. 2020, 9, e014628. [Google Scholar] [CrossRef]
- Mestroni, L.; Brun, F.; Spezzacatene, A.; Sinagra, G.; Taylor, M.R. Genetic Causes of Dilated Cardiomyopathy. Prog. Pediatr. Cardiol. 2014, 37, 13–18. [Google Scholar] [CrossRef]
- Zolty, R. The Role of Neurohormonal Systems, Inflammatory Mediators and Oxydative Stress in Cardiomyopathy; IntechOpen: London, UK, 2021. [Google Scholar]
- Zhang, Y.; Liu, C.; Liu, M.; Liu, T.; Lin, H.; Huang, C.B.; Ning, L. Attention is all you need: Utilizing attention in AI-enabled drug discovery. Brief. Bioinform. 2023, 25. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Ren, L.; Zhan, M.; Sun, T.; Zou, Q.; Zhang, Y. Predicting intercellular communication based on metabolite-related ligand-receptor interactions with MRCLinkdb. BMC Biol. 2024, 22, 152. [Google Scholar] [CrossRef] [PubMed]
- Mascolo, A.; di Mauro, G.; Cappetta, D.; De Angelis, A.; Torella, D.; Urbanek, K.; Berrino, L.; Nicoletti, G.F.; Capuano, A.; Rossi, F. Current and future therapeutic perspective in chronic heart failure. Pharmacol. Res. 2022, 175, 106035. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Liu, M.; Gou, Y.; Yang, Y.; He, B.; Huang, J. Development and application of ribonucleic acid therapy strategies against COVID-19. Int. J. Biol. Sci. 2022, 18, 5070–5085. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.; Qiao, J.; Zeng, X.; Zou, Q.; Wei, L. Deep generative models in de novo drug molecule generation. J. Chem. Inf. Model. 2024, 64, 2174–2194. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, M. Holistic View of TCM on Cancer Integrative Therapy. Future Integr. Med. 2023, 2, 159–167. [Google Scholar] [CrossRef]
- Lai, L.; Liu, Y.; Song, B.; Li, K.; Zeng, X.J.A.C.S. Deep Generative Models for Therapeutic Peptide Discovery: A Comprehensive Review. ACM Comput. Surv. 2025, 57, 155. [Google Scholar] [CrossRef]
- Liu, X.; Ai, C.; Yang, H.; Dong, R.; Tang, J.; Zheng, S.; Guo, F. RetroCaptioner: Beyond attention in end-to-end retrosynthesis transformer via contrastively captioned learnable graph representation. Bioinformatics 2024, 40, btae561. [Google Scholar] [CrossRef]
- Chen, L.; Li, L. Prediction of Drug Pathway-based Disease Classes using Multiple Properties of Drugs. Curr. Bioinform. 2024, 19, 859–872. [Google Scholar] [CrossRef]
- Wang, X.; Wang, T.; Wang, Y.; Li, X.; Chen, Q.; Wang, Y.; Zhang, X.; Wang, H.; Zhao, H.; Mou, Y.; et al. Research progress on classical traditional Chinese medicine Taohong Siwu decoction in the treatment of coronary heart disease. Biomed. Pharmacother. 2022, 152, 113249. [Google Scholar] [CrossRef]
- Das, A.J.; Mohideen, H.S. Systems pharmacology–principles, methods and applications. In Systems Biology and In-Depth Applications for Unlocking Diseases; Elsevier: Amsterdam, The Netherlands, 2025. [Google Scholar]
- Zhu, H.; Hao, H.; Yu, L. Identification of microbe–disease signed associations via multi-scale variational graph autoencoder based on signed message propagation. BMC Biol. 2024, 22, 172. [Google Scholar] [CrossRef]
- Chen, X.; Zhong, X.; Luo, D.; Lei, Y.; Huang, R. Plasma SMOC2 Predicts Prognosis in Patients with Heart Failure: A Prospective Cohort. Int. J. Gen. Med. 2024, 17, 1651–1664. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Y.; Zhao, C. Integrated gene expression pro fi ling analysis reveals SERPINA3, FCN3, FREM1, MNS1 as candidate biomarkers in heart failure and their correlation with immune in fi ltration. J. Thorac. Dis. 2022, 14, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Peng, F.; Li, J.; Gong, H. Exploring novel biomarkers in dilated cardiomyopathy-induced heart failure by integrated analysis and in vitro experiments. Exp. Ther. Med. 2023, 26, 325. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Y.; Cheng, F.; Chen, D.; Zhang, H. Identification of signature genes and subtypes for heart failure diagnosis based on machine learning. Front. Cardiovasc. Med. 2025, 12, 1492192. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ning, B.; Zhang, Q.; Ma, J.; Zhao, L.; Lu, Q.; Zhang, D. Identification of Hub Diagnostic Biomarkers and Candidate Therapeutic Drugs in Heart Failure. Int. J. Gen. Med. 2022, 15, 623–635. [Google Scholar] [CrossRef]
- Cannavo, A.; Rengo, G.; Liccardo, D.; Pagano, G.; Zincarelli, C.; De Angelis, M.C.; Puglia, R.; Di Pietro, E.; Rabinowitz, J.E.; Barone, M.V.; et al. beta1-adrenergic receptor and sphingosine-1-phosphate receptor 1 (S1PR1) reciprocal downregulation influences cardiac hypertrophic response and progression to heart failure: Protective role of S1PR1 cardiac gene therapy. Circulation 2013, 128, 1612–1622. [Google Scholar] [CrossRef]
- Wang, X.; Rao, J.; Chen, X.; Wang, Z.; Zhang, Y. Identification of Shared Signature Genes and Immune Microenvironment Subtypes for Heart Failure and Chronic Kidney Disease Based on Machine Learning. J. Inflamm. Res. 2024, 17, 1873–1895. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, X.; Li, X.; Ma, L.; Li, Y.J.M.B. Identification of Co-diagnostic Genes for Heart Failure and Hepatocellular Carcinoma Through WGCNA and Machine Learning Algorithms. Mol. Biotechnol. 2024, 66, 1229–1245. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Basith, S.; Shin, T.H.; Wei, L.; Lee, G. Meta-4mCpred: A Sequence-Based Meta-Predictor for Accurate DNA 4mC Site Prediction Using Effective Feature Representation. Mol. Ther. Nucleic Acids 2019, 16, 733–744. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, D.; Li, Y.A.; Yang, S.; Liang, Y.; Xing, Y.; Zuo, Y. RaacFold: A webserver for 3D visualization and analysis of protein structure by using reduced amino acid alphabets. Nucleic Acids Res. 2022, 50, W633–W638. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, D.; Song, Y.; Liang, Y.; Yu, H.; Zuo, Y. Designing a structure-function alphabet of helix based on reduced amino acid clusters. Arch. Biochem. Biophys. 2024, 754, 109942. [Google Scholar] [CrossRef]
- Kong, R.; Bharadwaj, U.; Eckols, T.K.; Kolosov, M.; Wu, H.; Cruz-Pavlovich, F.J.S.; Shaw, A.; Ifelayo, O.I.; Zhao, H.; Kasembeli, M.M.; et al. Novel STAT3 small-molecule inhibitors identified by structure-based virtual ligand screening incorporating SH2 domain flexibility. Pharmacol. Res. 2021, 169, 105637. [Google Scholar] [CrossRef]
- Li, X.; Li, X.; Wang, L.; Hou, Y.; Liu, Y.; Mao, J.; Zhang, L.; Li, X. Advancing Traditional Chinese Medicine Research through Network Pharmacology: Strategies for Target Identification, Mechanism Elucidation and Innovative Therapeutic Applications. Am. J. Chin. Med. 2025, 53, 2021–2042. [Google Scholar] [CrossRef]
- Tian, S.; Li, Y.; Li, D.; Xu, X.; Wang, J.; Zhang, Q.; Hou, T. Modeling Compound–Target Interaction Network of Traditional Chinese Medicines for Type II Diabetes Mellitus: Insight for Polypharmacology and Drug Design. J. Chem. Inf. Model. 2013, 53, 1787–1803. [Google Scholar] [CrossRef]
- Xu, B.; Liu, D.; Wang, Z.; Tian, R.; Zuo, Y. Multi-substrate selectivity based on key loops and non-homologous domains: New insight into ALKBH family. Cell Mol. Life Sci. 2021, 78, 129–141. [Google Scholar] [CrossRef]
- Ahmad, H.; Khan, H.; Haque, S.; Ahmad, S.; Srivastava, N.; Khan, A. Angiotensin-Converting Enzyme and Hypertension: A Systemic Analysis of Various ACE Inhibitors, Their Side Effects, and Bioactive Peptides as a Putative Therapy for Hypertension. J. Renin-Angiotensin-Aldosterone Syst. 2023, 2023, 7890188. [Google Scholar] [CrossRef]
- Jaisankar, D.; Ramamoorthy, S.; Ramasamy, J. Therapeutic potential of flavonoid-enriched Chinese medicinal herbs in atherosclerosis, hypertension, myocardial infarction, and heart failure. Pharmacol. Res. Mod. Chin. Med. 2025, 17, 100701. [Google Scholar] [CrossRef]
- Li, X.; Yang, H.; Xiao, J.; Zhang, J.; Zhang, J.; Liu, M.; Zheng, Y.; Ma, L. Network pharmacology based investigation into the bioactive compounds and molecular mechanisms of Schisandrae Chinensis Fructus against drug-induced liver injury. Bioorganic Chem. 2020, 96, 103553. [Google Scholar] [CrossRef]
- Yu, X.; Qin, W.; Cai, H.; Ren, C.; Huang, S.; Lin, X.; Tang, L.; Shan, Z.; Al-Ameer, W.H.A.; Wang, L.; et al. Analyzing the molecular mechanism of xuefuzhuyu decoction in the treatment of pulmonary hypertension with network pharmacology and bioinformatics and verifying molecular docking. Comput. Biol. Med. 2024, 169, 107863. [Google Scholar] [CrossRef]
- Ren, X.; Wen, Y.; Li, C.; Yuan, M.; Zhang, J.; Li, S.; Zhang, X.; Wang, L.; Wang, S. Integrated network pharmacology and comprehensive bioinformatics identifying the mechanisms and molecular targets of Jieyu Anshen Granules for treating comorbidity with Alzheimer’s disease and depression. Arab. J. Chem. 2024, 17, 105485. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, Y.; Wang, W.; He, Y.; Zhong, H.; Zhou, X.; Chen, Y.; Cai, X.-J.; Liu, L.-Q. Mechanisms underlying the therapeutic effects of Qingfeiyin in treating acute lung injury based on GEO datasets, network pharmacology and molecular docking. Comput. Biol. Med. 2022, 145, 105454. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, X.; Huang, W. Exploring the mechanism of Radix Bupleuri in the treatment of depression combined with SARS-CoV-2 infection through bioinformatics, network pharmacology, molecular docking, and molecular dynamic simulation. Metab. Brain Dis. 2025, 40, 105. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yao, Y.; Liu, Y.; Zhang, R.; Wen, C.; Zhou, Q.; Xu, Y.; Wang, W.; Jiang, H.; Tao, Z.; et al. IKKα-STAT3-S727 axis: A novel mechanism in DOX-induced cardiomyopathy. Cell. Mol. Life Sci. 2024, 81, 406. [Google Scholar] [CrossRef]
- Gogiraju, R.; Xu, X.; Bochenek, M.L.; Steinbrecher, J.H.; Lehnart, S.E.; Wenzel, P.; Kessel, M.; Zeisberg, E.M.; Dobbelstein, M.; Schäfer, K. Endothelial p53 Deletion Improves Angiogenesis and Prevents Cardiac Fibrosis and Heart Failure Induced by Pressure Overload in Mice. J. Am. Heart Assoc. 2015, 4, e001770. [Google Scholar] [CrossRef]
- Men, H.; Cai, H.; Cheng, Q.; Zhou, W.; Wang, X.; Huang, S.; Zheng, Y.; Cai, L. The regulatory roles of p53 in cardiovascular health and disease. Cell. Mol. Life Sci. 2021, 78, 2001–2018. [Google Scholar] [CrossRef]
- Zhai, Y.; Yang, J.; Zhang, J.; Yang, J.; Li, Q.; Zheng, T. Src-family Protein Tyrosine Kinases: A promising target for treating Cardiovascular Diseases. Int. J. Med. Sci. 2021, 18, 1216–1224. [Google Scholar] [CrossRef]
- Jubaidi, F.F.; Zainalabidin, S.; Taib, I.S.; Hamid, Z.A.; Budin, S.B. The Potential Role of Flavonoids in Ameliorating Diabetic Cardiomyopathy via Alleviation of Cardiac Oxidative Stress, Inflammation and Apoptosis. Int. J. Mol. Sciences 2021, 22, 5094. [Google Scholar] [CrossRef]
- Zhou, P.; Hua, F.; Wang, X.; Huang, J.-L. Therapeutic potential of IKK-β inhibitors from natural phenolics for inflammation in cardiovascular diseases. Inflammopharmacology 2020, 28, 19–37. [Google Scholar] [CrossRef]
- Mokra, D.; Joskova, M.; Mokry, J. Therapeutic Effects of Green Tea Polyphenol (–)-Epigallocatechin-3-Gallate (EGCG) in Relation to Molecular Pathways Controlling Inflammation, Oxidative Stress, and Apoptosis. Int. J. Mol. Sci. 2023, 24, 340. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.X.; Li, D.; Xu, S.Q.; He, F.; Xia, X.; Xu, S. Pharmacological properties of Humulus scandens: Insights from the TCMSP database and existing understanding. Pharmacol. Res. Mod. Chin. Med. 2024, 13, 100518. [Google Scholar] [CrossRef]
- Mu, C.; Sheng, Y.; Wang, Q.; Amin, A.; Li, X.; Xie, Y. Dataset of potential Rhizoma Polygonati compound-druggable targets and partial pharmacokinetics for treatment of COVID-19. Data Brief. 2020, 33, 106475. [Google Scholar] [CrossRef]
- Sun, J.; Ni, Q.; Jiang, F.; Liu, B.; Wang, J.; Zhang, L.; Huang, J.J.B.R.I. Discovery and validation of traditional chinese and western medicine combination antirheumatoid arthritis drugs based on machine learning (random forest model). BioMed Res. Int. 2023, 2023, 6086388. [Google Scholar] [CrossRef]
- Zhou, C.; Zhou, Z.-M.; Hu, L.; Yang, Y.-Y.; Meng, X.-W.; Yang, X.-S. Identification and Verification of the Effect of miR-143 on the Cardioprotective Role of Phosphocreatine in Doxorubicin-Induced Cardiotoxicity by Integrated Bioinformatics Analysis. Research Square 2021. [Google Scholar] [CrossRef]
- Barshir, R.; Fishilevich, S.; Iny-Stein, T.; Zelig, O.; Mazor, Y.; Guan-Golan, Y.; Safran, M.; Lancet, D. GeneCaRNA: A Comprehensive Gene-centric Database of Human Non-coding RNAs in the GeneCards Suite. J. Mol. Biol. 2021, 433, 166913. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Fu, Y.; Tan, T.; Hu, J.; Li, F.; Liao, Z.; Peng, J. Screening of lncRNA-miRNA-mRNA Coexpression Regulatory Networks Involved in Acute Traumatic Coagulation Dysfunction Based on CTD, GeneCards, and PharmGKB Databases. Oxid. Med. Cell Longev. 2022, 2022, 7280312. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Qiu, Y.; Chen, Y. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef]
- Alameer, A.; Chicco, D. geoCancerPrognosticDatasetsRetriever: A bioinformatics tool to easily identify cancer prognostic datasets on Gene Expression Omnibus (GEO). Bioinformatics 2022, 38, 1761–1763. [Google Scholar] [CrossRef]
- Tong, Y. The comparison of limma and DESeq2 in gene analysis. E3s Web Conf. 2021, 271, 03058. [Google Scholar] [CrossRef]
- Yu, J.W.; Wang, L.; Bao, L.D. Exploring the active compounds of traditional Mongolian medicine in intervention of novel coronavirus (COVID-19) based on molecular docking method. J. Funct. Foods 2020, 71, 104016. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Qian, X.X.; Zhang, X.M.; Jiang, T.; Li, X.J.; Sun, W. ETNK2 Low-Expression Predicts Poor Prognosis in Renal Cell Carcinoma with Immunosuppressive Tumor Microenvironment. J. Oncol. 2023, 2023, 1743357. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Franz, M.; Lopes, C.T.; Fong, D.; Kucera, M.; Cheung, M.; Siper, M.C.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js 2023 update: A graph theory library for visualization and analysis. Bioinformatics 2023, 39, btad031. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, Y.; Ding, Y.; Zou, Q. SBSM-Pro: Support bio-sequence machine for proteins. Sci. China-Inf. Sci. 2024, 67, 212106. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Sanner, M.F.; Olson, A.J.; Forli, S. The AutoDock suite at 30. Protein Sci. 2021, 30, 31–43. [Google Scholar] [CrossRef]
- Ahmad, B.; Saeed, A.; Castrosanto, M.A.; Amir Zia, M.; Farooq, U.; Abbas, Z.; Khan, S. Identification of natural marine compounds as potential inhibitors of CDK2 using molecular docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2023, 41, 8506–8516. [Google Scholar] [CrossRef]
- Abdullahi, M.; Adeniji, S.E.J.C.A. In-silico molecular docking and ADME/pharmacokinetic prediction studies of some novel carboxamide derivatives as anti-tubercular agents. Chem. Afr. 2020, 3, 989–1000. [Google Scholar] [CrossRef]
- Tripathy, S.; Sahu, S.K.; Azam, M.A.; Jupudi, S. Computer-aided identification of lead compounds as Staphylococcal epidermidis FtsZ inhibitors using molecular docking, virtual screening, DFT analysis, and molecular dynamic simulation. J. Mol. Model. 2019, 25, 360. [Google Scholar] [CrossRef]
- Pasala, C.; Katari, S.K.; Nalamolu, R.M.; Bitla, A.R.; Amineni, U. Hierarchical-Clustering, Scaffold-Mining Exercises and Dynamics Simulations for Effectual Inhibitors Against Lipid-A Biosynthesis of Helicobacter pylori. Cell Mol. Bioeng. 2019, 12, 255–274. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Lee, G. Unveiling local and global conformational changes and allosteric communications in SOD1 systems using molecular dynamics simulation and network analyses. Comput. Biol. Med. 2024, 168, 107688. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. A Molecular Dynamics Approach to Explore the Intramolecular Signal Transduction of PPAR-alpha. Int. J. Mol. Sci. 2019, 20, 1666. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. Mapping the intramolecular communications among different glutamate dehydrogenase states using molecular dynamics. Biomolecules 2021, 11, 798. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pang, Y.; Liu, B. BioSeq-BLM: A platform for analyzing DNA, RNA, and protein sequences based on biological language models. Nucleic Acids Res. 2021, 49, e129. [Google Scholar] [CrossRef]
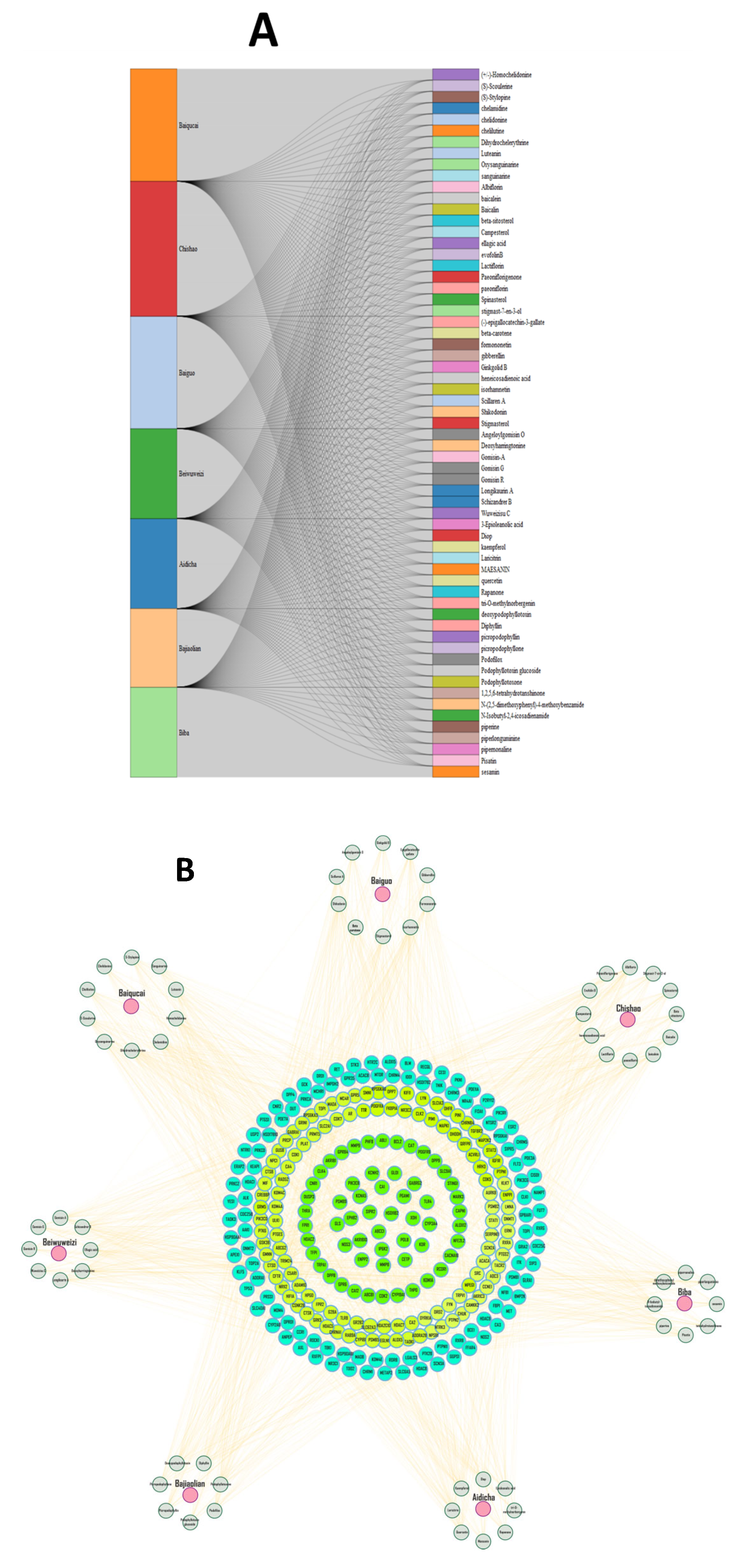
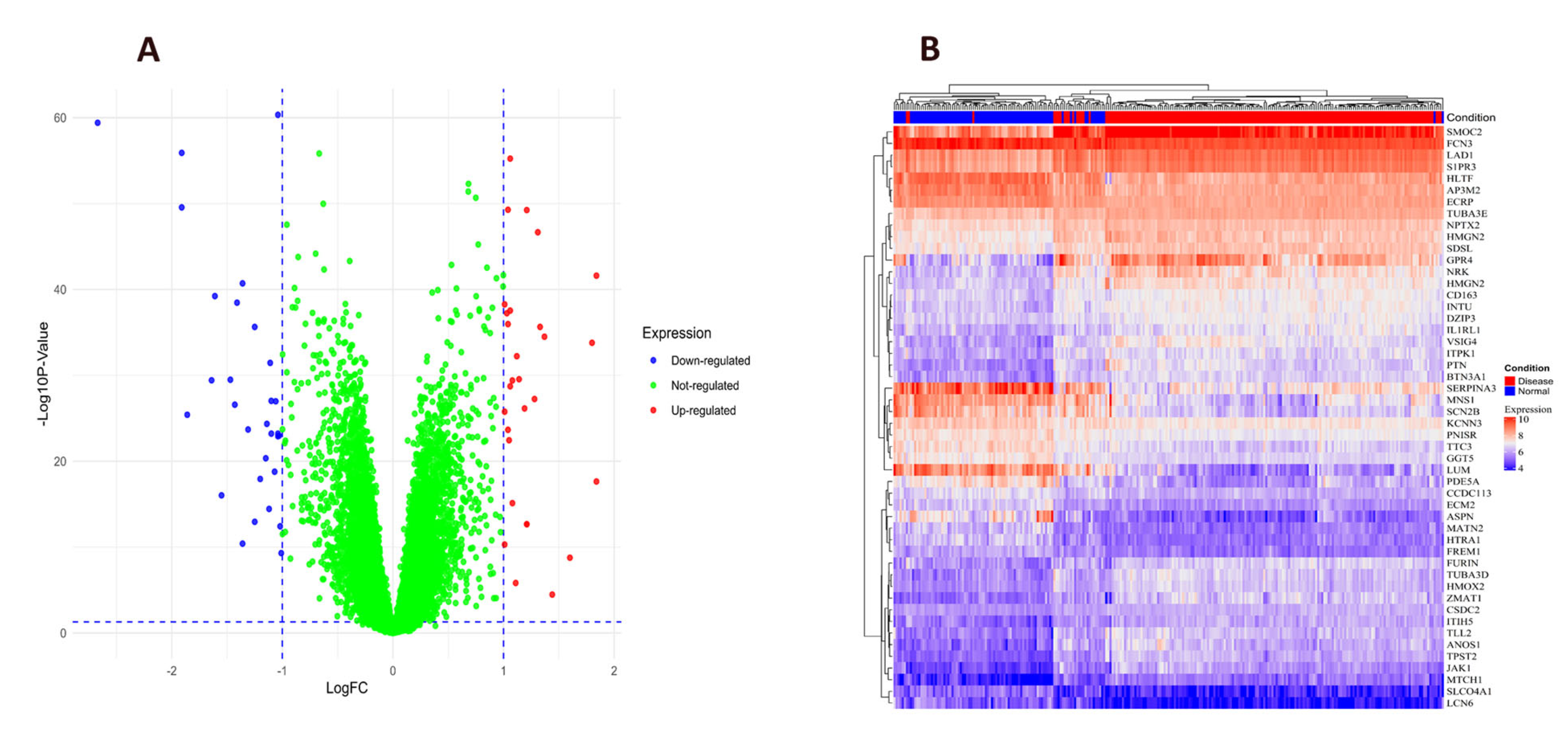

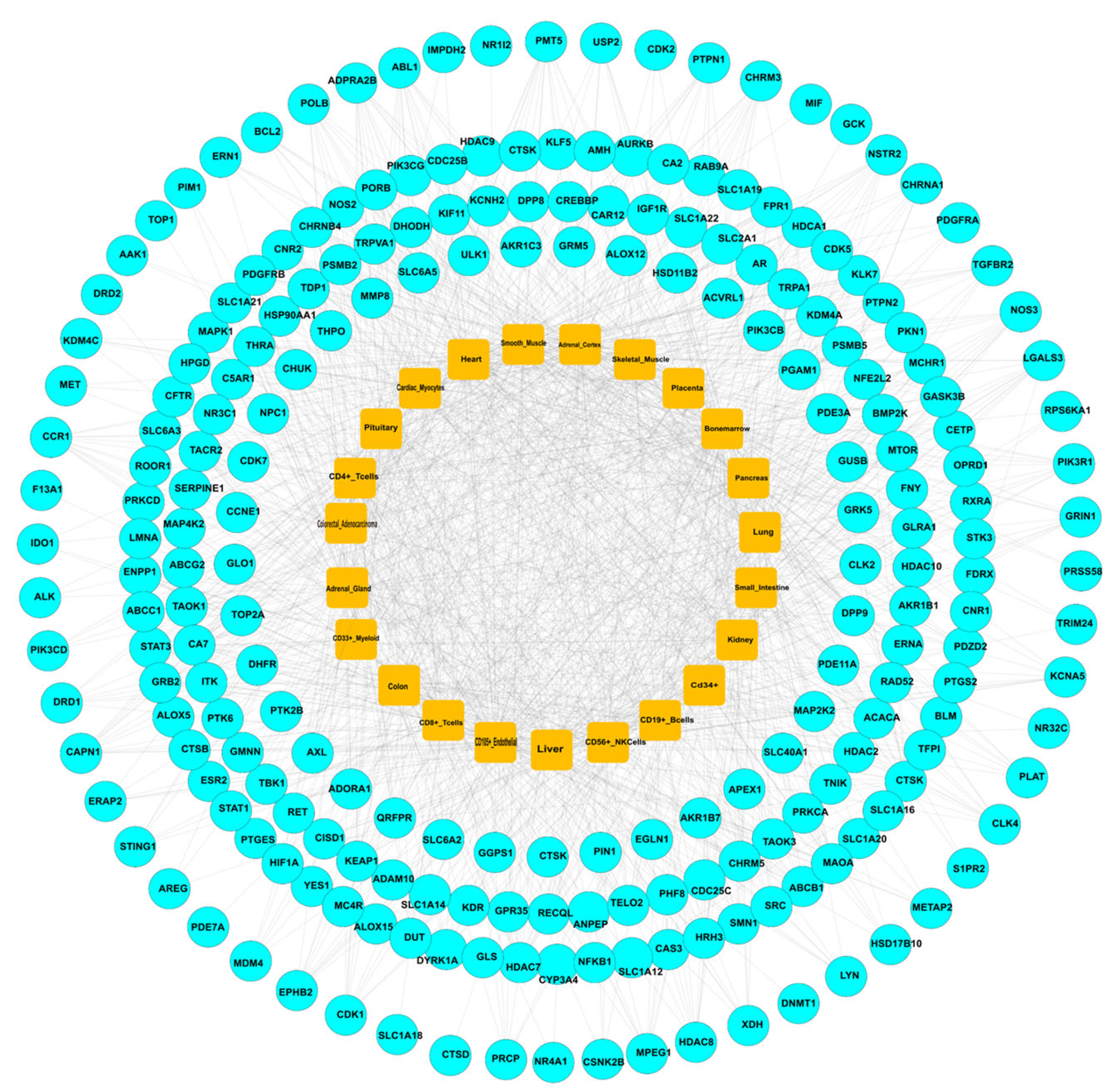
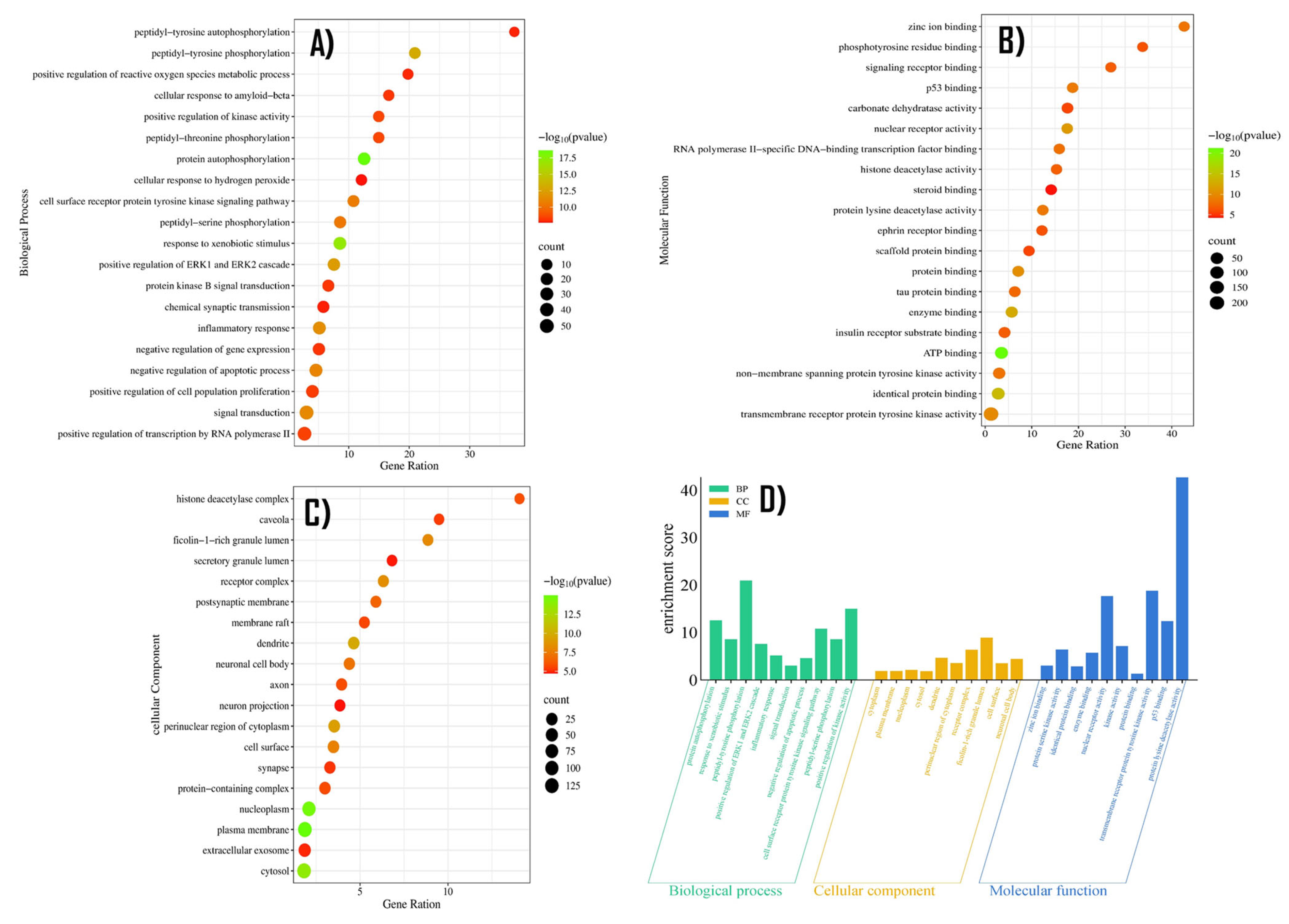
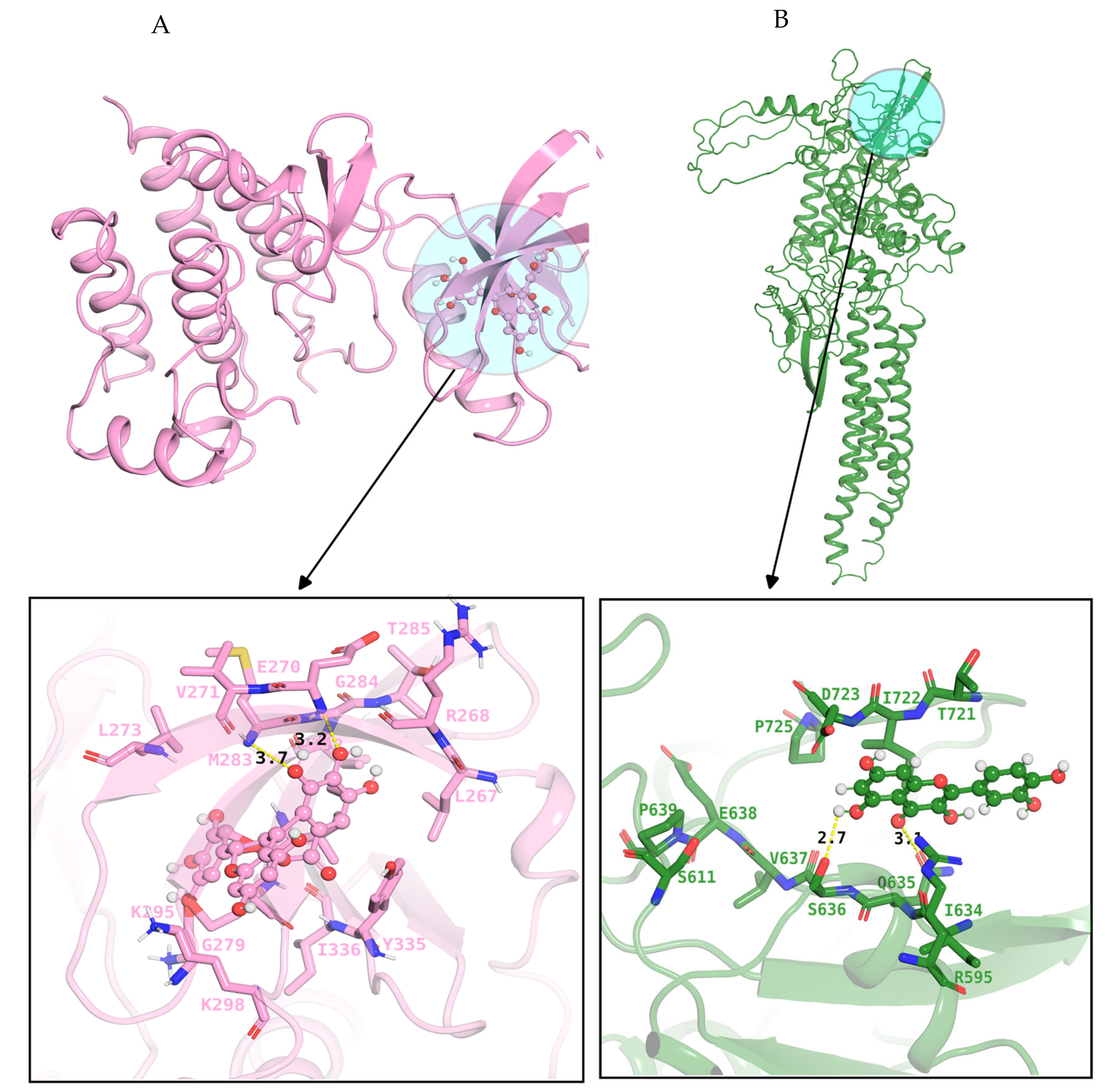
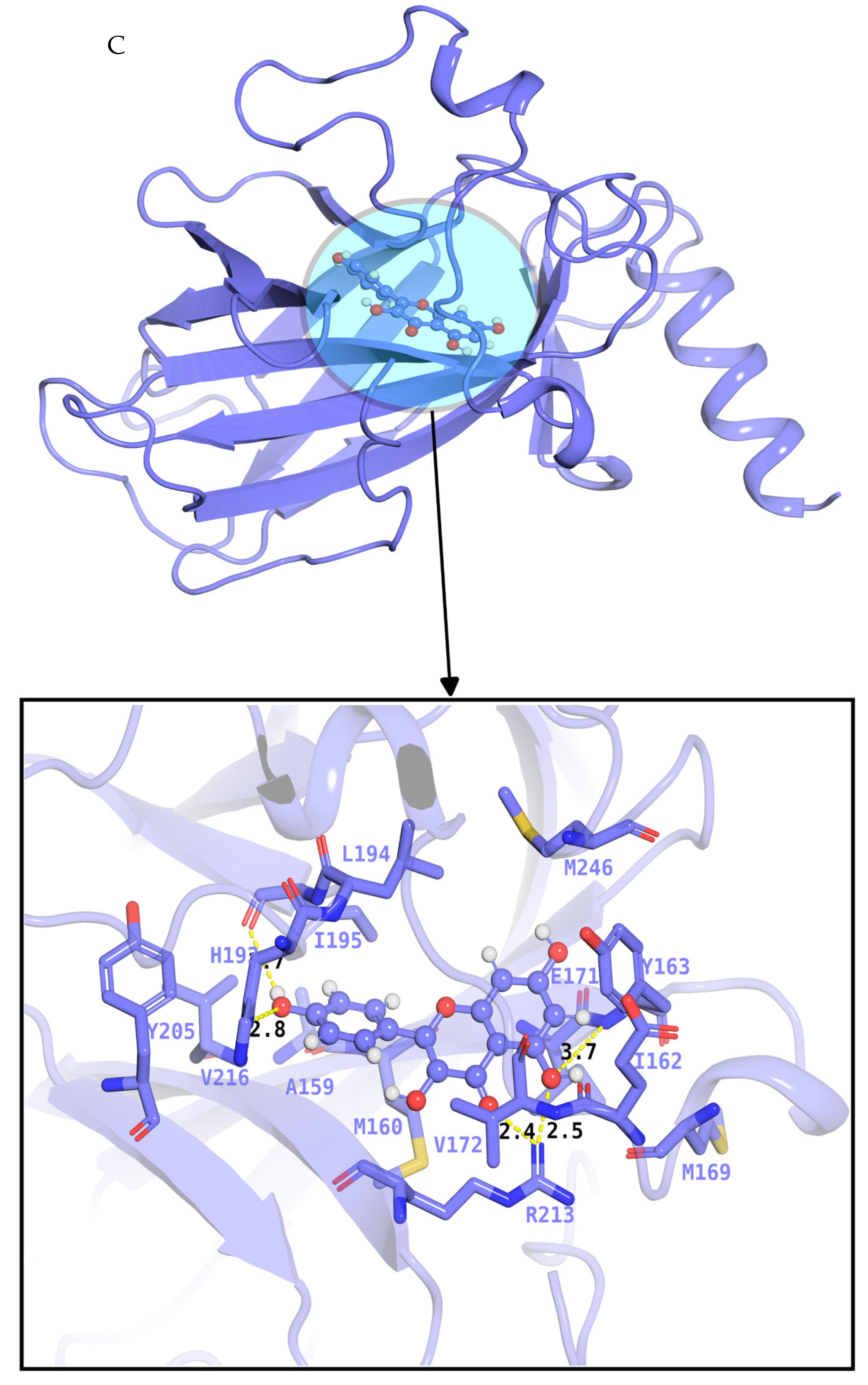
| Gene Symbol | Protein Name | logFC | t | p Value | adj. p Val | B | Change |
|---|---|---|---|---|---|---|---|
| TUBA3E | Tubulin Alpha 3E | −1.04 | −21.8 | 4.68 × 10−61 | 1.56 × 10−56 | 128.0436 | DR |
| SERPINA3 | Alpha-1-antichymotrypsin | −2.67 | −21.5 | 3.97 × 10−60 | 6.61 × 10−56 | 125.9362 | DR |
| FCN3 | Ficolin 3 | −1.91 | −20.5 | 1.26 × 10−56 | 1.24 × 10−52 | 117.9906 | DR |
| FREM1 | FRAS1 Related Extracellular Matrix protein 1 | 1.06 | 20.3 | 5.86 × 10−56 | 3.90 × 10−52 | 116.4756 | UR |
| HMGN2 | Non-histone chromosomal protein HMG-17 | 0.683 | 19.4 | 5.01 × 10−53 | 2.78 × 10−49 | 109.818 | UR |
| ZMAT1 | Zinc finger matrin-type protein 1 | 0.749 | 19 | 2.13 × 10−51 | 8.85 × 10−48 | 106.1184 | DR |
| FURIN | Furin | −0.63 | −18.8 | 1.07 × 10−50 | 3.96 × 10−47 | 104.524 | DR |
| IL1RL1 | Interleukin 1 Receptor Like 1 | −1.91 | −18.7 | 2.79 × 10−50 | 9.30 × 10−47 | 103.577 | DR |
| MNS1 | Meiosis Specific Nuclear structural 1 | 1.04 | 18.6 | 5.28 × 10−50 | 1.59 × 10−46 | 102.9497 | UR |
| SMOC2 | SPARC Related Modular Calcium Binding protein 2 | 1.21 | 18.6 | 5.75 × 10−50 | 1.59 × 10−46 | 102.865 | UR |
| LCN6 | Epididymal-specific lipocalin-6 | −0.959 | −18.1 | 2.88 × 10−48 | 7.38 × 10−45 | 99.0001 | DR |
| LUM | Lumican | 1.31 | 17.8 | 2.20 × 10−47 | 5.23 × 10−44 | 96.99357 | UR |
| KCNN3 | Small conductance calcium-activated potassium channel protein 3 | 0.771 | 17.4 | 5.84 × 10−46 | 1.30 × 10−42 | 93.75647 | UR |
| LAD1 | Ladinin 1 | −0.699 | −17.1 | 6.86 × 10−45 | 1.43 × 10−41 | 91.32271 | UR |
| GGT5 | Glutathione hydrolase 5 proenzyme | −0.857 | −17 | 1.68 × 10−44 | 3.29 × 10−41 | 90.43837 | DR |
| MTCH1 | Mitochondrial carrier homolog 1 | −0.391 | −16.9 | 4.91 × 10−44 | 9.09 × 10−41 | 89.37772 | DR |
| AP3M2 | AP-3 complex subunit mu-2 | 0.529 | 16.8 | 1.39 × 10−43 | 2.44 × 10−40 | 88.35005 | UR |
| ITIH5 | Inter-alpha-trypsin inhibitor heavy chain H5 | 0.85 | 16.7 | 2.88 × 10−43 | 4.80 × 10−40 | 87.62974 | DR |
| S1PR3 | Sphingosine 1-phosphate receptor 3 | −0.623 | −16.6 | 4.81 × 10−43 | 7.62 × 10−40 | 87.12467 | UR |
| ECM2 | Extracellular matrix protein 2 | 0.998 | 16.4 | 2.19 × 10−42 | 3.31 × 10−39 | 85.62849 | DR |
| ASPN | Asporin | 1.84 | 16.4 | 2.49 × 10−42 | 3.60 × 10−39 | 85.50125 | UR |
| SLCO4A1 | Solute carrier organic anion transporter family member 4A1 | 0.936 | −16.2 | 1.97 × 10−41 | 2.63 × 10−38 | 83.45463 | DR |
| PDE5A | cGMP-specific 3′,5′-cyclic phosphodiesterase | −1.36 | 16.1 | 4.56 × 10−41 | 5.84 × 10−38 | 82.62749 | DR |
| NPTX2 | Neuronal pentraxin-2 | 0.996 | −16 | 6.75 × 10−41 | 8.33 × 10−38 | 82.23937 | UR |
| HLTF | Helicase-like transcription factor | −0.89 | 16 | 7.67 × 10−41 | 9.12 × 10−38 | 82.11379 | UR |
| TTC3 | E3 ubiquitin-protein ligase TTC3 | 0.574 | 15.9 | 1.20 × 10−40 | 1.38 × 10−37 | 81.66974 | DR |
| PNISR | Arginine/serine-rich protein PNISR | 0.406 | 15.9 | 2.26 × 10−40 | 2.51 × 10−37 | 81.04558 | UR |
| CD163 | Scavenger receptor cysteine-rich type 1 protein M130 | 0.355 | −15.7 | 5.97 × 10−40 | 6.41 × 10−37 | 80.08657 | DR |
| SDSL | Serine dehydratase-like | −1.61 | 15.7 | 6.18 × 10−40 | 6.43 × 10−37 | 80.05264 | UR |
| CSDC2 | Cold shock domain-containing protein C2 | 0.753 | −15.6 | 2.13 × 10−39 | 2.15 × 10−36 | 78.82751 | DR |
| VSIG4 | V-set and immunoglobulin domain-containing protein 4 | −0.862 | −15.5 | 3.48 × 10−39 | 3.40 × 10−36 | 78.34529 | DR |
| ITPK1 | Inositol-tetrakisphosphate 1-kinase | −1.41 | −15.5 | 4.95 × 10−39 | 4.71 × 10−36 | 77.99655 | DR |
| NRK | Nik-related protein kinase | −0.43 | 15.5 | 5.49 × 10−39 | 5.08 × 10−36 | 77.89357 | UR |
| ECRP | Eosinophil cationic-related protein | 1.01 | −15.4 | 1.17 × 10−38 | 1.05 × 10−35 | 77.14903 | UR |
| TUBA3E | Tubulin alpha-3E chain | −0.909 | −15.4 | 1.28 × 10−38 | 1.12 × 10−35 | 77.05474 | UR |
| MATN2 | Matrilin-2 | −0.881 | 15.4 | 1.36 × 10−38 | 1.16 × 10−35 | 76.99504 | DR |
| ANOS1 | Anosmin-1 | 0.899 | 15.3 | 2.04 × 10−38 | 1.70 × 10−35 | 76.59485 | DR |
| DZIP3 | E3 ubiquitin-protein ligase DZIP3 | 0.78 | 15.3 | 2.88 × 10−38 | 2.29 × 10−35 | 76.25551 | DR |
| TLL2 | Tolloid-like protein 2 | 0.568 | 15.3 | 2.89 × 10−38 | 2.29 × 10−35 | 76.25287 | UR |
| CCDC113 | Cilia- and flagella-associated protein 263 | 1.06 | 15.3 | 3.20 × 10−38 | 2.48 × 10−35 | 76.15123 | DR |
| TPST2 | Protein-tyrosine sulfotransferase 2 | 0.781 | −15.2 | 4.66 × 10−38 | 3.53 × 10−35 | 75.78051 | DR |
| GPR4 | G-protein coupled receptor 4 | −0.424 | −15.2 | 5.40 × 10−38 | 3.99 × 10−35 | 75.63523 | UR |
| PTN | Pleiotrophin | −0.727 | 15.2 | 5.88 × 10−38 | 4.25 × 10−35 | 75.55116 | UR |
| HTRA1 | Serine protease HTRA1 | 1.03 | 15.1 | 8.40 × 10−38 | 5.95 × 10−35 | 75.19841 | DR |
| JAK1 | Tyrosine-protein kinase JAK1 | 0.58 | −15.1 | 9.85 × 10−38 | 6.83 × 10−35 | 75.04061 | DR |
| BTN3A1 | Butyrophilin subfamily 3 members A1 | −0.309 | 15.1 | 1.15 × 10−37 | 7.79 × 10−35 | 74.89022 | DR |
| SCN2B | Sodium channel regulatory subunit beta-2 | 0.699 | 15 | 1.85 × 10−37 | 1.23 × 10−34 | 74.41531 | UR |
| INTU | Protein inturned | 0.86 | 15 | 2.29 × 10−37 | 1.50 × 10−34 | 74.20487 | UR |
| No | Uniport IDs | Gene Symbol | Protein Name |
|---|---|---|---|
| 1 | P04637 | TP53 | Cellular tumor antigen p53 |
| 2 | P40763 | STAT3 | Signal transducer and activator of transcription 3 |
| 3 | P12931 | SRC | Proto-oncogene tyrosine kinase |
| 4 | P07900 | HSP90AA1 | Heat shock protein HSP90-alpha |
| 5 | P08238 | HSP90AB1 | Heat shock protein HSP90-beta |
| 6 | P27986 | PIK3R1 | Phosphatidylinositol 3-kinase |
| 7 | Q16665 | HIF1A | Hypoxia-inducible factor 1-alpha |
| 8 | P62993 | GRB2 | Growth factor receptor bound protein |
| 9 | P10415 | BCL2 | Apoptosis Regulator BcI-2 |
| 10 | P42224 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta |
| Name | MW(g/mol) | GI | BBB | PGP | BS | nHBA | nHBD | TPSA(Å) | iLOGP | WLOG | nLV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| quercetin | 302 | high | No | no | 0.55 | 7 | 5 | 131.36 | 1.63 | 1.99 | 0 |
| kaempferol | 286 | high | No | no | 0.57 | 6 | 4 | 111.13 | 1.7 | 2.28 | 0 |
| epigallocatechin | 458 | high | No | no | 0.63 | 9 | 5 | 146.14 | 1.87 | 1.91 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, B.; Ma, C.-Y.; Bakanina Kissanga, G.-M.; Temesgen, S.A.; Fida, H.; Lin, H.; Huang, C.-B. Unveiling the Therapeutic Mechanisms of Chinese Herbs in Heart Failure: Integrating Network Pharmacology, Molecular Docking, and Simulation Analysis. Pharmaceuticals 2025, 18, 1648. https://doi.org/10.3390/ph18111648
Ahmad B, Ma C-Y, Bakanina Kissanga G-M, Temesgen SA, Fida H, Lin H, Huang C-B. Unveiling the Therapeutic Mechanisms of Chinese Herbs in Heart Failure: Integrating Network Pharmacology, Molecular Docking, and Simulation Analysis. Pharmaceuticals. 2025; 18(11):1648. https://doi.org/10.3390/ph18111648
Chicago/Turabian StyleAhmad, Basharat, Cai-Yi Ma, Grace-Mercure Bakanina Kissanga, Sebu Aboma Temesgen, Huma Fida, Hao Lin, and Cheng-Bing Huang. 2025. "Unveiling the Therapeutic Mechanisms of Chinese Herbs in Heart Failure: Integrating Network Pharmacology, Molecular Docking, and Simulation Analysis" Pharmaceuticals 18, no. 11: 1648. https://doi.org/10.3390/ph18111648
APA StyleAhmad, B., Ma, C.-Y., Bakanina Kissanga, G.-M., Temesgen, S. A., Fida, H., Lin, H., & Huang, C.-B. (2025). Unveiling the Therapeutic Mechanisms of Chinese Herbs in Heart Failure: Integrating Network Pharmacology, Molecular Docking, and Simulation Analysis. Pharmaceuticals, 18(11), 1648. https://doi.org/10.3390/ph18111648








