Abstract
Background/Objectives: Glioblastoma (GBM), the most aggressive primary malignant brain tumor, has a dismal prognosis and limited treatment options. The dried rhizome of Ligusticum chuanxiong Hort. (Chuanxiong, CX) is a traditional Chinese medicinal herb frequently prescribed in formulas intended to invigorate blood circulation. CX also exhibits anti-glioma activity, but its molecular mechanisms remain incompletely understood. Methods: In this study, we combined transcriptomics and Raman spectroscopy to investigate the effects of reconstituted CX-dispensing granules (hereafter referred to as CXG solution) on U87MG cells, suggesting their dual role in promoting cell death and modulating collagen deposition and lipid metabolism. Results: Mechanistically, we demonstrated that the CXG solution downregulates hsa-miR-10a-5p, which directly targets BCL2L11, known to induce pro-apoptotic effects, as validated by qPCR and dual-luciferase reporter assays. Furthermore, the CXG solution and hsa-miR-10a-5p suppress lipid metabolism through a coherent feed-forward loop via targeting transcription factors SREBF1 and E2F1. An electrophoretic mobility shift assay (EMSA) confirmed E2F1 binds to the hsa-miR-29a promoter, leading to the synergistic repression of hsa-miR-29a-3p by SREBF1 and E2F1. Network pharmacology analysis combined with molecular docking suggested that the ferulic acid and adenosine in CX potentially modulate EGFR-the E2F1-hsa-miR-10a-5p axis. Conclusions: These findings elucidate CX’s multi-target anti-GBM mechanisms and propose a novel therapeutic strategy combining metabolic intervention with miRNA-targeted therapy, providing novel insights into feed-forward loop regulation in miRNA networks.
1. Introduction
Glioblastoma (GBM) is the most common and most aggressive primary malignant tumor of the central nervous system, characterized by rapid progression, high invasiveness, and a high rate of recurrence. The overall 2-year survival rate of GBM patients is 18–33%, regardless of advanced treatment that includes maximal surgical resection, followed by radiotherapy plus concomitant and maintenance temozolomide chemotherapy [1,2,3]. One of the major challenges in GBM treatment lies in the development of therapeutic resistance and frequent tumor relapse. Emerging evidence suggests that metabolic reprogramming, particularly involving lipid metabolism [4] and extracellular matrix remodeling, such as collagen deposition, plays critical roles in tumor progression, therapy resistance, and poor prognosis [5].
Traditional Chinese medicine (TCM), with its multi-target and system-level therapeutic approach, has gained increasing attention for its potential in treating complex diseases, such as GBM, and those related to metabolic syndrome. Classified as spicy-warm and entering the liver, gallbladder, and pericardium meridians, CX disperses wind-cold and mobilizes both qi and blood; consequently, practitioners prescribe it alone or in classic formulas such as Chuanxiong Cha-Tiao-San for disorders ranging from migraines and cerebrovascular events to contusions, oedema, and menstrual cramps. Modern pharmacological studies reveal that extracts and purified constituents of the herb act on the brain, cardiovascular, hematological, and nervous systems, offering antioxidant, neuroprotective, anti-inflammatory, and analgesic effects that together highlight its pronounced adaptogenic potential [6,7]. It has also recently been explored for its anti-tumor effects in brain malignancies. Accumulating evidence suggests that CX and its hallmark constituents hinder glioma progression through multiple mechanisms [8]. Notably, CX is a key ingredient in the herbal formula “Pingliu Keli,” which has been reported to inhibit glioma cell proliferation and induce apoptosis [9]. Active compounds extracted from CX, such as tetramethylpyrazine, n-butylidenephthalide, ferulic acid, and ligustilide, have demonstrated the ability to suppress GBM growth by inhibiting proliferation, tumor migration, invasion, and sensitizing GBM to temozolomide treatment targeting CXCR4, Rho GTPases, PI3K/Akt, ERK1/2, and Nur77 (NR4A1), thereby highlighting its potential as a therapeutic agent [10,11,12,13,14,15,16,17,18,19].
In parallel, microRNA (miRNA) has been increasingly recognized as playing a role in tumor cell proliferation, metastasis, invasiveness, angiogenesis, self-renewal, differentiation, and chemotherapy resistance, which may lead to treatment failure and tumor recurrence [20]. miR-10b was reported to be overexpressed in GBM and shows a pertinent role in GBM progression grading through the downstream targeting of HOXD10/MMP-14/uPAR and the RhoC axis [21,22]. Applying a miR-17-3p, miR-222, and miR-340 combinatorial overexpression efficiently induced cell death and attenuated tumor growth in three GBM subtypes [23].
More recently, attention has turned to the intersection between miRNA regulation and metabolic pathways. For instance, miR-29 was found to regulate the sterol regulatory element-binding protein 1 (SREBP1) [24], encoded by the SREBF1 gene, in GBM to inhibit SREBP-dependent cholesterol synthesis and uptake, which limits cell survival and tumor formation [25]. Upstream regulation of miRNA is also essential in GBM. miR-29/SREBF1 forms a negative feedback loop with upstream EGFR signals. It enhances SCAP-SREBF1 expression, and hence SREBF1, as a transcription factor, upregulates the expression of miR-29 [26]. Moreover, the scope of miRNA regulation extends further upstream to non-coding RNAs. For instance, the knockdown of long intergenic non-coding RNA 511 (LINC00511) suppressed miR-15a-5p/AEBP1 axis in malignant glioma [27]. Additionally, miR-424 and miR-503 can downregulate miR-9, promoting terminal differentiation in acute myeloid leukemia [28].
However, while the critical role of miRNAs in GBM has been established, studies investigating the impact of TCM, particularly CX, on miRNA-mediated regulatory networks remain scarce. Whether CX modulates miRNA expression to influence downstream pathways related to lipid metabolism, collagen deposition, and apoptosis in GBM remains unknown. To address this gap, the present study aims to elucidate the regulatory mechanisms through which CX affects GBM progression, focusing on its influence on miRNA expression and the associated metabolic and apoptotic targets and pathways in combination with transcriptomic analysis, Raman spectroscopy analysis, miRNA network construction, and experimental validation (Figure 1). By uncovering this regulation, we hope to provide new insights into the anti-tumor potential of TCM and highlight novel molecular targets for GBM therapy.
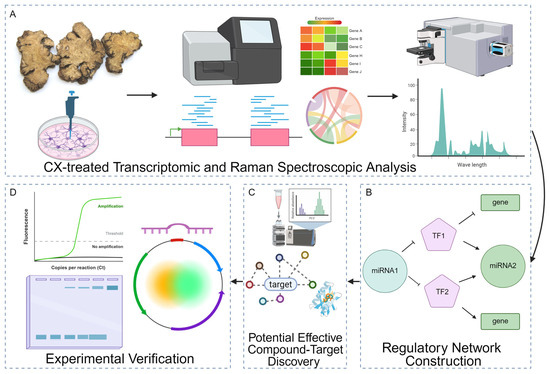
Figure 1.
Integrated workflow for dissecting the molecular effects of CX. (A) CXG solution-treated transcriptomic and Raman spectroscopic analysis. Cells are treated with the CXG solution, followed by RNA- and small RNA-seq to quantify differential gene expression and Raman microscopy to capture biochemical fingerprints. (B) Differentially expressed transcription factors (TFs, pentagons), miRNAs (circles), and target genes (rectangles) are assembled into a mixed TF/miRNA/gene interaction network to prioritize key regulatory motifs. Solid arrows indicate activation and T-bars denote repression. (C) Potential effective compounds in the CXG solution were found by UHPLC-MS and network pharmacology in combination with molecular docking. (D) Selected molecular axes are validated by quantitative RT-PCR (amplification curve), gel electrophoresis of PCR products, and dual-luciferase reporter assays using plasmid constructs containing miRNA binding sites or TF-responsive promoters.
2. Results
2.1. CXG Solution May Promote Glioblastoma Cell Apoptosis and Suppresses Extracellular Matrix Organization
To ensure the quality of the CXG solution, The HPLC-UV method quantified major compounds using commercially available standards, including ferulic acid, senkyunolide A, senkyunolide I, senkyunolide H, and ligustilide. In contrast, UHPLC-MS profiling revealed additional metabolites such as bayogenin, pipecolic acid derivatives, nicotinic acid, and cucurbitacin IIb (Table 1 and Figure S1), which is in concordance with previous studies [6,7]. The dose-dependent effect of the CXG solution on the proliferation of the U87MG cell line was tested by a cell viability assay (Figure 2A). CXG exhibited a clear cytotoxic effect on U87MG cells in a dose-dependent manner. Notably, the curve revealed a biphasic response: at lower concentrations, CXG promoted cell viability, whereas at higher concentrations it markedly reduced viability. As a result, 72.0 (IC50, CH), 52.1 (IC30, CM), and 31.2 mg/mL (IC10, CL) of the CXG solution were used at a series of concentrations in the subsequent experiments. Based on the quantification by HPLC-UV absorption, in CXG, the concentration of ferulic acid in the CXG solution is in the range of CL: 80.277–86.047 μM, CM: 133.802–143.42 μM, and CH: 185.345–198.667 μM with the content of other components shown in Table 2. Among these, ferulic acid and senkyunolide I were the most abundant, whereas ligustilide and levistilide A were present only at trace levels. The morphology of U87MG cells is shown in Figure 2B. With increasing CXG solution concentration, cell density decreased. Cells began to exhibit reduced cell–cell contact and display retraction, resulting in a more rounded morphology.

Table 1.
UHPLC-MS reveals the main components in the CXG solution.
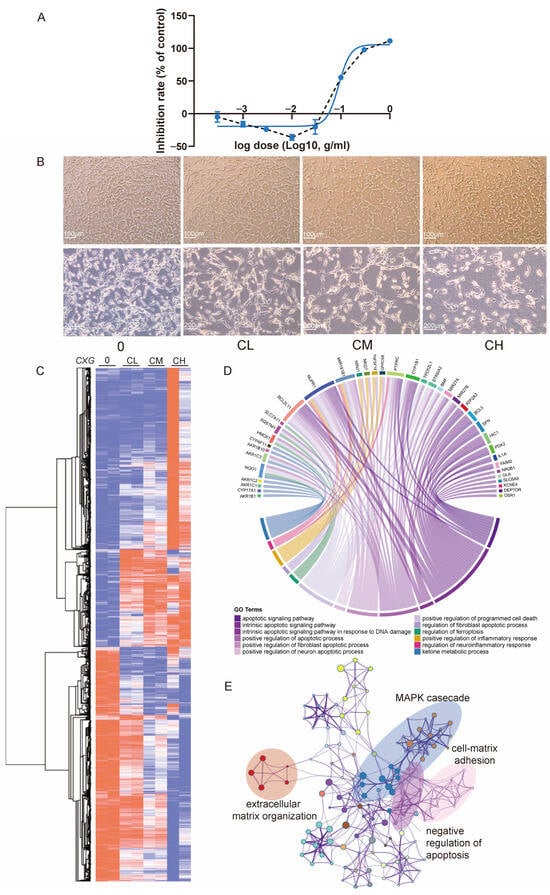
Figure 2.
CXG induces cell death and transcriptional reprogramming in U87MG cells. (A) Dose–response curve of U87MG cell viability after CXG treatment for 24 h measured by CCK-8 assay (mean ± SD, n = 6). Blue curve: non-linear regression of inhibition rate vs. log10(dose); black dashed line: line graph connecting the data points. (B) Representative phase-contrast images showing cell morphology under control (0), low (CL), medium (CM), and high (CH) concentrations of CXG. Scale bars: 100 μm (upper panels) and 200 μm (lower panels). (C) Heatmap and hierarchical clustering of DEGs in all three concentrations compared to the non-treated control. Colors represent row-wise normalized expression values (row min, row max; blue–white–red scale). (D) Enrichment analysis for downregulated genes and the pathways in which they are involved. Apoptosis-related pathways are denoted in purple. (E) Enriched pathways of upregulated genes.

Table 2.
HPLC–UV absorption characteristics and molar concentrations of the main compounds in CXG solution.
With the treatment of the CXG solution in a concentration gradient, there were 2168 upregulated and 1842 downregulated differentially expressed genes among CXG solution-treated samples compared with non-treated control (Figure 2C and Figure S2A,B). Genes differentially expressed under two concentrations (594 upregulated and 802 downregulated) were collected for enrichment analysis (Figure S2C). Major pathways for upregulated genes are apoptosis-related pathways (Figure 2D), with the most related gene being BCL2L11. The CXG solution also upregulates pathways related to ketone metabolism and the regulation of ferroptosis. Meanwhile, pathways associated with the negative regulation of apoptosis were downregulated. Other downregulated pathways included oncogenic MAPK cascades and tumor metastasis pathways, such as cell–matrix adhesion and extracellular matrix (ECM) organization (Figure 2E). Survival analysis revealed that lower expression of genes involved in ECM organization, such as PLAUR, TGM2, ADAMTSL1, FN1, and PXN, is associated with improved overall survival (Figure S3). Notably, TGM2 and HMGA2 are also genes involved in lipid metabolism. These results demonstrate the efficacy of the CXG solution in inhibiting extracellular matrix organization and inducing tumor cell apoptosis in the glioblastoma cell line. Previous studies have reported that collagen deposition and ECM remodeling enhance GBM invasion, therapy resistance, and correlate with poor prognosis [29,30]. Lipid metabolism reprogramming, particularly through SREBF1 [26,31], has also been recognized as a hallmark of GBM progression [25]. Our results are consistent with these observations.
2.2. The CXG Solution Effectively Downregulates the Collagen and Lipid Synthesis in Glioblastoma Cells
The linear discriminant analysis (LDA) plot demonstrated distinct clustering of Raman spectra from glioblastoma cells treated with increasing CXG solution concentrations (CL, CM, CH) compared with the non-treated control. Each point represents an individual cell spectrum, and ellipses indicate each group’s 95% confidence interval (Figure 3A). The progressive separation of clusters indicated dose-dependent biochemical alterations induced by the treatment. Raman spectroscopy revealed that glioblastoma cells treated with increasing drug concentrations exhibited a progressive decrease in the intensities of key Raman peaks (937, 1002, 1033, and 1657 cm−1, Figure 3B) corresponding to significant alterations in collagen composition [32]. Single peak integral intensity was quantified and compared in a boxplot (Figure 3C–F), visualizing the signal intensity as Figure 3G. The intensity of the bond vibration under different wavelengths in each condition was detected in a randomly selected cell. Notably, Raman imaging demonstrated a reduction in cytoplasmic projections and a redistribution of intracellular components, suggesting impaired structural integrity and metabolic remodeling under treatment stress (Figure S4).
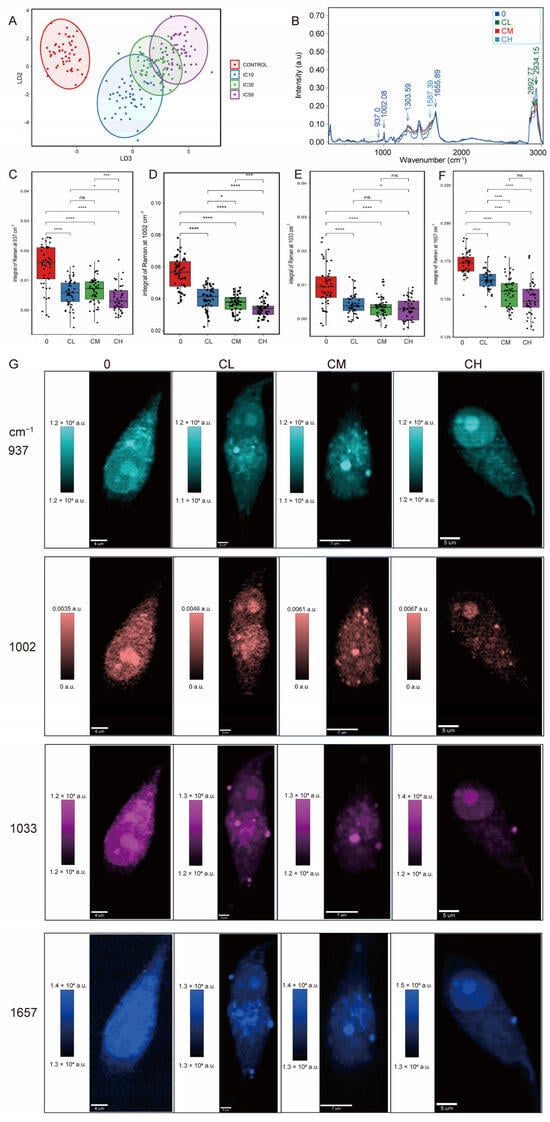
Figure 3.
Raman spectroscopic analysis reveals a dose-dependent reduction in collagen synthesis in glioblastoma cells. (A) Linear discriminant analysis (LDA) of Raman spectroscopic analysis. (B) Mean Raman spectra of different treatment groups. (C–F) Quantitative comparison of Raman peak (937, 1002, 1033, and 1657 cm−1) intensities. Statistical significance was determined by one-way ANOVA. ns, not significant; * p < 0.05; *** p < 0.001; **** p < 0.0001, n = 50 cells. (G) Raman mapping of individual cells at selected vibrational frequencies.
Raman intensity maps of individual cells also highlighted alterations in lipid metabolism and spatial redistribution. Peaks corresponding to CH2 symmetric stretching vibrations, characteristic markers of lipid alkyl chains (Figure 4A,B) at 2892 cm−1, and CH fatty acid chains (Figure 4C,D) at 2934 cm−1 showed a dose-dependent reduction from control (0) to low (CL), medium (CM), and high (CH) drug concentration groups. Results suggested a gradual decrease in saturated lipid content with increasing CXG solution concentration.
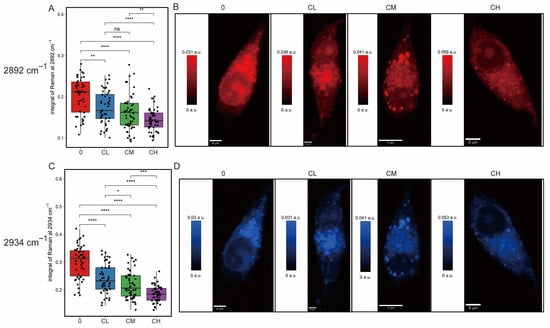
Figure 4.
Raman mapping of lipid-associated CH-stretching vibrations in glioblastoma cells under increasing drug treatment. (A) Boxplot of integral intensity at 2892 cm−1 and (B) corresponding Raman map. (C) Boxplot of integral intensity at 2934 cm−1 and (D) corresponding map. Statistical significance was determined by one-way ANOVA. ns, not significant; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001, n = 50 cells.
Similar phenotypic suppression of ECM remodeling and lipid metabolism has been linked to reduced aggressiveness and increased therapy sensitization in GBM models [33,34]. Taken together, these findings confirm that CXG solution treatment leads to significant alterations in cellular collagen deposition and lipid composition and spatial distribution, supporting the notion that its anti-tumor effect is mediated, at least in part, through the inhibition of collagen deposition and lipid metabolism.
2.3. The CXG Solution May Promote Pro-Apoptotic Effects and Reduce Lipid Metabolism Through hsa-miR-10a-5p and hsa-miR-29a-3p
Small RNA-seq was performed to explore how CX regulates miRNAs. The results showed that the CXG solution downregulated eight miRNAs and upregulated four miRNAs (Figure 5A). The TF-miRNA-target regulatory network is shown in Figure 5B. Within the network there are 12 pairs of TF-miRNA regulations and 2172 pairs of miRNA-target regulations. This network indicates that a feed-forward loop regulation exists between hsa-miR-10a-5p and hsa-miR-29a-3p, mediated through targeting E2F1 and SREBF1. The expressions of hsa-miR-10a-5p and hsa-miR-29a-3p were verified by qPCR. The CXG solution significantly downregulated their expression, which was consistent with the sequencing data (Figure 5C,D). To place these findings in context, we next compared them with the previously reported roles of miR-10a-5p and miR-29a-3p in glioblastoma. It has been reported that miR-10a-5p is frequently overexpressed in GBM and promotes proliferation and migration [35,36], whereas miR-29a-3p acts as a tumor suppressor and inhibits lipogenesis [37]. Our results, therefore, align with these previous findings, supporting the role of CX in modulating miRNA-driven pro-apoptotic effects and metabolic reprogramming. Among them, BCL2L11, E2F1, and SREBF1 were confirmed to be significantly regulated by the CXG solution. The downregulation of hsa-miR-10a-5p was accompanied by increased BCL2L11, while E2F1 and SREBF1 were suppressed, and the regulation of hsa-miR-29a-3p was subsequently verified. Because miRNA-profiling pointed to an E2F1-centered transcriptional program controlling the downregulated lipid-metabolic miRNAs, we next searched the network for proteins that (i) are directly modulated by the detected compounds, (ii) lie immediately upstream of, and (iii) negatively modulate E2F1. For each compound, we queried compound–target interaction (CTI) pairs from ChEMBL (v35), BindingDB (July 2025 release), PubChem BioAssay, and a literature-sourced CTI dataset [38]. After de-duplication, we obtained 406 unique human CTI pairs (Table S1). CTIs and E2F1-interacting genes data were assembled into a bipartite network (Figure S5). Hub analysis highlighted nicotinic acid (k = 17) and ferulic acid (k = 12) as the most multi-target phytochemicals. At the same time, MAPK1, EGFR, ESR1, and PPARG were the four highest-degree protein nodes (k ≥ 6), indicating that the CXG solution may converge on classic growth-factor and nuclear-receptor signaling axes. EGFR emerged as the sole node that satisfied all criteria, which is inhibited by adenosine and ferulic acid according to ChEMBL bioassay data. The inhibition of EGFR was reported to inhibit the activity of E2F1 in cancers [39]. Molecular docking results show that the Vina score of binding to EGFR is −7.3 kcal/mol for adenosine and −6.3 kcal/mol for ferulic acid (Figure 5E,F).
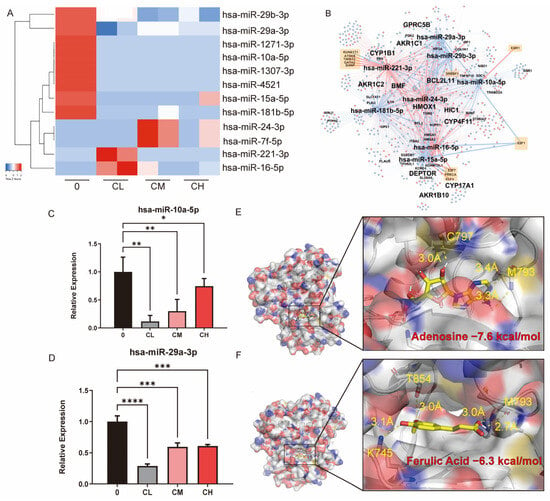
Figure 5.
The CXG solution induce cell death and alterations in lipid metabolism in glioblastoma cells via miRNAs. (A) Heatmap of mature miRNA expression in control (0), IC10 (CL), IC30 (CM), and IC50 (CH). (B) TF-miRNA-target interaction network. Red color nodes represent upregulated genes in CXG solution-treated conditions, while blue nodes represent downregulated genes. Orange squares denote transcription factors. (C,D) qPCR result of hsa-miR-10a-5p and hsa-miR-29a-3p. (E,F) molecular docking results and the demonstration of binding pockets of adenosine and ferulic acid to EGFR. Statistical significance was determined by one-way ANOVA. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
The expression levels of key apoptosis- and lipid metabolism-related molecules in U87MG glioblastoma cells following treatment with the CXG solution were evaluated. qPCR results indicated elevated levels of pro-apoptotic genes (i.e., BCL2L11) alongside the decreased expression of anti-apoptotic markers (i.e., BCL2) with increasing CXG solution dosage, consistent with the activation of cell death-associated transcriptional programs (Figure 6A,B). Figure 6C,D highlight the suppression of critical lipid metabolism regulators, such as SREBF1 and E2F1, suggesting that the CXG solution impairs fatty acid or cholesterol biosynthesis. Notably, BCL2 and SREBF1 mRNA levels decreased already at the lowest CXG concentration (CL), where cell viability was largely unaffected, indicating a lack of strict CXG dose-dependence. A dual-luciferase reporter assay validated the miRNA-mRNA interactions of hsa-miR-10a-5p/BCL2L11, and hsa-miR-29a-3p/SREBF1. There was a significant reduction in luciferase activity when the wild-type 3′-UTR was present with the miRNA mimic introduced, indicating direct binding to the predicted site (Figure 6E,F). These data provide supporting evidence that hsa-miR-10a-5p directly regulates BCL2L11 as a pro-apoptotic mediator, which is in concordance with previous studies [40,41], highlighting the regulatory mechanism of the CXG solution through the miRNA regulatory network.
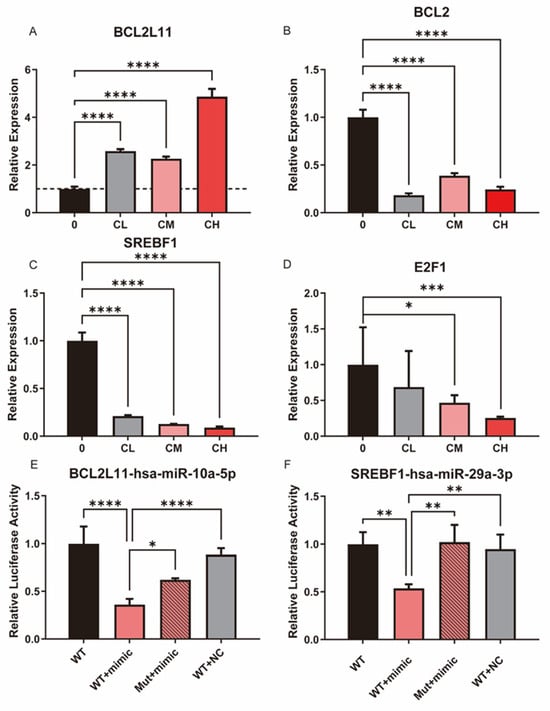
Figure 6.
The CXG solution may promote apoptosis and suppress lipid metabolism through a miRNA-involved regulatory network. (A–D) qPCR results of BCL2L11, BCL2, SREBF1, and E2F1 following CXG solution treatment. The dashed line indicates a relative expression level of 1 in the qPCR results. (E,F) Dual-luciferase reporter assay validated miRNA-mRNA interactions of hsa-miR-10a-5p/BCL2L11 and hsa-miR-29a-3p/SREBF1. Statistical significance was determined by one-way ANOVA. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
As the CXG solution downregulated hsa-miR-10a-5p, the regulation between hsa-miR-10a-5p and BCL2L11 was further verified by the restoration of hsa-miR-10a-5p. qPCR results showed that when hsa-miR-10a-5p was restored (Figure 7A), expression of BCL2L11 was downregulated in the U87MG cell line (Figure 7B). Notably, E2F1, SREBF1, and hsa-miR-29a-3p expressions were also downregulated (Figure 7C–E). As associated in the TF-miRNA-target regulatory network, E2F1 and SREBF1 are also downstream targets of miR-10a-5p, which, to our knowledge, are validated for the first time by a dual-luciferase reporter assay (Figure 7F,G). By an EMSA, we further assessed the binding of the E2F1 protein as a transcription factor in the promoter region of hsa-miR-29a-3p, as predicted. Figure 7H shows an evident shift in the probe when the protein concentration was larger than 1.6 μM, whereas the mutated probe did not show a shift at 1.6 μM. It was not significant at 3.2 μM (Figure 7H,I). Figure 7J shows a sharp shift gradient as the positive control of the probe was input, which contains a known E2F1-binding region [42]. As the regulation of SREBP1 by hsa-miR-29a has been previously reported [26], the hypothesis was proposed that the expression of hsa-miR-29a-3p can be suppressed by the CXG solution and hsa-miR-10a-5p through the downregulation of its transcription factors, SREBF1 and E2F1, in a coherent feed-forward loop, thereby orchestrating pro-apoptotic effects and metabolic reprogramming.
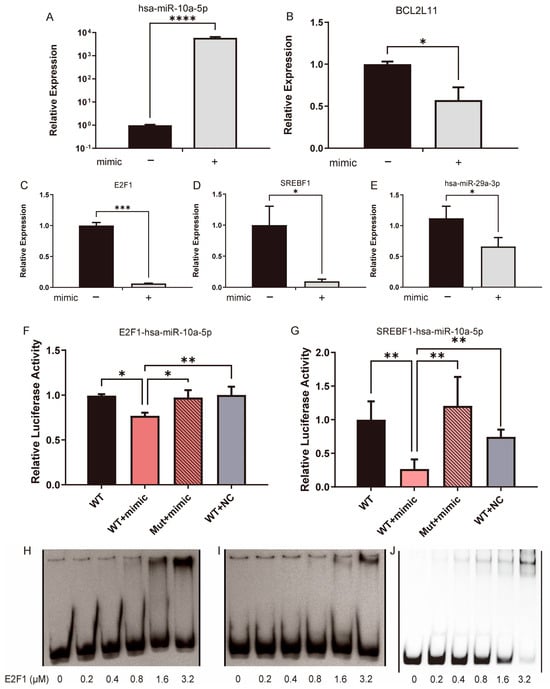
Figure 7.
The CXG solution and hsa-miR-10a-5p downregulated hsa-miR-29a-3p by the coherent feed-forward loop. (A–E) qPCR result compared gene expression with or without the transfection of hsa-miR-10a-5p. (F,G) Dual-luciferase reporter assay verifies MTI hsa-miR-10a-5p/E2F1 and hsa-miR-10a-5p/SREBF1. Statistical significance was determined by one-way ANOVA. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. (H) An EMSA shift verified the interaction between E2F1 and the hsa-miR-29a promoter region. (I) The shift is not apparent when the promoter region is mutated. (H,I) are derived from the same gel. One irrelevant lane between the WT probe (H) and mutant probe (I), and one empty irrelevant lane at the left and right, respectively, were removed. A thin black line indicates the splice. (J) An EMSA shift in positive control. One irrelevant empty lane on the left and eight irrelevant empty lanes on the right were removed and are indicated by a thin black line.
3. Discussion
In this study, we demonstrated that the CXG solution exerts anti-tumor effects on GBM cells, manifested primarily through the potential induction of apoptosis and the suppression of lipid metabolism and ECM organization. Our multi-omics approach, including transcriptomics and small RNA sequencing, and Raman spectroscopy, provided convergent evidence of the CXG solution’s efficacy in targeting GBM via key molecular axes, particularly the hsa-miR-10a-5p/BCL2L11/BCL2 and CX/SREBF1/E2F1/hsa-miR-29a-3p feed-forward loop. BCL2L11 encodes BIM, a pro-apoptotic BH3-only protein that directly antagonizes BCL2 and promotes the activation of Bax/Bak, thereby linking our observed transcriptional changes in BCL2L11 and BCL2 to the intrinsic apoptotic pathway, while our intention was not to imply the direct transcriptional regulation of BCL2 by BCL2L11, but rather to illustrate a potential regulatory axis through protein–protein interactions suggested by the transcriptomic data. Previous studies have reported on the anti-glioma activities of CX and its constituents, including tetramethylpyrazine, ligustilide, and ferulic acid, which inhibit proliferation, invasion, and sensitize GBM to temozolomide via CXCR4, Rho GTPases, and PI3K/Akt pathways [10,11,12,13,14,15,16,17,18,19]. From a mechanistic viewpoint, this research brought the role of miRNAs in mediating CX’s anti-tumor activities into sharper focus (Figure 8A).
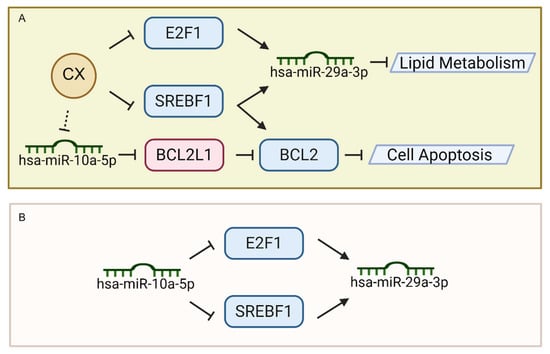
Figure 8.
Regulatory mechanism of CX in promoting cell death and suppressing lipid metabolism by feed-forward loop. (A) Mechanism of CX in regulating lipid metabolism and cell apoptosis. (B) Feed-forward loop of hsa-miR-10a-5p in suppressing hsa-miR-29a-3p by targeting transcription factors.
Notably, this feed-forward loop not only underscores the central role of hsa-miR-10a-5p in modulating lipid metabolism and apoptosis-related factors (e.g., SREBF1, E2F1) [43,44] but also highlights how upstream miRNA regulation can ultimately control the expression of another downstream miRNA (hsa-miR-29a-3p). However, the importance of higher-level regulation, particularly in miRNA-TF-miRNA loops, has often been underappreciated. However, prior studies have extensively investigated miRNA-mRNA interactions. Our findings demonstrate that hsa-miR-10a-5p serves as an upstream regulator in a coherent feed-forward loop (Figure 8B), paving the way for a more nuanced understanding of how multiple layers of non-coding RNAs and transcription factors synergistically govern glioblastoma cell fate.
The CXG solution significantly reduces the expression of ECM-related genes (e.g., FN1, TGM2, and PXN) and collagen synthesis pathways, as suggested by RNA-seq data and Raman spectroscopy. Survival analyses in our cohort further showed that a lower expression of ECM-related genes correlated with prolonged survival, consistent with the literature [45,46]. Our survival analysis further showed that a lower expression of ECM-related genes correlates with prolonged overall survival in GBM, underscoring the potential therapeutic value of inhibiting ECM organization and remodeling. CXG solution treatment thus limits cell proliferation and diminishes invasive capacities by disrupting the ECM.
Another important aspect of this work is the explicit demonstration of the CXG solution-mediated suppression of lipid metabolism. Emerging evidence suggests that lipid-based therapeutic molecules are promising research targets for unraveling novel drugs in GBM [47]. The SREBF1 gene encodes the nuclear transcription factors SREBP-1a and SREBP-1c, which are released from the endoplasmic reticulum membrane when cellular sterol levels decrease or insulin/carbohydrate signals increase. Once in the nucleus, SREBP-1 binds sterol-response elements and potently upregulates the core enzymes of de novo fatty-acid and triglyceride synthesis, thereby coordinating lipid storage, very-low-density lipoprotein assembly, and adipocyte differentiation [48]. As SREBP plays a key role in GBM sterol lipid regulation, its inhibitor fatostatin has been reported to induce ferroptosis in GBM [49]. Additionally, it was reported that E2F1 promotes the proliferation and metastasis of clear cell renal cell carcinoma by activating SREBP1-dependent fatty acid biosynthesis [50]. Our findings indicate that the CXG solution effectively downregulates key lipid regulatory genes, including SREBF1, via an FFL mediated by the hsa-miR-10a-5p/E2F1 axis. Raman spectrometric mapping corroborated these transcriptomic results, revealing dose-dependent decreases in lipid-associated signals in GBM cells treated with the CXG solution. Our findings that the CXG solution diminishes lipid synthesis genes and lipid-associated signals are therefore consistent with and extend these prior reports.
Despite these promising findings, several limitations warrant consideration. First, the current study was limited to in vitro experiments in a single GBM cell line (U87MG). The intricate heterogeneity of GBM necessitates validation in multiple cell lines, patient-derived primary cell lines, organoids, or animal models before clinical relevance can be firmly established. Second, although we identified multiple bioactive compounds in CX, the specific contribution of each constituent to the observed anti-GBM effects remains to be elucidated. Network pharmacology and molecular docking results indicate that adenosine and ferulic acid bind to EGFR, acting as an upstream suppressor of E2F1 and hsa-miR-10a-5p. In our study, the concentration of the CXG solution was calculated based on the original mass of CX. While the IC50 values indeed appear high compared to many purified phytochemicals, quantitative HPLC-UV data (Table 2) revealed that ferulic acid and senkyunolide I are relatively abundant (tens to hundreds of μM), whereas ligustilide and levistilide A are present at trace levels. For instance, ligustilide was detected in CXG at 0–8.6 µM, whereas previous studies have reported that ligustilide inhibits the migration of T98G glioma cells at 5 µM without inducing cell proliferation or apoptosis [51], and exerts cytotoxic effects with an IC50 of approximately 20 μg/mL in U251 glioma cells [19]. Similarly, ferulic acid was quantified in our CXG solution at 80–199 μM, which is higher than the reported concentration that induces apoptosis in GBM cells (~36 μM [18]) yet far below the reported IC50 concentration of 4.706 mM for U87MG cells [52]. These comparisons suggest that the nominal concentrations used in our study are within a biologically relevant range for its key constituents, once their relative abundance in the crude extract is considered. The results highlight that only a small fraction of CXG contributes to its anti-GBM activity. Interestingly, within the CXG mixture in concentration of IC50, the measured concentrations of key constituents such as ferulic acid and ligustilide were below or close to the IC50 values reported for these compounds when tested individually in glioma cells. This observation suggests that CXG exerts its bioactivity through additive or synergistic interactions among multiple components, allowing the overall preparation to achieve cytotoxic effects at concentrations where the individual compounds would otherwise be sub-effective. Such synergy aligns with the TCM principle that therapeutic efficacy results from the coordinated modulation of multiple pathways rather than from single-target inhibition.
Third, while our rationale in this work was to evaluate CXG in its clinically used composite form, consistent with the Traditional Chinese medicine (TCM) principle of considering the overall pharmacological effect of the preparation rather than isolated compounds. Consequently, the concentrations in our study were expressed as raw herb equivalents of Chuanxiong rhizome (mg/mL), which appear higher than those reported for lyophilized ethanol/water extracts, extracts fractions, and hydrolyzed extracted powder. For example, Hu et al. reported that the IC50 values of CX fractions against HepG2 and SMMC7721 cells, as determined by an in vitro MTT assay, were approximately 100 μg/mL for CX polysaccharide fractions in HepG2 and SMMC7721 cells, corresponding to 0.5–1 mg of crude herb equivalent [53]. In the same study, IC50 values of CX lyophilized polysaccharide fractions in other cancer cell lines, such as HCT-116 and A549, were reported to be higher, reaching ~500–1000 μg/mL (2.5–5 mg/mL CX equivalent), indicating that the effective concentrations of CX fractions vary by cancer type [53]. These complexities underscored the need for fractionation and more targeted studies to pinpoint the most potent chemical entities.
Fourth, pharmacokinetic constraints must be considered. While we have not yet measured the plasma levels of ferulic acid after the administration of CXG in animal models or human subjects, published pharmacokinetic studies of ferulic acid or its precursors/metabolites showed that in vivo plasma concentrations are many orders of magnitude lower than the cell culture concentrations used. Ferulic acid was rapidly absorbed following oral administration with a mean time to peak plasma concentration (Tmax) of 0.03 h. The corresponding maximum plasma concentration (Cmax) and the area under the concentration–time curve (AUC) were 8.175 μg/mL (42.1 ± 5.2 μM) and 2.595 μg·h /mL, respectively [54]. Further in vivo pharmacokinetics and formulation strategies should be addressed to enhance bioavailability and blood–brain barrier penetration in future work. To better reflect the pharmacologically relevant conditions, future studies could evaluate combinations of major CXG constituents at their experimentally determined plasma concentrations or in ratios corresponding to their abundance in CXG extracts. This strategy would enable the simulation of the integrated, multi-component pharmacology of TCM and help bridge in vitro efficacy with in vivo exposure. Moreover, systematic testing of these physiologically relevant combinations could clarify whether the enhanced potency of CXG arises from compound synergy, complementary target engagement, or improved cellular uptake, providing a framework for PK-PD modeling and translational optimization. In addition, we acknowledge the absence of non-cancerous proliferating cell controls, which would strengthen conclusions regarding selectivity. Previous work reported that the in vitro IC50 in mouse embryonic fibroblasts (3T3) of Chuanxiong Rhizoma decoction was 9.39 mg/mL, and the IC50 for mouse embryonic stem cell D3 was 18.78 mg/mL [55]. Although such experiments were not performed here, we propose to address this in future work, particularly by testing hematopoietic or neural progenitor cell models.
Furthermore, the observed effects may represent a biphasic, concentration-dependent response (hormesis). Low concentrations promote metabolic activity or proliferation, while higher concentrations induce cell death. This dual behavior is consistent with the traditional TCM concept of “huo xue xing qi” (“invigorating blood circulation and promoting energy flow”), which emphasizes regulatory rather than exclusively inhibitory functions. In addition, as shown in Figure 6B,C, BCL2 and SREBF1 expression were already significantly reduced at the lowest CXG concentration (CL), while cell viability was largely unaffected. This apparent lack of dose-response, together with the biphasic effect, is likely attributable to the complex and potentially opposing actions of multiple components within CXG. Accordingly, the present manuscript focuses primarily on elucidating the cytotoxic mechanisms of CXG at high concentrations, while the cytoprotective or growth-promoting effects observed at lower concentrations represent an interesting and valuable direction for future investigation. We also recognize that elevated concentrations raise concerns regarding nonspecific or nonselective effects. In the case of CXG, this may also reflect its complex, multi-component nature, where different constituents can exert opposing activities (e.g., cytoprotective versus proapoptotic) depending on the concentration. Thus, what appears as nonspecific cytotoxicity may instead represent the overlapping effects of multiple active molecules, rather than technical assay interference, highlighting that the complex pharmacology of TCM formulations inherently involves multi-target interactions, and systematic investigations remain necessary.
Finally, while our data suggest a regulatory role of miRNAs in lipid metabolism and apoptosis, additional apoptosis-specific assays, protein-level analyses, and in vivo functional assays are needed to further strengthen mechanistic interpretation. Accordingly, the proposed hsa-miR-10a-5p/BCL2L11/BCL2 and CX/SREBF1/E2F1/hsa-miR-29a-3p axes should be regarded as hypothesis-generating models, and we explicitly recognize that protein-level validation will be essential in future studies to establish causality. From a translational standpoint, CX compounds may complement existing GBM therapies, particularly by modulating lipid biosynthesis and ECM remodeling pathways that are increasingly recognized as therapeutic vulnerabilities. By fine-tuning miRNA networks and transcriptional regulators, CX may offer a valuable complementary approach to standard-of-care treatments, such as temozolomide or radiotherapy. Future research should address the pharmacokinetics, blood–brain barrier permeability, and synergistic potential of CX-derived compounds in combination with current GBM therapeutics.
4. Materials and Methods
4.1. HPLC-UV and LC-MS/MS Analysis of the CXG Solution
Chuanxiong dispensing granules (CXG) were purchased from E-Fang Pharmaceutical Co., Ltd. (Foshan, China). According to the company’s statement and national guidelines, CXG was produced from authenticated crude CX. The manufacturing process involves (i) water decoction under controlled conditions, (ii) extraction of essential oils by steam distillation, (iii) recombination of volatile oil with the aqueous extract, (iv) concentration and spray-drying to obtain dry extract powder, and (v) dry granulation to yield instant granules. This procedure follows the Chinese Pharmacopoeia requirements for dispensing granules and ensures chemical equivalence to the corresponding traditional decoction. According to the manufacturer’s equivalence, 0.217 g CXG corresponds to 1 g of dried rhizome of Ligusticum chuanxiong (as per the Chinese Pharmacopoeia standard). CXG was dissolved in serum-free DMEM medium to reach a final concentration 0.01 g/mL of CXG (around 46 mg/mL of crude CX). The suspension was incubated at 37 °C with gentle shaking and centrifuged at 12,000 rpm for 5 min. The supernatant was collected and designated as the CXG solution, which was used for HPLC-UV and LC-MS/MS and all in vitro experiments. The system for HPLC-UV absorption consisted of a Shimadzu LC-20AT equipped with a UV detector (SPD-20A, Shimadzu, Kyoto, Japan). Separation was carried out on a C18-A column (4.6 × 250 mm, 5 µm, Cat# DC952505-0, Agela Technologies, Tianjin, China) maintained at 30 °C. The mobile phase consisted of solvent A (water with 0.1% phosphoric acid, Cat# J2210679, Aladdin, Shanghai, China) and solvent B (acetonitrile, 99.90%, Cat# CAEQ-4-003306-4000, CNW, Shanghai, China), with the following gradient program: 0.00–1.00 min, 8% B; 1.00–7.00 min, 8–12% B; 7.00–12.00 min, 12–18% B; 12.00–20.00 min, 18–30% B; 20.00–24.00 min, 30% B; 24.00–32.00 min, 30–45% B; 32.00–40.00 min, 45–60% B; 40.00–47.00 min, 60–75% B; 47.00–50.00 min, 75–80% B; and 50.00–60.00 min, 80% B, followed by re-equilibration at 8% B for 8 min (60–68 min). The flow rate was set at 1.0 mL/min, and the injection volume was 10 μL. Detection wavelengths were selected according to the maximum absorbance of each compound, including 320 nm for ferulic acid, 325 nm for ligustilide, 277 nm for senkyunolide I, 278 nm for senkyunolide A, 276 nm for senkyunolide H, and 275 nm for levistilide A. Quantification was performed using external calibration curves constructed with authentic standards (ferulic acid (Cat# DST-DY0009-0020, Desite, Chengdu, China. The same supplier was used for all other standard compounds.), senkyunolide A (Cat# DST-DY0008), senkyunolide I (Cat# DST-DY0009-0020), senkyunolide H (Cat# DST-DY0182-0020), and ligustilide (Cat# DST-DH0007), and levistilide A (Cat# DST-DO0001-0020)). Each calibration curve exhibited good linearity (R2 > 0.999) within the tested concentration range. The compound contents in the samples were calculated from the corresponding calibration equations and expressed as mg/g of extract with 95% confidence intervals (95% CI).
LC-MS and MS/MS analyses of the CXG solution were performed using a high-resolution tandem mass spectrometer Q-Exactive connected to a Vanquish Flex UHPLC system (Thermo Fisher Scientific, Bremen, Germany). An ACQUITY UPLC T3 column (100 mm × 2.1 mm, 1.7 µm, Waters, Milford, MA, USA) was used for the reversed-phase separation. The flow rate for UHPLC was 0.3 mL/min, and the mobile phase consisted of solvent A (water, 0.1% formic acid) and solvent B (Acetonitrile, 0.1% formic acid). Gradient elution conditions were set as follows: 0–0.8 min, 2% B; 0.8–2.8 min, 2–70% B; 2.8–5.6 min, 70–90% B; 5.6–6.4 min, 90–100% B; 6.4–8 min, 100% B; 8–8.1 min, 100–2% B; and 8.1–10 min, 2%B. The Q-Exactive was operated in both positive and negative ion modes.
4.2. Cell Culture and Cell Viability Assay
A human glioblastoma-like astrocytoma U87MG cell line (Cat# CC1703, Cellcook, Guangzhou, China) was purchased and cultured in high-glucose Dulbecco’s Modified Eagle Medium (Cat# 11965092, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Cat# A5256701, Gibco) and 1% penicillin/streptomycin (Cat# 15140122, Gibco) at 37 °C with 5% CO2. The cell line was originally established from a female patient with glioblastoma and is widely used as a representative in vitro model for GBM research. U87MG cells display high proliferative and invasive capacities, closely mimicking the aggressive nature of GBM. The anti-tumor effects of the CXG solution were evaluated using a cell viability assay. The U87MG cell line was seeded at a density of 1.0 × 104 cells/well in 96-well plates. After 24 h incubation, the suspension was replaced with fresh medium (control group, 0 μg/mL) or drug solution (treatment groups) at a series of doses of herbal decoction pieces (0.0003, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 g/mL). After another 24 h of incubation, the suspension was replaced with 100 μL/well CCK-8 (Cat# C0038, Beyotime, Shanghai, China) solution at a ratio of 1:9 and incubated for 1 h at 37 °C. The microplate reader (BioTek Epoch 2, BioTek, Winooski, VT, USA) measured the absorbance at a wavelength of 450 nm. For the subsequent sequencing experiments in this study, the CXG solution was used at a series of concentrations according to the cell viability inhibition rate: 72.0 mg/mL (IC50, CH), 52.1 mg/mL (IC30, CM), and 31.2 mg/mL (IC10, CL) (Table 3). IC50 (half-maximal inhibitory concentration) was used to measure the efficacy of the CXG solution’s efficacy, which describes the amount of CX needed to inhibit the growth of cancer cells by half.

Table 3.
Concentration equivalent of the CXG solution and crude CX.
4.3. Raman Spectroscopy
Raman spectroscopy was performed on U87MG glioblastoma cells treated with CXG at different concentrations (0, CL, CM, and CH). A micro-Raman Spectrometer (Deuterium Peak Medical Instrument Co., Ltd., Shanghai, China, Model: CD100) was applied with a 532 nm laser, grating: 600 g/mm, spectral range: −203 to 3724 cm−1, laser power: 20 mW, integration time: 3 s, 63× water immersion objective, and spot size: approximately 300 nm. Before data collection, the instrument was calibrated using a silicon wafer, adjusting the silicon signal to 520.7 cm−1.
4.4. Transcriptomic Sequencing and Data Analysis
When cells reached 70–80% confluency, total RNA from U87MG was harvested using the TRIzol agent (Cat# 15596026, Invitrogen, Carlsbad, CA, USA) and assessed for integrity using a 2100 Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA) before library preparation. A spectrophotometer (Implen NanoPhotometer® N60/N50, Munich, Germany) was used to measure the concentration of RNA. The sequencing was performed as paired-end reads of 100 bp (PE100) on the BGISEQ-500 platform for subsequent data analysis. The sequencing depth was approximately 6 Gb.
Small RNA was first extracted using a total RNA extraction kit (Cat# DP761, TIANGEN, Beijing, China). Libraries were constructed using the MGIEasy Small RNA Library Prep Kit (MGI, Shenzhen, China) following the manufacturer’s instructions. Briefly, 18–30 nt small RNAs were size-selected by PAGE gel purification and ligated to 5′ and 3′ adapters. Reverse transcription was performed to generate cDNA, followed by PCR amplification and purification. The resulting libraries were quantified, pooled, and sequenced on the BGISEQ-500 platform (BGI, Shenzhen, China) to generate single-end 50 bp reads. The sequencing depth was around 10 M.
RNA-seq data processing: Low-quality reads and adaptor sequences were filtered and trimmed using Trim Galore! v0.6.8. After removing ribosomal RNA using SortMeRNA [56], the cleaned reads were aligned to the reference genome hg38 using bowtie2. An mRNA count matrix was generated to perform differential expression analysis. The matrix was normalized by count per million (CPM) within samples to reduce the inter-sample difference. Differentiation analysis was performed by the R (version 4.2) package DESeq2 (version 3.15) [24] with |log2FC| ≥ 1. The p-value was calculated and adjusted with the Benjamini–Hochberg algorithm, selecting differentially expressed genes (DEGs) with FDR ≤ 0.1.
Small RNA-seq data processing: the low-quality reads and adaptor sequences in raw reads were filtered and trimmed using Trim Galore!. Next, the cleaned reads were aligned against miRbase [57], mature miRNA, and hairpin using Bowtie. The expression matrix of mature miRNA expression was obtained, and the low-expression reads with a summary CPM less than 10 in all samples were removed. Differentially expressed miRNA (DEmiRs) was performed using DESeq2 with |log2FC| ≥ 1. The p-value was calculated using the default method and adjusted with the Benjamini–Hochberg algorithm, selecting DEmiRs with an FDR ≤ 0.1.
4.5. miRNA-Target Prediction
To further analyze the miRNA and mRNA profiles in our study, we explored the relationship between the screened DEGs and DEmiRs through miRTarBase [58] and the prediction programs TargetScan [59], miRDB [60], miRWalk [61], and miRTarBase [58].
4.6. Transcription Factors Prediction
Human transcription factors were retrieved from the TRANSFAC Professional database [62] and intersected with DEGs and the predicted miRNA targets. miRNA transcription factors were obtained from TransmiR2.0 [63].
4.7. Compound–Target Interaction Network Construction and Molecular Docking
Pairs of compound–target interactions were collected from ChEMBL (v35) [64], BindingDB (July 2025 release) [65], PubChem BioAssay [66], and a literature-sourced CTI dataset [38]. The direct targets of the compounds were then subjected to a protein–protein interaction network construction by STRING v12.0 [67]. Molecular docking analysis at the canonical binding pocket was conducted by AutoDockTools 1.5.6 (ADT) [68], specifically AutoDock Vina. The protein structure of EGFR (PDB: 1XKK, resolution 2.4 Å) was retrieved from the Protein Data Bank (RCSB PDB, www.rcsb.org/). The 3D coordinates of anticipated active compounds were retrieved from the PubChem database (Pubchem CID: adenosine: 60961; ferulic acid: 445858).
4.8. Regulatory Network Construction
A TF-miRNA-target regulatory network was constructed by combining transcription factors in DEGs and DEmiRs. Gephi v10 [69] was used for network visualization.
4.9. Gene Functional Enrichment Analysis
Gene Ontology (GO) analysis on biological processes was performed by Metascape [70] and Metacore (Clarivate, Philadelphia, PA, USA) to observe the enriched biological pathways of DEGs between the control and CXG solution to identify the pathways regulated. FDR ≤ 0.05 was adopted.
4.10. Survival Analysis
GEPIA2 [33] is an interactive web server that contains 9736 tumor samples and 8587 normal samples from the TCGA and GTEx projects, providing differential expression analysis between tumor and normal tissues. Survival analysis of identified DEGs in 82 GBM tumors was obtained with 50 percent quantile and log-rank p-value < 0.01 as the cutoff values.
4.11. Quantitative Reverse Transcriptase-PCR (qPCR)
Total RNAs of U87MG were harvested when cells reached 70–80% confluency by TRIzol agent (Cat# 15596026, Invitrogen). Total RNA was extracted per the manufacturer’s instructions using the Direct-zolTM RNA miniprep kit (Cat# R2052, Zymo Research, Irvine, CA, USA). A spectrophotometer (Implen NanoPhotometer® N60/N50, Munich, Germany) was used to measure the concentration of RNA. Next, first-strand cDNA was generated with 1 μg total RNA using Super Script III Reverse Transcriptase (Cat# 12574026, Invitrogen) in a 20 μL PCR system. A volume of 2 μL of the product was subjected to amplification using Applied Biosystems QuantStudioTM 6 with PowerUp SYBR Green Master Mix (Cat# A25742, Applied Biosystems™, Waltham, MA, USA). The primers used are listed in Table 4.

Table 4.
Primers used for qPCR validation.
4.12. Dual-Luciferase Reporter Assay
A dual-luciferase reporter assay was performed to validate the miRNA-mRNA interaction. miRWalk [71] was used to predict the binding side. HEK-293 cells (Cat# CC4002, cellcook) in 24-well plates were transfected with 0.1 μM miRNA mimics and 0.5 μg pmiRGLO using Lipofectamine 2000 (Cat# 11668019, Invitrogen). HEK-293 cells were lysed after 24 h, and activity was determined using a dual-luciferase assay kit (Cat# RG089M, Beyotime) on FlexStation® 3 (Molecular Devices, San Jose, CA, USA).
4.13. Electrophoretic Mobility Shift Assay
An electrophoretic mobility shift assay (EMSA) was performed to assess the DNA-binding activity of the protein of interest. The binding sites of E2F1 in the promoter region of miR-29a (2000 bp upper-stranded and 500 bp down-stranded) were predicted by Tomtom [72] in the MEME Suite [73] against the known binding motif of E2F1 in the JASPAR database [74]. Double-stranded DNA probes were inserted into the pBR322 vector between the XbaI and BamHI cutting sites. Oligonucleotide probes were designed and amplified from plasmids with PCR (Cat# K0172, Thermo Scientific™). E2F1 protein was purchased from AtaGenix (~40.9kDa, Cat# YA6315H, Wuhan, China). Binding reactions were carried out in a total volume of 10 μL, containing E2F1 protein (0, 0.2, 0.4, 0.8, 1.6, and 3.2 μM) and ~280 ng double-stranded DNA probes, as provided by Table 5, with the binding site and mutated binding site highlighted. The reaction mixture was incubated at 37 °C for 30 min and 4 °C for 10 min. After incubation, the reaction mixtures were loaded on a 6% non-denaturing polyacrylamide gel in 0.5× TAE buffer with 6× novel juice fluorescent nucleotide dye (Cat# P001S1, Solarbio, Beijing, China). The electrophoresis was then carried out at 4 °C under a constant voltage of 80 V for 100 min. Bands corresponding to DNA–protein complexes were detected under UV light (BG-gdsAUTO 550, Baygene, Beijing, China).

Table 5.
Sequence of double-stranded DNA probes. Bold text indicates the positions of (mutated) E2F1 binding sites.
4.14. Statistical Analysis
Data are presented as mean ± standard deviation (SD). Statistical comparisons among groups were performed using One-way ANOVA. Significance levels were defined as * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. Analyses were conducted using GraphPad Prism 10.0. Other data were visualized by R packages ggplot2 [75] and circos [76].
5. Conclusions
In conclusion, our study demonstrates that the CXG solution exerts multi-faceted anti-glioblastoma effects by simultaneously inducing apoptosis through the hsa-miR-10a-5p/BCL2L11/BCL2 axis, suppressing extracellular matrix organization, and disrupting lipid metabolism via the SREBF1/E2F1/hsa-miR-29a-3p regulatory network. By coordinately downregulating critical lipid metabolism genes and modulating key tumor-promoting miRNAs, the CXG solution effectively targets multiple survival pathways in GBM cells. These findings not only provide mechanistic insights into CX’s anti-tumor activity but also highlight the therapeutic potential of combining miRNA-targeted approaches with metabolic intervention for GBM treatment. To translate these findings, activate phytochemical–target–miRNA regulatory network construction, in vivo models’ verification, and combination therapeutic strategy avenues merit priority. Pursuing these focused lines of inquiry will clarify pharmacodynamics, optimize delivery across the blood–brain barrier, and accelerate the clinical development of CX-derived agents for glioblastoma.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph18101553/s1. Figure S1: UPHPLC-MS profile of the CXG solution, showing four major components in CXG; Figure S2: Differentially expressed genes regulated by CXG and their functional enrichment analysis. (A) Venn diagram of upregulated differentially expressed genes (DEGs) across CL, CM, and CH treatment groups compared with control. (B) Venn diagram of downregulated DEGs across the same comparisons. (C) MetaCore pathway enrichment analysis of all DEGs, with the top significantly enriched pathways (ranked by −log10 p-value) shown. Figure S3: Survival analysis shows that the lower expression of genes involved in extracellular matrix organization and lipid metabolism improves overall survival. Figure S4: Raman single peak intensity under different conditions. Four treatment conditions are shown from left to right: 0 (untreated control), CL (CXG low dose), CM (CXG medium dose), and CH (CXG high dose; doses as defined in Methods). For each condition, three rows are provided: White (top row): Bright-field micrographs. The red rectangle marks the region of interest that was raster-scanned for Raman mapping. Scale bar: 20 µm. Raman (middle row): Representative Raman mapping images reconstructed from integrated spectral intensity. These maps show the overall signal distribution and scanned area, confirming that Raman spectra were acquired across the full cell body. The consistent intensity range (1.6 × 104–1.9 × 104 a.u.) ensures comparability between treatments. Merge (bottom row): Composite overlays to visualize single-band maps of selected characteristic spectral bands and assign pseudo-colors, where different biochemical components can be visualized simultaneously within the same cell. Figure S5: Bipartite compound–target interaction network after filtering direct E2F1 modulators. Table S1: A list of CTI pairs collected from databases. Raw reads of both RNA and small RNA sequencing data were deposited on the Sequence Read Archive (SRA) with accession number PRJNA1260138.
Author Contributions
Conceptualization, H.-D.H.; Data Curation, X.-X.C., H.-Y.H. and Y.-Y.X.; Formal Analysis, X.-X.C.; Funding Acquisition, H.-L.Z., H.-Y.H., H.-D.H. and Y.-C.-D.L.; Investigation, X.-X.C. and Y.-Y.X.; Methodology, X.-X.C., D.Z., H.-D.H. and Y.-C.-D.L.; Project Administration, Y.-C.-D.L.; Resources, H.-L.Z., H.-D.H. and Y.-C.-D.L.; Software, X.-X.C. and H.-Y.H.; Supervision, H.-D.H. and Y.-C.-D.L.; Validation, X.-X.C., L.-P.L., J.N., P.-S.W. and X.-Y.X.; Visualization, X.-X.C. and J.L.; Writing—Original Draft, X.-X.C.; Writing—Review and Editing, X.-X.C., H.-L.Z., J.L., H.-D.H. and Y.-C.-D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32070674; Shenzhen Science and Technology Program, grant number JCYJ20220530143615035; the Warshel Institute for Computational Biology funding from Shenzhen City and Longgang District, grant number LGKCSDPT2024001; the Shenzhen–Hong Kong Cooperation Zone for Technology and Innovation, grant number HZQB-KCZYB-2020056 and P2-2022-HDH-001-A; the Guangdong Young Scholar Development Fund of Shenzhen Ganghong Group Co., Ltd., grant number 2022E0035; and the Guangdong S&T program, grant number 2024A0505050001 and 2024A0505050002.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in the Sequence Read Archive at PRJNA1260138.
Acknowledgments
This research benefited significantly from the interdisciplinary research environment and cutting-edge instrumentation maintained by the Experimental Platform of the Warshel Institute for Computational Biology. Their ongoing commitment to research infrastructure development has been crucial to our scientific endeavors. The authors are also grateful to the library of The Chinese University of Hong Kong, Shenzhen, for providing effective database service.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ADAMTSL1 | A disintegrin and metalloproteinase with thrombospondin motifs like 1 (gene) |
| AEBP1 | Adipocyte enhancer binding protein 1 |
| BCL2 | B-cell lymphoma 2 (gene) |
| BCL2L11 | BCL2-like 11 (gene) |
| CX | Ligusticum Chuanxiong Hort. |
| CXG | CX dispensing granules |
| E2F1 | Early region two binding factor transcription factor 1 (gene, protein) |
| ECM | Extracellular matrix |
| EGFR | Epithelial growth factor receptor (gene) |
| FN1 | Fibronectin 1 (gene) |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase (gene) |
| GBM | Glioblastoma |
| GO | Gene Ontology |
| HMGA2 | High mobility group AT-hook 2 (gene) |
| HOXD10 | Homeobox D10 (gene) |
| IC | Inhibitory concentration |
| LC | Liquid chromatography |
| LDA | Linear discriminant analysis |
| MAPK | Mitogen-activated protein kinase |
| miRNA | microRNA |
| MMP-14 | Matrix metalloproteinase-14 |
| MS | Mass spectrometry |
| Nur77 | Nuclear receptor 4A1 (gene) |
| PI3K/Akt | Phosphoinositide 3-kinase/Protein kinase B |
| PLAUR | Plasminogen activator, urokinase receptor (gene) |
| PXN | Paxillin (gene) |
| qPCR | Quantitative polymerase chain reaction |
| RhoC | Ras homolog family member C (gene) |
| SCAP | SREBF chaperone |
| SREBF1 | Sterol regulatory element-binding factor 1 (gene) |
| SREBP1 | Sterol regulatory element-binding protein 1 |
| TCM | Traditional Chinese medicine |
| TGM2 | Transglutaminase 2 (gene) |
| U6 | U6 small nuclear RNA (gene) |
| UHPLC | Ultra-high-performance liquid chromatography |
| uPAR | Urokinase receptor |
References
- Fernandes, C.; Costa, A.; Osório, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Current Standards of Care in Glioblastoma Therapy. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Poon, M.T.C.; Sudlow, C.L.M.; Figueroa, J.D.; Brennan, P.M. Longer-term (≥2 years) survival in patients with glioblastoma in population-based studies pre- and post-2005: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 11622. [Google Scholar] [CrossRef] [PubMed]
- Miska, J.; Chandel, N.S. Targeting fatty acid metabolism in glioblastoma. J. Clin. Investig. 2023, 133, e163448. [Google Scholar] [CrossRef] [PubMed]
- Pointer, K.B.; Clark, P.A.; Schroeder, A.B.; Salamat, M.S.; Eliceiri, K.W.; Kuo, J.S. Association of collagen architecture with glioblastoma patient survival. J. Neurosurg. 2017, 126, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Ma, L.; Fau-Peng, C.; Peng, C.; Fau-Zhang, H.; Zhang, H.; Fau-Qin, L.-P.; Qin, L.P. Ligusticum chuanxiong Hort: A review of chemistry and pharmacology. Pharm. Biol. 2011, 49, 1180–1189. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, L.; Wang, H.; Jiang, M.; Chen, Y.; Zheng, X.; Li, L.; Yin, Q.; Han, L.; Bai, L.; et al. Ligusticum chuanxiong: A chemical, pharmacological and clinical review. Front. Pharmacol. 2025, 16, 1523176. [Google Scholar] [CrossRef]
- Li, D.; Long, Y.; Yu, S.; Shi, A.; Wan, J.; Wen, J.; Li, X.; Liu, S.; Zhang, Y.; Li, N.; et al. Research Advances in Cardio-Cerebrovascular Diseases of Ligusticum chuanxiong Hort. Front. Pharmacol. 2022, 12, 832673. [Google Scholar] [CrossRef]
- Cao, P.; Cai, X.; Lu, W.; Zhou, F.; Huo, J. Growth Inhibition and Induction of Apoptosis in SHG-44 Glioma Cells by Chinese Medicine Formula “Pingliu Keli”. Evid.-Based Complement. Altern. Med. 2011, 2011, 958243. [Google Scholar] [CrossRef]
- Shuai, S.Y.; Liu, S.S.; Liu, X.J.; Zhang, G.S.; Zheng, Q.; Yue, P.F.; Yang, M.; Hu, P.Y. Essential oil of Ligusticum chuanxiong Hort. Regulated P-gp protein and tight junction protein to change pharmacokinetic parameters of temozolomide in blood, brain and tumor. J. Ethnopharmacol. 2022, 298, 115646. [Google Scholar] [CrossRef]
- Lin, P.C.; Chen, Y.L.; Chiu, S.C.; Yu, Y.L.; Chen, S.P.; Chien, M.H.; Chen, K.Y.; Chang, W.L.; Lin, S.Z.; Chiou, T.W.; et al. Orphan nuclear receptor, Nurr-77 was a possible target gene of butylidenephthalide chemotherapy on glioblastoma multiform brain tumor. J. Neurochem. 2008, 106, 1017–1026. [Google Scholar] [CrossRef]
- Chang, L.F.; Lin, P.C.; Ho, L.I.; Liu, P.Y.; Wu, W.C.; Chiang, I.P.; Chang, H.W.; Lin, S.Z.; Harn, Y.C.; Harn, H.J.; et al. Overexpression of the orphan receptor Nur77 and its translocation induced by PCH4 may inhibit malignant glioma cell growth and induce cell apoptosis. J. Surg. Oncol. 2011, 103, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.G.; Ma, L.; Zhang, S.Y.; Zhu, Z.Z.; Zhang, H.; Qin, L.P.; Wei, Y.J. Essential oil from rhizomes of Ligusticum chuanxiong induces apoptosis in hypertrophic scar fibroblasts. Pharm. Biol. 2011, 49, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Niu, T.; Li, C.; Li, X.; Liang, H.; Zhao, K. Mechanism of Ferulic Acid in PI3K/AKT Pathway and Research in Glioblastoma. Altern. Ther. Health Med. 2024, 30, 104–109. [Google Scholar] [PubMed]
- Li, Z.Q.; Zhang, G.S.; Liu, R.Q.; Shuai, S.Y.; Hu, P.Y.; Zheng, Q.; Xiao, S.H. Anti-Glioma Effects of Ligustilide or n-Butylphthalide on Their Own and the Synergistic Effects with Temozolomide via PI3K/Akt Signaling Pathway. Onco Targets Ther. 2023, 16, 983–994. [Google Scholar] [CrossRef]
- Yu, K.; Chen, Z.; Pan, X.; Yang, Y.; Tian, S.; Zhang, J.; Ge, J.; Ambati, B.; Zhuang, J. Tetramethylpyrazine-mediated suppression of C6 gliomas involves inhibition of chemokine receptor CXCR4 expression. Oncol. Rep. 2012, 28, 955–960. [Google Scholar] [CrossRef]
- Yu, N.; Zhang, Z.; Chen, P.; Zhong, Y.; Cai, X.; Hu, H.; Yang, Y.; Zhang, J.; Li, K.; Ge, J.; et al. Tetramethylpyrazine (TMP), an Active Ingredient of Chinese Herb Medicine Chuanxiong, Attenuates the Degeneration of Trabecular Meshwork through SDF-1/CXCR4 Axis. PLoS ONE 2015, 10, e0133055. [Google Scholar] [CrossRef]
- Grasso, R.; Dell’Albani, P.; Carbone, C.; Spatuzza, M.; Bonfanti, R.; Sposito, G.; Puglisi, G.; Musumeci, F.; Scordino, A.; Campisi, A. Synergic pro-apoptotic effects of Ferulic Acid and nanostructured lipid carrier in glioblastoma cells assessed through molecular and Delayed Luminescence studies. Sci. Rep. 2020, 10, 4680. [Google Scholar] [CrossRef]
- Ke, G.; Zhang, M.; Hu, P.; Zhang, J.; Naeem, A.; Wang, L.; Xu, H.; Liu, Y.; Cao, M.; Zheng, Q. Exploratory Study on Nanoparticle Co-Delivery of Temozolomide and Ligustilide for Enhanced Brain Tumor Therapy. Pharmaceutics 2025, 17, 191. [Google Scholar] [CrossRef]
- Mafi, A.; Rahmati, A.; Babaei Aghdam, Z.; Salami, R.; Salami, M.; Vakili, O.; Aghadavod, E. Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell. Mol. Biol. Lett. 2022, 27, 65. [Google Scholar] [CrossRef]
- Sun, L.; Yan, W.; Wang, Y.; Sun, G.; Luo, H.; Zhang, J.; Wang, X.; You, Y.; Yang, Z.; Liu, N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011, 1389, 9–18. [Google Scholar] [CrossRef]
- Malzkorn, B.; Wolter, M.; Liesenberg, F.; Grzendowski, M.; Stühler, K.; Meyer, H.E.; Reifenberger, G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010, 20, 539–550. [Google Scholar] [CrossRef]
- Bassot, A.; Dragic, H.; Haddad, S.A.; Moindrot, L.; Odouard, S.; Corlazzoli, F.; Marinari, E.; Bomane, A.; Brassens, A.; Marteyn, A.; et al. Identification of a miRNA multi-targeting therapeutic strategy in glioblastoma. Cell Death Dis. 2023, 14, 630. [Google Scholar] [CrossRef]
- Ru, P.; Guo, D. microRNA-29 mediates a novel negative feedback loop to regulate SCAP/SREBP-1 and lipid metabolism. RNA Dis. 2017, 4, e1525. [Google Scholar]
- Lu, L.; Zhang, Y.; Yang, Y.; Jin, M.; Ma, A.; Wang, X.; Zhao, Q.; Zhang, X.; Zheng, J.; Zheng, X. Lipid metabolism: The potential therapeutic targets in glioblastoma. Cell Death Discov. 2025, 11, 107. [Google Scholar] [CrossRef]
- Ru, P.; Hu, P.; Geng, F.; Mo, X.; Cheng, C.; Yoo, J.Y.; Cheng, X.; Wu, X.; Guo, J.Y.; Nakano, I.; et al. Feedback Loop Regulation of SCAP/SREBP-1 by miR-29 Modulates EGFR Signaling-Driven Glioblastoma Growth. Cell Rep. 2016, 16, 1527–1535. [Google Scholar] [CrossRef]
- Liu, Z.; Tao, B.; Li, L.; Liu, P.; Xia, K.; Zhong, C. LINC00511 knockdown suppresses glioma cell malignant progression through miR-15a-5p/AEBP1 axis. Brain Res. Bull. 2021, 173, 82–96. [Google Scholar] [CrossRef]
- Forrest, A.R.; Kanamori-Katayama, M.; Tomaru, Y.; Lassmann, T.; Ninomiya, N.; Takahashi, Y.; de Hoon, M.J.; Kubosaki, A.; Kaiho, A.; Suzuki, M.; et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2010, 24, 460–466. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Chang, Y.-C.; Li, C.-H.; Chan, M.-H.; Chen, C.-L.; Tsai, W.-C.; Hsiao, M. Type V collagen alpha 1 chain promotes the malignancy of glioblastoma through PPRC1-ESM1 axis activation and extracellular matrix remodeling. Cell Death Discov. 2021, 7, 313. [Google Scholar] [CrossRef]
- Cha, J.; Ding, E.A.; Carvalho, E.M.; Fowler, A.; Aghi, M.K.; Kumar, S. Collagen VI deposition primes the glioblastoma microenvironment for invasion through mechanostimulation of β-catenin signaling. PNAS Nexus 2024, 3, pgae355. [Google Scholar] [CrossRef]
- Geng, F.; Zhong, Y.; Su, H.; Lefai, E.; Magaki, S.; Cloughesy, T.F.; Yong, W.H.; Chakravarti, A.; Guo, D. SREBP-1 upregulates lipophagy to maintain cholesterol homeostasis in brain tumor cells. Cell Rep. 2023, 42, 112790. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Shazza, R.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Faisal, S.M.; Comba, A.; Varela, M.L.; Argento, A.E.; Brumley, E.; Abel, C.; Castro, M.G.; Lowenstein, P.R. The complex interactions between the cellular and non-cellular components of the brain tumor microenvironmental landscape and their therapeutic implications. Front. Oncol. 2022, 12, 1005069. [Google Scholar] [CrossRef]
- Duman, C.; Di Marco, B.; Nevedomskaya, E.; Ulug, B.; Lesche, R.; Christian, S.; Alfonso, J. Targeting fatty acid oxidation via Acyl-CoA binding protein hinders glioblastoma invasion. Cell Death Dis. 2023, 14, 296. [Google Scholar] [CrossRef]
- Wang, R.; Wang, J.; Wang, Y.; Yang, L. lncRNA TUSC7 sponges miR-10a-5p and inhibits BDNF/ERK pathway to suppress glioma cell proliferation and migration. Aging 2023, 15, 3021–3034. [Google Scholar] [CrossRef]
- Yan, Y.; Yan, H.; Wang, Q.; Zhang, L.; Liu, Y.; Yu, H. MicroRNA 10a induces glioma tumorigenesis by targeting myotubularin-related protein 3 and regulating the Wnt/β-catenin signaling pathway. FEBS J. 2019, 286, 2577–2592. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, W.; Kim, T.-M.; Jung, Y.; Menon, L.G.; Xing, H.; Li, H.; Carroll, R.S.; Park, P.J.; Yang, H.W.; et al. MicroRNA-29a activates a multi-component growth and invasion program in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 36. [Google Scholar] [CrossRef]
- Chen, Y.; Ji, X.; Zhang, Z.; Zhou, Y.; Lin, Y.-C.-D.; Huang, H.-Y.; Zhang, T.; Lai, Y.; Chen, K.; Su, C.; et al. SCOPE-DTI: Semi-Inductive Dataset Construction and Framework Optimization for Practical Usability Enhancement in Deep Learning-Based Drug Target Interaction Prediction. arXiv 2025, arXiv:2503.09251. [Google Scholar]
- Wheeler, D.L.; Dunn, E.F.; Harari, P.M. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat. Rev. Clin. Oncol. 2010, 7, 493–507. [Google Scholar] [CrossRef]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M.; et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef]
- Shea, A.; Harish, V.; Afzal, Z.; Chijioke, J.; Kedir, H.; Dusmatova, S.; Roy, A.; Ramalinga, M.; Harris, B.; Blancato, J.; et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016, 5, 1917–1946. [Google Scholar] [CrossRef]
- Dick, F.A.; Dyson, N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 2003, 12, 639–649. [Google Scholar] [CrossRef]
- Seo, Y.K.; Chong, H.K.; Infante, A.M.; Im, S.S.; Xie, X.; Osborne, T.F. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc. Natl. Acad. Sci. USA 2009, 106, 13765–13769. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, T.; Yan, H.; Guo, K.; Liu, Z.; Wei, L.; Lu, W.; Qiu, C.; Jiang, J. Fatostatin reverses progesterone resistance by inhibiting the SREBP1-NF-κB pathway in endometrial carcinoma. Cell Death Dis. 2021, 12, 544. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- So, J.-S.; Kim, H.; Han, K.-S. Mechanisms of Invasion in Glioblastoma: Extracellular Matrix, Ca2+ Signaling, and Glutamate. Front. Cell. Neurosci. 2021, 15, 663092. [Google Scholar] [CrossRef]
- Darwish, A.; Pammer, M.; Gallyas, F.; Vígh, L.; Balogi, Z.; Juhász, K. Emerging Lipid Targets in Glioblastoma. Cancers 2024, 16, 397. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002, 109, 1125–1131. [Google Scholar] [CrossRef]
- Cai, J.; Ye, Z.; Hu, Y.; Ye, L.; Gao, L.; Wang, Y.; Sun, Q.; Tong, S.; Zhang, S.; Wu, L.; et al. Fatostatin induces ferroptosis through inhibition of the AKT/mTORC1/GPX4 signaling pathway in glioblastoma. Cell Death Dis. 2023, 14, 211. [Google Scholar] [CrossRef]
- Shen, D.; Gao, Y.; Huang, Q.; Xuan, Y.; Yao, Y.; Gu, L.; Huang, Y.; Zhang, Y.; Li, P.; Fan, Y.; et al. E2F1 promotes proliferation and metastasis of clear cell renal cell carcinoma via activation of SREBP1-dependent fatty acid biosynthesis. Cancer Lett. 2021, 514, 48–62. [Google Scholar] [CrossRef]
- Yin, J.; Wang, C.; Mody, A.; Bao, L.; Hung, S.H.; Svoronos, S.A.; Tseng, Y. The Effect of Z-Ligustilide on the Mobility of Human Glioblastoma T98G Cells. PLoS ONE 2013, 8, e66598. [Google Scholar] [CrossRef]
- Jin, P.; Zhao, Z.; Li, X.; Mei, Q.; Liu, D.; Liu, M.; Zhao, K. Effects of Ferulic Acid Regulation of the PI3k/Akt Signaling Pathway on the Proliferation, Migration or Apoptosis of U87-MG Cells Based on Bioinformatics. IJP 2025, 21, 510–520. [Google Scholar] [CrossRef]
- Hu, J.; Jia, X.; Fang, X.; Li, P.; He, C.; Chen, M. Ultrasonic extraction, antioxidant and anticancer activities of novel polysaccharides from Chuanxiong rhizome. Int. J. Biol. Macromol. 2016, 85, 277–284. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Zhang, Y.; Mi, S.; Wang, N. Pharmacokinetics of ferulic acid and potential interactions with Honghua and clopidogrel in rats. J. Ethnopharmacol. 2011, 137, 562–567. [Google Scholar] [CrossRef]
- Wang, H.; Bao, Q.; Yi, H.; Xia, Q. The evaluation of embryotoxicity of Ligusticum chuanxiong on mice and embryonic stem cells. J. Ethnopharmacol. 2019, 239, 111895. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Matys, V.; Fricke, E.; Geffers, R.; Gossling, E.; Haubrock, M.; Hehl, R.; Hornischer, K.; Karas, D.; Kel, A.E.; Kel-Margoulis, O.V.; et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003, 31, 374–378. [Google Scholar] [CrossRef]
- Tong, Z.; Cui, Q.; Wang, J.; Zhou, Y. TransmiR v2.0: An updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019, 47, D253–D258. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).