The Other Side of the Same Coin: Beyond the Coding Region in Amyotrophic Lateral Sclerosis
Abstract
1. Introduction
2. Transposable Elements in the Human Genome
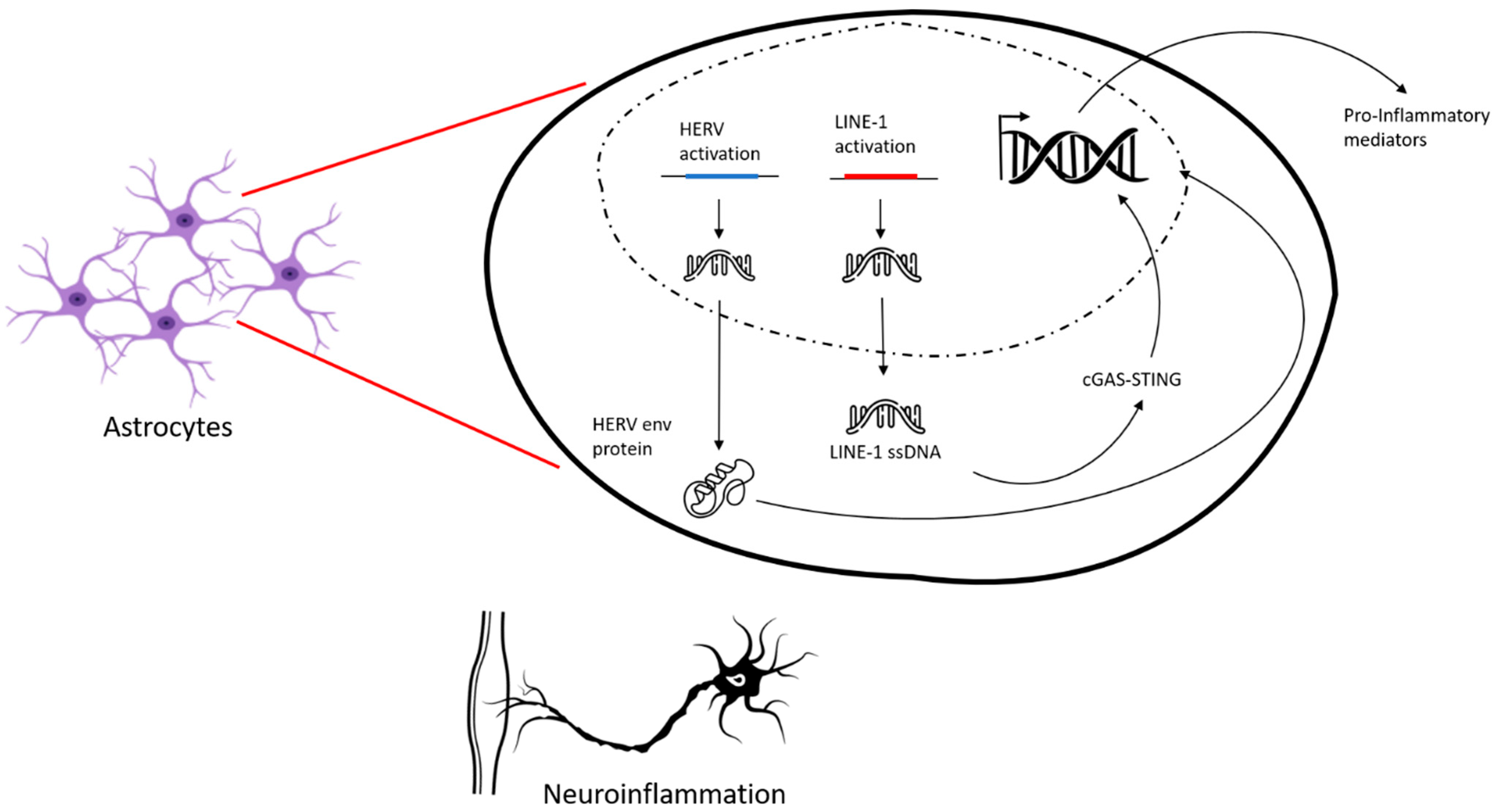
3. ALS and Transposable Elements
4. Epigenetic Regulation of Transposable Elements
5. Transposable Elements as Therapeutic Targets
6. Biochemical Parameters and Transposable Elements in ALS Patients
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | Antiretroviral Backbone Combination (part of Triumeq therapy) |
| AD | Alzheimer’s Disease |
| ALS | Amyotrophic Lateral Sclerosis |
| Alu | Short Interspersed Nuclear Element (type of SINE) |
| ART | Antiretroviral Therapy |
| AZT | Zidovudine |
| cDNA | Complementary DNA |
| cGAS | Cyclic GMP-AMP Synthase |
| CNS | Central Nervous System |
| DNMT | DNA Methyltransferase |
| dsRNA | Double-Stranded RNA |
| EED | Embryonic Ectoderm Development |
| EN | Endonuclease |
| ERV | Endogenous Retrovirus |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FTD | Frontotemporal Dementia |
| FUS | Fused in Sarcoma |
| fALS | Familial ALS |
| FKTN | Fukutin |
| HD | Huntington’s Disease |
| HDAC | Histone Deacetylase |
| H3K27me3 | Histone H3 Lysine 27 Trimethylation |
| H3K9me3 | Histone H3 Lysine 9 Trimethylation |
| HERV | Human Endogenous Retrovirus |
| HP1 | Heterochromatin Protein 1 |
| IFN-I | Type I Interferon |
| IL-6 | Interleukin 6 |
| KRAB-ZFP | Krüppel-Associated Box Zinc Finger Protein |
| KAP1 | KRAB-associated protein 1 |
| LINE-1/L1 | Long Interspersed Nuclear Element-1 |
| LTR | Long Terminal Repeat |
| lncRNA | Long Non-Coding RNA |
| MAPT | Microtubule-Associated Protein Tau |
| m6A | N6-Methyladenosine |
| MeCP2 | Methyl-CpG-binding protein 2 |
| miRNA | MicroRNA |
| MS | Multiple Sclerosis |
| NfH | Neurofilament Heavy Chain |
| NfL | Neurofilament Light Chain |
| PBMC | Peripheral Blood Mononuclear Cells |
| PD | Parkinson’s Disease |
| piRNA | PIWI-interacting RNA |
| PRC2 | Polycomb Repressive Complex 2 |
| RIG-I | Retinoic acid-Inducible Gene I |
| RLR | RIG-I-like Receptor |
| RT | Reverse Transcriptase |
| SAM | S-adenosylmethionine |
| SETDB1 | SET Domain Bifurcated 1 (a H3K9 methyltransferase) |
| siRNA | Small Interfering RNA |
| SINE | Short Interspersed Nuclear Element |
| SMARCAD1 | SWI/SNF-related Matrix-Associated Actin-Dependent Regulator of Chromatin A1 |
| SOD1 | Superoxide Dismutase 1 |
| ssDNA | Single-Stranded DNA |
| STING | Stimulator of Interferon Genes |
| SUZ12 | Suppressor of Zeste 12 |
| SVA | SINE-VNTR-Alu |
| sALS | Sporadic ALS |
| SWI/SNF | SWItch/Sucrose Non-Fermentable (chromatin remodeling complex) |
| TAD | Topologically Associating Domain |
| TAF1 | TATA-Box Binding Protein Associated Factor 1 |
| TE | Transposable Element |
| TDP-43 | TAR DNA-binding Protein 43 |
| TNF-α | Tumor Necrosis Factor Alpha |
| UTR | Untranslated Region |
| XDP | X-linked Dystonia-Parkinsonism |
References
- Talbott, E.O.; Malek, A.M.; Lacomis, D. The epidemiology of amyotrophic lateral sclerosis. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 138, pp. 225–238. [Google Scholar] [CrossRef]
- Masrori, P.; Van Damme, P. Amyotrophic lateral sclerosis: A clinical review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Longinetti, E.; Fang, F. Epidemiology of amyotrophic lateral sclerosis: An update of recent literature. Curr. Opin. Neurol. 2019, 32, 771–776. [Google Scholar] [CrossRef]
- Wolfson, C.; Gauvin, D.E.; Ishola, F.; Oskoui, M. Global Prevalence and Incidence of Amyotrophic Lateral Sclerosis: A Systematic Review. Neurology 2023, 101, e613–e623. [Google Scholar] [CrossRef]
- Parobkova, E.; Matej, R. Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degenerations: Similarities in Genetic Background. Diagnostics 2021, 11, 509. [Google Scholar] [CrossRef]
- Oskarsson, B.; Horton, D.K.; Mitsumoto, H. Potential Environmental Factors in Amyotrophic Lateral Sclerosis. Neurol. Clin. 2015, 33, 877–888. [Google Scholar] [CrossRef]
- Ruffo, P.; Traynor, B.J.; Conforti, F.L. Advancements in genetic research and RNA therapy strategies for amyotrophic lateral sclerosis (ALS): Current progress and future prospects. J. Neurol. 2025, 272, 233. [Google Scholar] [CrossRef] [PubMed]
- Rizea, R.E.; Corlatescu, A.D.; Costin, H.P.; Dumitru, A.; Ciurea, A.V. Understanding Amyotrophic Lateral Sclerosis: Pathophysiology, Diagnosis, and Therapeutic Advances. Int. J. Mol. Sci. 2024, 25, 9966. [Google Scholar] [CrossRef] [PubMed]
- Ruffo, P.; Traynor, B.J.; Conforti, F.L. Unveiling the regulatory potential of the non-coding genome: Insights from the human genome project to precision medicine. Genes Dis. 2025, 12, 101652. [Google Scholar] [CrossRef]
- Han, M.; Perkins, M.H.; Novaes, L.S.; Xu, T.; Chang, H. Advances in transposable elements: From mechanisms to applications in mammalian genomics. Front. Genet. 2023, 14, 1290146. [Google Scholar] [CrossRef]
- Chenais, B. Transposable Elements and Human Diseases: Mechanisms and Implication in the Response to Environmental Pollutants. Int. J. Mol. Sci. 2022, 23, 2551. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Rubanova, N.; Singh, D.; Barolle, L.; Chalvet, F.; Netter, S.; Poidevin, M.; Servant, N.; Bardin, A.J.; Siudeja, K. An endogenous retroviral element co-opts an upstream regulatory sequence to achieve somatic expression and mobility. Nucleic Acids Res. 2025, 53, gkaf485. [Google Scholar] [CrossRef]
- Tang, W.; Liang, P. The identification of retro-DNAs in primate genomes as DNA transposons mobilizing via retrotransposition. F1000Research 2023, 12, 255. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Piegu, B.; Bire, S.; Arensburger, P.; Bigot, Y. A survey of transposable element classification systems—A call for a fundamental update to meet the challenge of their diversity and complexity. Mol. Phylogenet. Evol. 2015, 86, 90–109. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Ardeljan, D.; Taylor, M.S.; Ting, D.T.; Burns, K.H. The Human Long Interspersed Element-1 Retrotransposon: An Emerging Biomarker of Neoplasia. Clin. Chem. 2017, 63, 816–822. [Google Scholar] [CrossRef]
- Gianfrancesco, O.; Geary, B.; Savage, A.L.; Billingsley, K.J.; Bubb, V.J.; Quinn, J.P. The Role of SINE-VNTR-Alu (SVA) Retrotransposons in Shaping the Human Genome. Int. J. Mol. Sci. 2019, 20, 5977. [Google Scholar] [CrossRef] [PubMed]
- Prokopov, D.; Tunbak, H.; Leddy, E.; Drylie, B.; Camera, F.; Deniz, O. Transposable elements as genome regulators in normal and malignant haematopoiesis. Blood Cancer J. 2025, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.N.; Friedman, R.Z.; Wang, J.T.; Jang, H.S.; Zhuo, X.; Wang, T. Co-opted transposons help perpetuate conserved higher-order chromosomal structures. Genome Biol. 2020, 21, 16. [Google Scholar] [CrossRef]
- Bragg, D.C.; Mangkalaphiban, K.; Vaine, C.A.; Kulkarni, N.J.; Shin, D.; Yadav, R.; Dhakal, J.; Ton, M.L.; Cheng, A.; Russo, C.T.; et al. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc. Natl. Acad. Sci. USA 2017, 114, E11020–E11028. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kobayashi, K.; Jin, F.; Park, K.S.; Yamada, T.; Tokunaga, K.; Toda, T. Founder SVA retrotransposal insertion in Fukuyama-type congenital muscular dystrophy and its origin in Japanese and Northeast Asian populations. Am. J. Med. Genet. A 2005, 138, 344–348. [Google Scholar] [CrossRef]
- Cheng, Y.; Saville, L.; Gollen, B.; Veronesi, A.A.; Mohajerani, M.; Joseph, J.T.; Zovoilis, A. Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes. EMBO Rep. 2021, 22, e52255. [Google Scholar] [CrossRef]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef]
- Floreani, L.; Ansaloni, F.; Mangoni, D.; Agostoni, E.; Sanges, R.; Persichetti, F.; Gustincich, S. Analysis of LINE1 Retrotransposons in Huntington’s Disease. Front. Cell. Neurosci. 2021, 15, 743797. [Google Scholar] [CrossRef]
- Prudencio, M.; Gonzales, P.K.; Cook, C.N.; Gendron, T.F.; Daughrity, L.M.; Song, Y.; Ebbert, M.T.W.; van Blitterswijk, M.; Zhang, Y.J.; Jansen-West, K.; et al. Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum. Mol. Genet. 2017, 26, 3421–3431. [Google Scholar] [CrossRef]
- Li, W.; Lee, M.H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef] [PubMed]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Muotri, A.R.; Marchetto, M.C.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature 2010, 468, 443–446. [Google Scholar] [CrossRef]
- Morandi, E.; Tanasescu, R.; Tarlinton, R.E.; Constantinescu, C.S.; Zhang, W.; Tench, C.; Gran, B. The association between human endogenous retroviruses and multiple sclerosis: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0172415. [Google Scholar] [CrossRef]
- Tamouza, R.; Meyer, U.; Foiselle, M.; Richard, J.R.; Wu, C.L.; Boukouaci, W.; Le Corvoisier, P.; Barrau, C.; Lucas, A.; Perron, H.; et al. Correction: Identification of inflammatory subgroups of schizophrenia and bipolar disorder patients with HERV-W ENV antigenemia by unsupervised cluster analysis. Transl. Psychiatry 2021, 11, 447. [Google Scholar] [CrossRef]
- Zhao, B.; Nguyen, M.A.; Woo, S.; Kim, J.; Yu, T.W.; Lee, E.A. Contribution and therapeutic implications of retroelement insertions in ataxia telangiectasia. Am. J. Hum. Genet. 2023, 110, 1976–1982. [Google Scholar] [CrossRef]
- Luth, T.; Labeta, J.; Schaake, S.; Wohlers, I.; Pozojevic, J.; Jamora, R.D.G.; Rosales, R.L.; Bruggemann, N.; Saranza, G.; Diesta, C.C.E.; et al. Elucidating Hexanucleotide Repeat Number and Methylation within the X-Linked Dystonia-Parkinsonism (XDP)-Related SVA Retrotransposon in TAF1 with Nanopore Sequencing. Genes 2022, 13, 126. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Ivancevic, A.M.; Kortschak, R.D.; Bertozzi, T.; Adelson, D.L. LINEs between Species: Evolutionary Dynamics of LINE-1 Retrotransposons across the Eukaryotic Tree of Life. Genome Biol. Evol. 2016, 8, 3301–3322. [Google Scholar] [CrossRef]
- Comer, A.L.; Jinadasa, T.; Sriram, B.; Phadke, R.A.; Kretsge, L.N.; Nguyen, T.P.H.; Antognetti, G.; Gilbert, J.P.; Lee, J.; Newmark, E.R.; et al. Increased expression of schizophrenia-associated gene C4 leads to hypoconnectivity of prefrontal cortex and reduced social interaction. PLoS Biol. 2020, 18, e3000604. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.S.; Fablet, M.; Larue, A.; Vallier, A.; Carareto, C.M.A.; Rebollo, R.; Vieira, C. ChimeraTE: A pipeline to detect chimeric transcripts derived from genes and transposable elements. Nucleic Acids Res. 2023, 51, 9764–9784. [Google Scholar] [CrossRef]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef]
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 2014, 81, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Huang, Q.; Boeke, J.D. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 2011, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Adler, G.L.; Le, K.; Fu, Y.; Kim, W.S. Human Endogenous Retroviruses in Neurodegenerative Diseases. Genes 2024, 15, 745. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Dellaire, G. LINE-1: An emerging initiator of cGAS-STING signalling and inflammation that is dysregulated in disease. Biochem. Cell Biol. 2024, 102, 38–46. [Google Scholar] [CrossRef]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef]
- Fukuda, S.; Varshney, A.; Fowler, B.J.; Wang, S.B.; Narendran, S.; Ambati, K.; Yasuma, T.; Magagnoli, J.; Leung, H.; Hirahara, S.; et al. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2021, 118, e2022751118. [Google Scholar] [CrossRef]
- Li, W.; Pandya, D.; Pasternack, N.; Garcia-Montojo, M.; Henderson, L.; Kozak, C.A.; Nath, A. Retroviral Elements in Pathophysiology and as Therapeutic Targets for Amyotrophic Lateral Sclerosis. Neurotherapeutics 2022, 19, 1085–1101. [Google Scholar] [CrossRef]
- Hughes, L.S.; Frohlich, A.; Pfaff, A.L.; Bubb, V.J.; Quinn, J.P.; Koks, S. Exploring SVA Insertion Polymorphisms in Shaping Differential Gene Expressions in the Central Nervous System. Biomolecules 2024, 14, 358. [Google Scholar] [CrossRef]
- Valdebenito-Maturana, B.; Rojas-Tapia, M.I.; Carrasco, M.; Tapia, J.C. Dysregulated Expression of Transposable Elements in TDP-43(M337V) Human Motor Neurons That Recapitulate Amyotrophic Lateral Sclerosis In Vitro. Int. J. Mol. Sci. 2022, 23, 16222. [Google Scholar] [CrossRef]
- Pfaff, A.L.; Kõks, S. Retrotransposition-competent L1s are increased in the genomes of individuals with amyotrophic lateral sclerosis. Exp. Biol. Med. 2025, 250, 10575. [Google Scholar] [CrossRef]
- Macia, A.; Munoz-Lopez, M.; Cortes, J.L.; Hastings, R.K.; Morell, S.; Lucena-Aguilar, G.; Marchal, J.A.; Badge, R.M.; Garcia-Perez, J.L. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol. Cell. Biol. 2011, 31, 300–316. [Google Scholar] [CrossRef]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.M.; Chen, D.; Yin, J.; et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020, 11, 3355. [Google Scholar] [CrossRef]
- Levinsky, A.J.; McEdwards, G.; Sethna, N.; Currie, M.A. Targets of histone H3 lysine 9 methyltransferases. Front. Cell Dev. Biol. 2022, 10, 1026406. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J., 3rd. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafo, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell. Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Schwaemmle, H.; Soldati, H.; Lykoskoufis, N.M.R.; Docquier, M.; Hainard, A.; Braun, S.M.G. CRISPR screen decodes SWI/SNF chromatin remodeling complex assembly. Nat. Commun. 2025, 16, 5011. [Google Scholar] [CrossRef]
- Abakir, A.; Giles, T.C.; Cristini, A.; Foster, J.M.; Dai, N.; Starczak, M.; Rubio-Roldan, A.; Li, M.; Eleftheriou, M.; Crutchley, J.; et al. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 2020, 52, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Savage, A.L.; Schumann, G.G.; Breen, G.; Bubb, V.J.; Al-Chalabi, A.; Quinn, J.P. Retrotransposons in the development and progression of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 284–293. [Google Scholar] [CrossRef]
- Burg, T.; Rossaert, E.; Moisse, M.; Van Damme, P.; Van Den Bosch, L. Histone Deacetylase Inhibition Regulates Lipid Homeostasis in a Mouse Model of Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2021, 22, 11224. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Li, L.; Wang, Q.; Le, W. Decreased level of 5-methyltetrahydrofolate: A potential biomarker for pre-symptomatic amyotrophic lateral sclerosis. J. Neurol. Sci. 2010, 293, 102–105. [Google Scholar] [CrossRef]
- Lanciano, S.; Philippe, C.; Sarkar, A.; Pratella, D.; Domrane, C.; Doucet, A.J.; van Essen, D.; Saccani, S.; Ferry, L.; Defossez, P.A.; et al. Locus-level L1 DNA methylation profiling reveals the epigenetic and transcriptional interplay between L1s and their integration sites. Cell Genom. 2024, 4, 100498. [Google Scholar] [CrossRef]
- Zoccolella, S.; Bendotti, C.; Beghi, E.; Logroscino, G. Homocysteine levels and amyotrophic lateral sclerosis: A possible link. Amyotroph. Lateral Scler. 2010, 11, 140–147. [Google Scholar] [CrossRef]
- Valentino, F.; Bivona, G.; Butera, D.; Paladino, P.; Fazzari, M.; Piccoli, T.; Ciaccio, M.; La Bella, V. Elevated cerebrospinal fluid and plasma homocysteine levels in ALS. Eur. J. Neurol. 2010, 17, 84–89. [Google Scholar] [CrossRef]
- Liu, J.; Luo, X.; Chen, X.; Shang, H. Lipid Profile in Patients With Amyotrophic Lateral Sclerosis: A Systematic Review and Meta-Analysis. Front. Neurol. 2020, 11, 567753. [Google Scholar] [CrossRef] [PubMed]
- Dodge, J.C.; Jensen, E.H.; Yu, J.; Sardi, S.P.; Bialas, A.R.; Taksir, T.V.; Bangari, D.S.; Shihabuddin, L.S. Neutral Lipid Cacostasis Contributes to Disease Pathogenesis in Amyotrophic Lateral Sclerosis. J. Neurosci. 2020, 40, 9137–9147. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, L.; Corcia, P.; Fergani, A.; Gonzalez De Aguilar, J.L.; Bonnefont-Rousselot, D.; Bittar, R.; Seilhean, D.; Hauw, J.J.; Lacomblez, L.; Loeffler, J.P.; et al. Dyslipidemia is a protective factor in amyotrophic lateral sclerosis. Neurology 2008, 70, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, E.P.; Williams, N.E.; Calvey, T.N. Differential effects of neuromuscular blocking agents on suxamethonium-induced fasciculations and myalgia. Br. J. Anaesth. 1988, 60, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Arru, G.; Galleri, G.; Deiana, G.A.; Zarbo, I.R.; Sechi, E.; Bo, M.; Cadoni, M.P.L.; Corda, D.G.; Frau, C.; Simula, E.R.; et al. HERV-K Modulates the Immune Response in ALS Patients. Microorganisms 2021, 9, 1784. [Google Scholar] [CrossRef] [PubMed]

| Associated Disorders | TEs Type | TEs Activity | References |
|---|---|---|---|
| AD | Alu LINEs | Increased Alu RNA processing in the brain → altered gene expression (e.g., genes related to synaptic transmission and inflammation). Accumulation of cytoplasmic Alu RNA correlates with oxidative stress and neuronal damage. Widespread activation of L1 and ERVs associated with neurofibrillary tangle burden. | [25,26] |
| HD | LINEs | Inverse correlation between L1 expression and CAG repeat length | [27] |
| ALS/FTD | Alu | Dysregulation of transcripts of Alu elements including AluYk12 and AluYa5 | [28] |
| ALS | HERVs LINEs | HERV-K potentially contributes to neurodegeneration due to toxic effects of HML-2 Envelope (Env) protein; Overexpression of the HERV-K(HML-2) envelope protein causes motor neuron toxicity and motor dysfunction in transgenic mice; L1 retrotransposition | [29,30] |
| Rett syndrome | LINEs, Alu | Methylated MeCP2 is involved in the control of L1 mobility in the nervous system; MeCP2 represses L1 expression and transposition; In MECP2-KO mouse models, L1 is derepressed and can transpose into the brain. | [31] |
| MS | HERVs | Increased expression of HERV-H, HERV-W families. | [32] |
| Schizophrenia | HERVs | Higher HERV-W env RNA expression | [33] |
| Ataxia telangiectasia | LINEs | L1 retrotransposition | [34] |
| XDP | SVA | Reduced TAF1 expression due to SVA element integration Hypermethylation of the TAF1 SVA insertion | [35] |
| Therapy/Strategy | Targeted TE | Mechanism of Action | Clinical Trial | Phase | Notes/Biomarkers |
|---|---|---|---|---|---|
| TPN-101 (Transposon Tx) | LINE-1 | Selective reverse transcriptase inhibition | NCT05136885 (HEALEY) | Phase 2/3 | ↓ NfL/NfH, ↓ IL-6, 50% slower SVC decline |
| TPN-101 | LINE-1 | Same mechanism, tested in C9orf72 ALS/FTD | NCT04993755 | Phase 2 | Completed. Promising biomarker effects; initial ALS-specific results encouraging |
| Triumeq (ART: ABC therapy) | HERV-K | Inhibits retroviral replication (RT inhibition + integrase block) | NCT06658977 (Lighthouse II) | Phase 3 | Terminated early (futility: inconsistent efficacy despite reduction of HERV-K env levels |
| Zidovudine (AZT) | HERV-K | Nucleoside RT inhibitor (legacy HIV drug) | Preclinical | N/A | ↓ HERV-K expression in vitro; limited CNS penetration |
| Lamivudine | LINE-1, HERVs | Broad RT inhibition | NCT03280198 | Phase 2 | TE suppression in vitro; no published results |
| HDAC inhibitors | Indirect (LINE-1/HERVs) | Epigenetic silencing via chromatin remodeling | NCT05021536 (PHOENIX trial) | Phase 3 | ↓ neuroinflammation, improved epigenetic repression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruffo, P.; Perrone, B.; Perrone, F.; De Amicis, F.; Iuliano, R.; Bucci, C.; Messina, A.; Conforti, F.L. The Other Side of the Same Coin: Beyond the Coding Region in Amyotrophic Lateral Sclerosis. Pharmaceuticals 2025, 18, 1573. https://doi.org/10.3390/ph18101573
Ruffo P, Perrone B, Perrone F, De Amicis F, Iuliano R, Bucci C, Messina A, Conforti FL. The Other Side of the Same Coin: Beyond the Coding Region in Amyotrophic Lateral Sclerosis. Pharmaceuticals. 2025; 18(10):1573. https://doi.org/10.3390/ph18101573
Chicago/Turabian StyleRuffo, Paola, Benedetta Perrone, Francesco Perrone, Francesca De Amicis, Rodolfo Iuliano, Cecilia Bucci, Angela Messina, and Francesca Luisa Conforti. 2025. "The Other Side of the Same Coin: Beyond the Coding Region in Amyotrophic Lateral Sclerosis" Pharmaceuticals 18, no. 10: 1573. https://doi.org/10.3390/ph18101573
APA StyleRuffo, P., Perrone, B., Perrone, F., De Amicis, F., Iuliano, R., Bucci, C., Messina, A., & Conforti, F. L. (2025). The Other Side of the Same Coin: Beyond the Coding Region in Amyotrophic Lateral Sclerosis. Pharmaceuticals, 18(10), 1573. https://doi.org/10.3390/ph18101573











