Interactions of Linalool and Linalyl Acetate with Selected Dog Cytochrome P450 (CYP) Proteins Identified by In Silico Drug Discovery Followed by Molecular Docking Analysis
Abstract
1. Introduction
2. Results
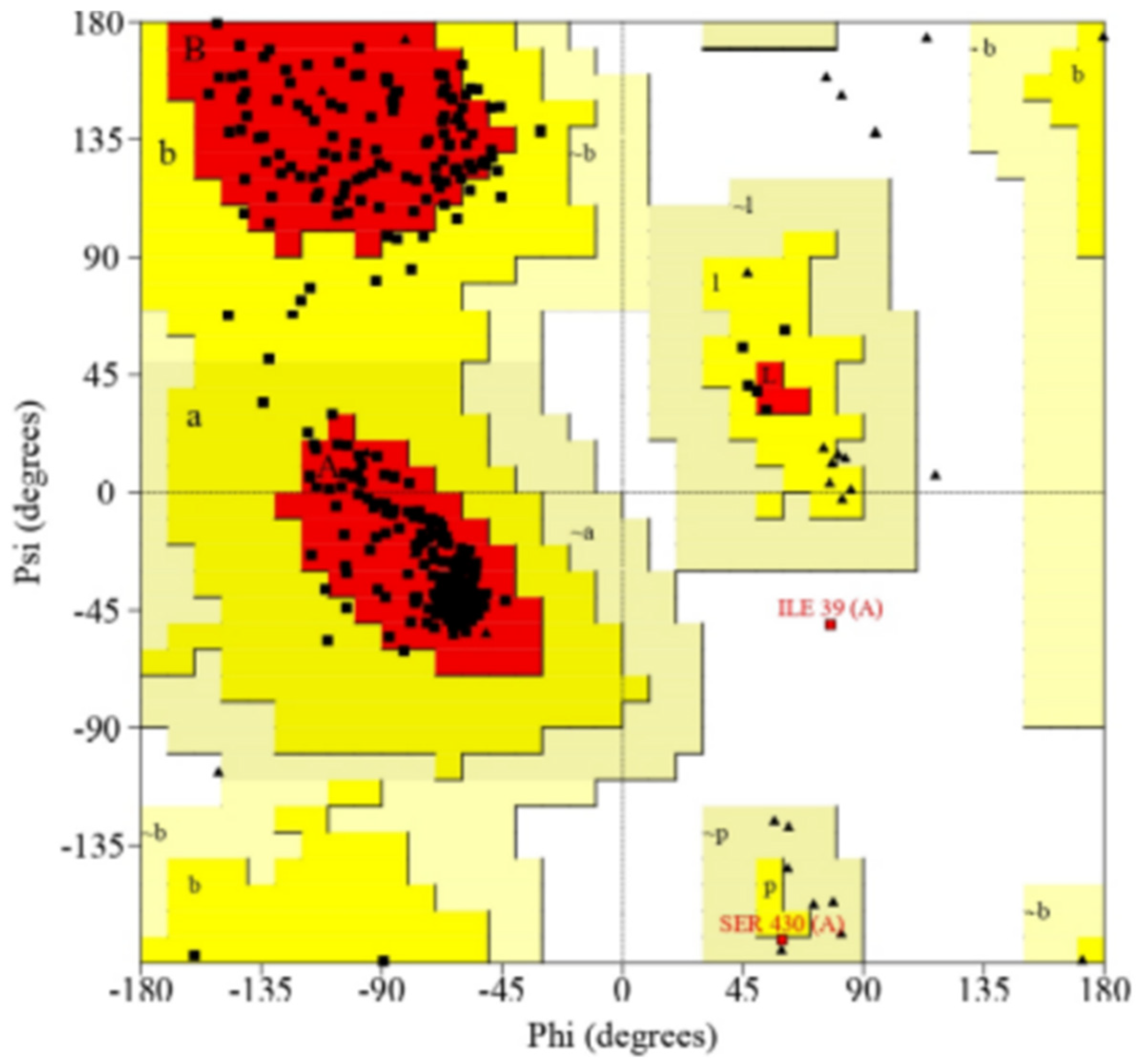
2.1. Molecular Modeling of the Target Proteins CYP2B11, CYP2C21, and CYP2D15
2.2. Molecular Docking Analysis
2.3. Ligand-Based Drug Design
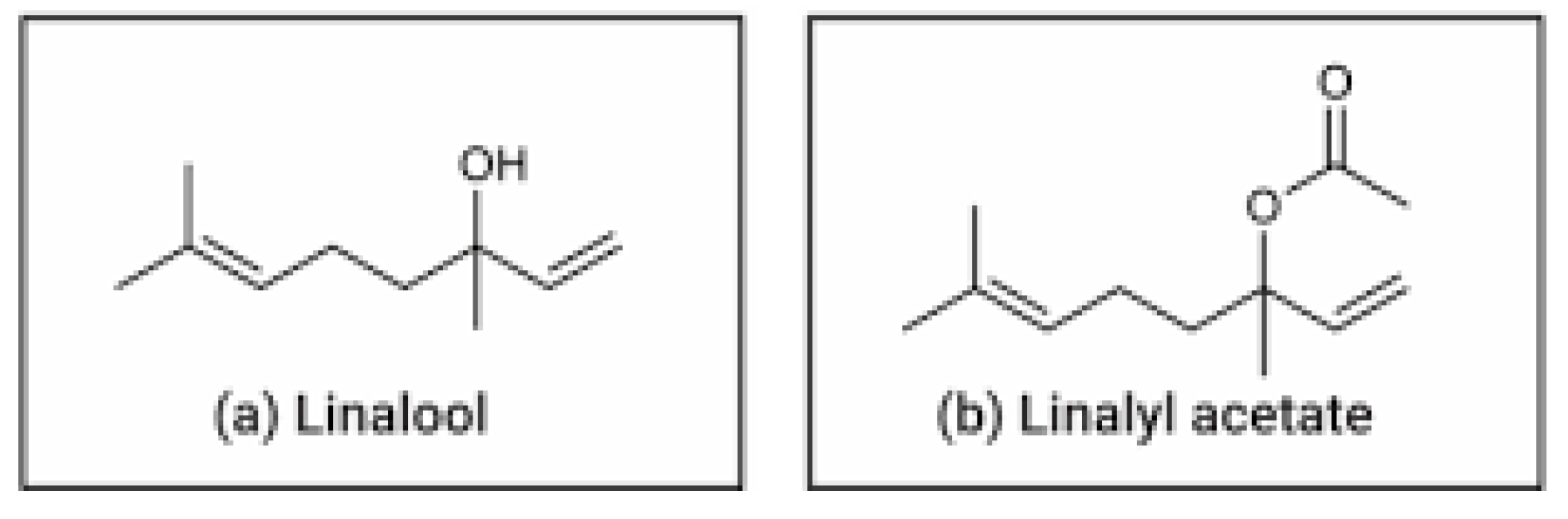
2.3.1. Pharmacophore Properties of Linalool and Linalyl Acetate
2.3.2. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Molecular Modeling of the Target Proteins
4.2. Structure-Based Drug Designing—Preparation of Proteins and Ligand Compound Molecules for Docking Analysis
4.3. Ligand-Based Drug Design
4.3.1. Molecular Docking
4.3.2. Ligands Preparation
4.3.3. Protein Preparation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| ADMET | Absorption, distribution, metabolism, excretion, and toxicity |
| ADT | AutoDock Tools |
| ARG | Arginine |
| ASN | Asparagine |
| ASP | Aspartic acid |
| AtomQ | Qualifying Atom |
| CYP450 | Cytochrome P450 |
| DonAcc | Dual donor/acceptor |
| F | Feature number |
| GBVI/WSA | Generalized-born volume integral/weighted surface area |
| GLN | Glutamine |
| GMQE | Global model quality estimate |
| Hyd | Hydrophobic region |
| ILE | Isoleucine |
| Km | Michaelis-Menten constant |
| LEU | Leucine |
| LIN | Linalool |
| LINAct | Linalyl acetate |
| MOE | Molecular Operating Environment |
| PHE | Phenylalanine |
| PSA | Polar surface area |
| RMS | Root mean square |
| RMSD | Root mean square deviation |
| SER | Serine |
| THR | Threonine |
| TYR | Tyrosine |
| VAL | Valine |
References
- Chen, J.; Lin, A.; Luo, P. Advancing pharmaceutical research: A comprehensive review of cutting-edge tools and technologies. Curr. Pharm. Anal. 2024, 21, 1–19. [Google Scholar] [CrossRef]
- Wooller, S.K.; Benstead-Hume, G.; Chen, X.; Ali, Y.; Pearl, F.M.G. Bioinformatics in translational drug discovery. Biosci. Rep. 2017, 37, BSR20160180. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.K.; Mariam, Z. Computer-aided drug design and drug discovery: A prospective analysis. Pharmaceuticals 2023, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Oselusi, S.O.; Dube, P.; Odugbemi, A.I.; Akinyede, K.A.; Ilori, T.L.; Egieyeh, E.; Sibuyi, N.R.; Meyer, M.; Madiehe, A.M.; Wyckoff, G.J.; et al. The role and potential of computer-aided drug discovery strategies in the discovery of novel antimicrobials. Comput. Biol. Med. 2024, 169, 107927. [Google Scholar] [CrossRef]
- Adelusi, T.I.; Oyedele, A.-Q.K.; Boyenle, I.D.; Ogunlana, A.T.; Adeyemi, R.O.; Ukachi, C.D.; Idris, M.O.; Olaoba, O.T.; Adedotun, I.O.; Kolawole, O.E.; et al. Molecular modeling in drug discovery. Inform. Med. Unlocked 2022, 29, 100880. [Google Scholar] [CrossRef]
- Bibow, A.; Oleszek, W. Essential oils as potential natural antioxidants, antimicrobial, and antifungal agents in active food packaging. Antibiotics 2024, 13, 1168. [Google Scholar] [CrossRef]
- Crișan, I.; Ona, A.; Vârban, D.; Muntean, L.; Vârban, R.; Stoie, A.; Mihăiescu, T.; Morea, A. Current trends for Lavender (Lavandula angustifolia Mill.) crops and products with emphasis on essential oil quality. Plants 2023, 12, 357. [Google Scholar] [CrossRef]
- Angulo, S.M.; Occhieppo, V.B.; Moya, C.; Crespo, R.; Bregonzio, C. Anxiolytic-like effect characterization of essential oil from local lavender cultivation. Pharmaceuticals 2025, 18, 624. [Google Scholar] [CrossRef]
- Genovese, A.G.; McLean, M.K.; Khan, S.A. Adverse reactions from essential oil-containing natural flea products exempted from Environmental Protection Agency regulations in dogs and cats. J. Vet. Emerg. Crit. Care 2012, 22, 470–475. [Google Scholar] [CrossRef]
- Gans, J.H.; Korson, R.; Cater, M.R.; Ackerly, C.C. Effects of short-term and long-term theobromine administration to male dogs. Toxicol. Appl. Pharmacol. 1980, 53, 481–496. [Google Scholar] [CrossRef]
- Villar, D.; Knight, M.J.; Hansen, S.R.; Buck, W.B. Toxicity of melaleuca oil and related essential oils applied topically on dogs and cats. Vet. Hum. Toxicol. 1994, 36, 139–142. [Google Scholar]
- Khan, S.A.; McLean, M.K.; Slater, M.R. Concentrated tea tree oil toxicosis in dogs and cats: 443 cases (2002–2012). J. Am. Vet. Med. Assoc. 2014, 244, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Sudekum, M.; Poppenga, R.H.; Raju, N.; Braselton, W.E., Jr. Pennyroyal oil toxicosis in a dog. J. Am. Vet. Med. Assoc. 1992, 200, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Ebani, V.V.; Mancianti, F. Use of essential oils in veterinary medicine to combat bacterial and fungal infections. Vet. Sci. 2020, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-driven deep learning techniques in protein structure prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Lohning, A.E.; Levonis, S.M.; Williams-Noonan, B.; Schweiker, S.S. A practical guide to molecular docking and homology modelling for medicinal chemists. Curr. Top. Med. Chem. 2017, 17, 2023–2040. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug Discov. 2016, 11, 137–148. [Google Scholar] [CrossRef]
- Muhammed, M.T.; Aki-Yalcin, E. Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chem. Biol. Drug Des. 2019, 93, 12–20. [Google Scholar] [CrossRef]
- Zawaira, A.; Ching, L.Y.; Coulson, L.; Blackburn, J.; Wei, Y.C. An expanded, unified substrate recognition site map for mammalian cytochrome P450s: Analysis of molecular interactions between 15 mammalian CYP450 isoforms and 868 substrates. Curr. Drug Metab. 2011, 12, 684–700. [Google Scholar] [CrossRef]
- Martiny, V.Y.; Carbonell, P.; Chevillard, F.; Moroy, G.; Nicot, A.B.; Vayer, P.; Villoutreix, B.O.; Miteva, M.A. Integrated structure- and ligand-based in silico approach to predict inhibition of cytochrome P450 2D6. Bioinformatics 2015, 31, 3930–3937. [Google Scholar] [CrossRef] [PubMed]
- Mestres, J. Structure conservation in cytochromes P450. Proteins 2005, 58, 596–609. [Google Scholar] [CrossRef]
- Szklarz, G.D.; Graham, S.E.; Paulsen, M.D. Molecular modeling of mammalian cytochromes P450: Application to study enzyme function. Vitam. Horm. 2000, 58, 53–87. [Google Scholar] [PubMed]
- Chothia, C.; Lesk, A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986, 5, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, reviews3003. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Ayinla, Z.A.; Ademakinwa, A.N.; Agboola, F.K. Comparative modelling, molecular docking and immobilization studies on Rhizopus oryzae lipase: Evaluation of potentials for fatty acid methyl esters synthesis. J. Biomol. Struct. Dyn. 2023, 41, 7235–7247. [Google Scholar] [CrossRef]
- Binbay, F.A.; Rathod, D.C.; George, A.A.P.; Imhof, D. Quality Assessment of selected protein structures derived from homology modeling and AlphaFold. Pharmaceuticals 2023, 16, 1662. [Google Scholar] [CrossRef]
- Chadha, A.; Madyastha, K.M. Omega-hydroxylation of acyclic monoterpene alcohols by rat lung microsomes. Biochem. Biophys. Res. Commun. 1982, 108, 1271–1277. [Google Scholar] [CrossRef]
- Chadha, A.; Madyastha, K.M. Metabolism of geraniol and linalool in the rat and effects on liver and lung microsomal enzymes. Xenobiotica 1984, 14, 365–374. [Google Scholar] [CrossRef]
- Meesters, R.J.; Duisken, M.; Hollender, J. Study on the cytochrome P450-mediated oxidative metabolism of the terpene alcohol linalool: Indication of biological epoxidation. Xenobiotica 2007, 37, 604–617. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.-J. Introduction to molecular recognition models. In Protein-Ligand Interactions: From Molecular Recognition to Drug Design; Böhm, H.-J., Schneider, G., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2003; pp. 21–50. [Google Scholar]
- IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature and symbolism for amino acids and peptides Recommendations 1983. Biochem. J. 1984, 219, 345–373. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, S. Molecular docking: A structure-based approach for drug repurposing. In Silico Drug Design; Roy, K., Ed.; Academic Press: London, UK, 2019; pp. 161–189. [Google Scholar]
- Miyazawa, M.; Sugie, A.; Shimada, T. Roles of human CYP2A6 and 2B6 and rat CYP2C11 and 2B1 in the 10-hydroxylation of (-)-verbenone by liver microsomes. Drug Metab. Dispos. 2003, 31, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, M.; Gyoubu, K. Metabolism of (-)-fenchone by CYP2A6 and CYP2B6 in human liver microsomes. Xenobiotica 2007, 37, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.A.; Kim, H.; Ku, H.Y.; Ahn, H.J.; Park, S.J.; Bae, S.K.; Shin, J.G.; Liu, K.H. The monoterpenoids citral and geraniol are moderate inhibitors of CYP2B6 hydroxylase activity. Chem. Biol. Interact. 2008, 174, 141–146. [Google Scholar] [CrossRef]
- Krihariyani, D.; Woelansari, E.D.; Haryanto, E.; Sasongkowati, R.; Handayati, A.; Astuti, S.S.E. In silico study of the potential of Brazilein sappan wood as a beta-lactamase inhibitor against extended-spectrum beta-lactamase-encoding genes. Malays. J. Med. Sci. 2024, 31, 107–116. [Google Scholar] [CrossRef]
- Liu, S. Enzymes. In Bioprocess Engineering, 2nd ed.; Liu, S., Ed.; Elsevier: Amsterdam, Netherlands, 2017; pp. 297–373. [Google Scholar]
- Shenouda, J.; Green, P.; Sultatos, L. An evaluation of the inhibition of human butyrylcholinesterase and acetylcholinesterase by the organophosphate chlorpyrifos oxon. Toxicol. Appl. Pharmacol. 2009, 241, 135–142. [Google Scholar] [CrossRef]
- Maurer, T.S.; Tabrizi-Fard, M.A.; Fung, H.L. Impact of mechanism-based enzyme inactivation on inhibitor potency: Implications for rational drug discovery. J. Pharm. Sci. 2000, 89, 1404–1414. [Google Scholar] [CrossRef]
- Yadav, S.; Mody, T.A.; Sharma, A.; Bachhawat, A.K. A genetic screen to identify genes influencing the secondary redox couple NADPH/NADP+ in the yeast Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2020, 10, 371–378. [Google Scholar] [CrossRef]
- Isin, E.M.; Guengerich, F.P. Substrte binding to cytochromes P450. Anal. Bioanal. Chem. 2008, 392, 1019–1030. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, S.; Wu, Y.; Jiang, T.; Wang, L.; Jiang, J.; Wen, J.; Deng, Y. Multiple CH/π interactions maintain the binding of aflatoxin B1 in the active cavity of human cytochrome P450 1A2. Toxins 2019, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Dudekula, D.B.; Rallabandi, V.P.S.; Srinivasan, S.; Natarajan, S.; Chung, H.; Park, J. Identification of potential cytochrome P450 3A5 inhibitors: An extensive virtual screening through molecular docking, negative image-based screening, machine learning and molecular dynamics simulation studies. Int. J. Mol. Sci. 2022, 23, 9374. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.; Kim, V.; Kim, C.; Lee, Y.B.; Kim, D. Structural insights into the interaction of terpenoids with Streptomyces avermitilis CYP107P2. Biomol. Ther. 2024, 32, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, K.; Kawaguchi, Y.; Watanabe, Y.; Masubuchi, Y.; Shinohara, Y.; Hori, H. Flexible structure of cytochrome P450: Promiscuity of ligand binding in the CYP3A4 heme pocket. Anticancer Res. 2009, 29, 935–942. [Google Scholar]
- Tie, Y.; McPhail, B.; Hong, H.; Pearce, B.A.; Schnackenberg, L.K.; Ge, W.; Buzatu, D.A.; Wilkes, J.G.; Fuscoe, J.C.; Tong, W.; et al. Modeling chemical interaction profiles: II. Molecular docking, spectral data-activity relationship, and structure-activity relationship models for potent and weak inhibitors of cytochrome P450 CYP3A4 isozyme. Molecules 2012, 17, 3407–3460. [Google Scholar] [CrossRef]
- Kirton, S.B.; Baxter, C.A.; Sutcliffe, M.J. Comparative modelling of cytochromes P450. Adv. Drug Deliv. Rev. 2002, 54, 385–406. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef]
- Cupp-Vickery, J.; Anderson, R.; Hatziris, Z. Crystal structures of ligand complexes of P450eryF exhibiting homotropic cooperativity. Proc. Natl. Acad. Sci. USA 2000, 97, 3050–3055. [Google Scholar] [CrossRef]
- Mast, N.; White, M.A.; Bjorkhem, I.; Johnson, E.F.; Stout, C.D.; Pikuleva, I.A. Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc. Natl. Acad. Sci. USA 2008, 105, 9546–9551. [Google Scholar] [CrossRef]
- Nair, P.C.; McKinnon, R.A.; Miners, J.O. Cytochrome P450 structure-function: Insights from molecular dynamics simulations. Drug Metab. Rev. 2016, 48, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Gay, S.C.; Roberts, A.G.; Halpert, J.R. Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by X-ray crystallography and NMR. Future Med. Chem. 2010, 2, 1451–1468. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, S.; Yesudhas, D.; Durai, P.; Anwar, M.A.; Gosu, V.; Choi, S. A combined molecular docking/dynamics approach to probe the binding mode of cancer drugs with cytochrome P450 3A4. Molecules 2015, 20, 14915–14935. [Google Scholar] [CrossRef] [PubMed]
- Sarkhel, S.; Desiraju, G.R. N-H…O, O-H…O, and C-H…O hydrogen bonds in protein-ligand complexes: Strong and weak interactions in molecular recognition. Proteins 2004, 54, 247–259. [Google Scholar] [CrossRef]
- Labute, P. The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008, 29, 1693–1698. [Google Scholar] [CrossRef]
- Chan, A.W.; Laskowski, R.A.; Selwood, D.L. Chemical fragments that hydrogen bond to Asp, Glu, Arg, and His side chains in protein binding sites. J. Med. Chem. 2010, 53, 3086–3094. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Xiao, T.; Wang, M.; Wei, G.W. Rigidity strengthening: A mechanism for protein-ligand binding. J. Chem. Inf. Model. 2017, 57, 1715–1721. [Google Scholar] [CrossRef]
- Lee, Y.; Park, H.G.; Kim, V.; Cho, M.A.; Kim, H.; Ho, T.H.; Cho, K.S.; Lee, I.S.; Kim, D. Inhibitory effect of α-terpinyl acetate on cytochrome P450 2B6 enzymatic activity. Chem. Biol. Interact 2018, 289, 90–97. [Google Scholar] [CrossRef]
- Austin, C.A.; Shephard, E.A.; Pike, S.F.; Rabin, B.R.; Phillips, I.R. The effect of terpenoid compounds on cytochrome P-450 levels in rat liver. Biochem. Pharmacol. 1988, 37, 2223–2229. [Google Scholar] [CrossRef]
- Kramlinger, V.M.; von Weymarn, L.B.; Murphy, S.E. Inhibition and inactivation of cytochrome P450 2A6 and cytochrome P450 2A13 by menthofuran, β-nicotyrine and menthol. Chem. Biol. Interact 2012, 197, 87–92. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Wadood, A.; Riaz, M.; Jamal, S.B.; Shah, M. Interactions of ketoamide inhibitors on HCV NS3/4A protease target: Molecular docking studies. Mol. Biol. Rep. 2014, 41, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Riasat, I.; Bakhtiar, S.M.; Faheem, M.; Jaiswal, A.K.; Naeem, M.; Khan, R.; Khan, A.U.; Khalil, A.A.K.; Haider, A.; Junaid, M.; et al. Application of pan genomics towards the druggability of Clostridium botulinum. Appl. Nanosci. 2022, 12, 3237–3249. [Google Scholar] [CrossRef]
- Wadood, A.; Jamal, S.B.; Riaz, M.; Mir, A. Computational analysis of benzofuran-2-carboxlic acids as potent Pim-1 kinase inhibitors. Pharm. Biol. 2014, 52, 1170–1178. [Google Scholar] [CrossRef]
- Faheem, M.; Jamal, S.B. Identification of Zika Virus NS5 novel inhibitors through virtual screening and docking studies. Life Sci. 2020, 1, 3–7. [Google Scholar] [CrossRef]




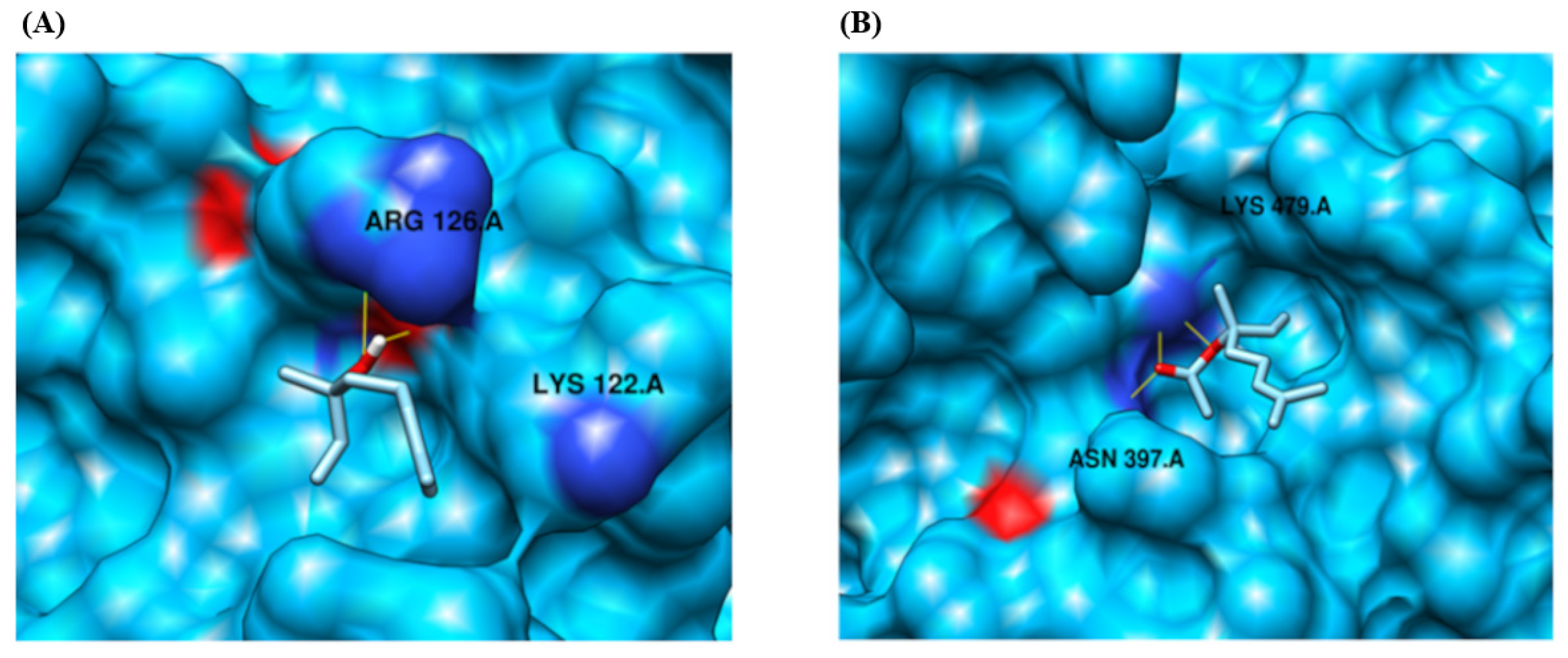

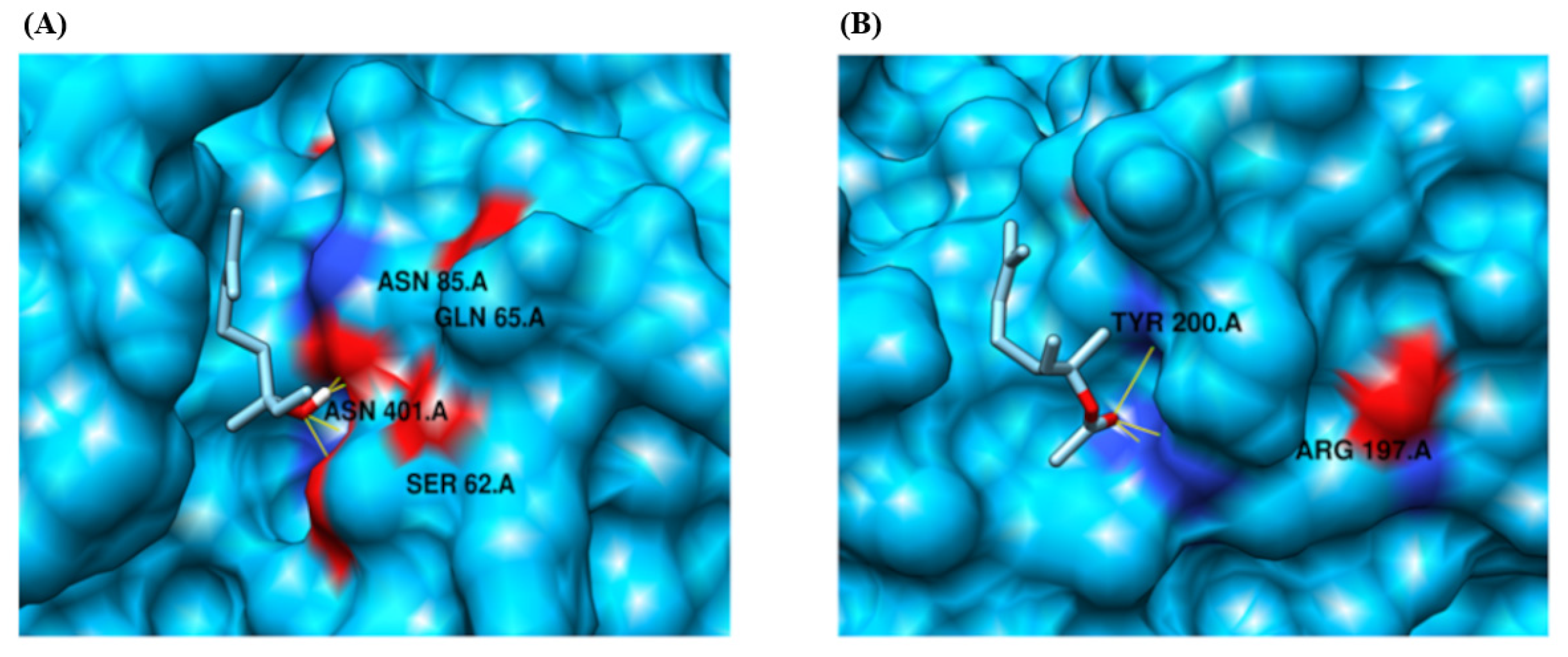

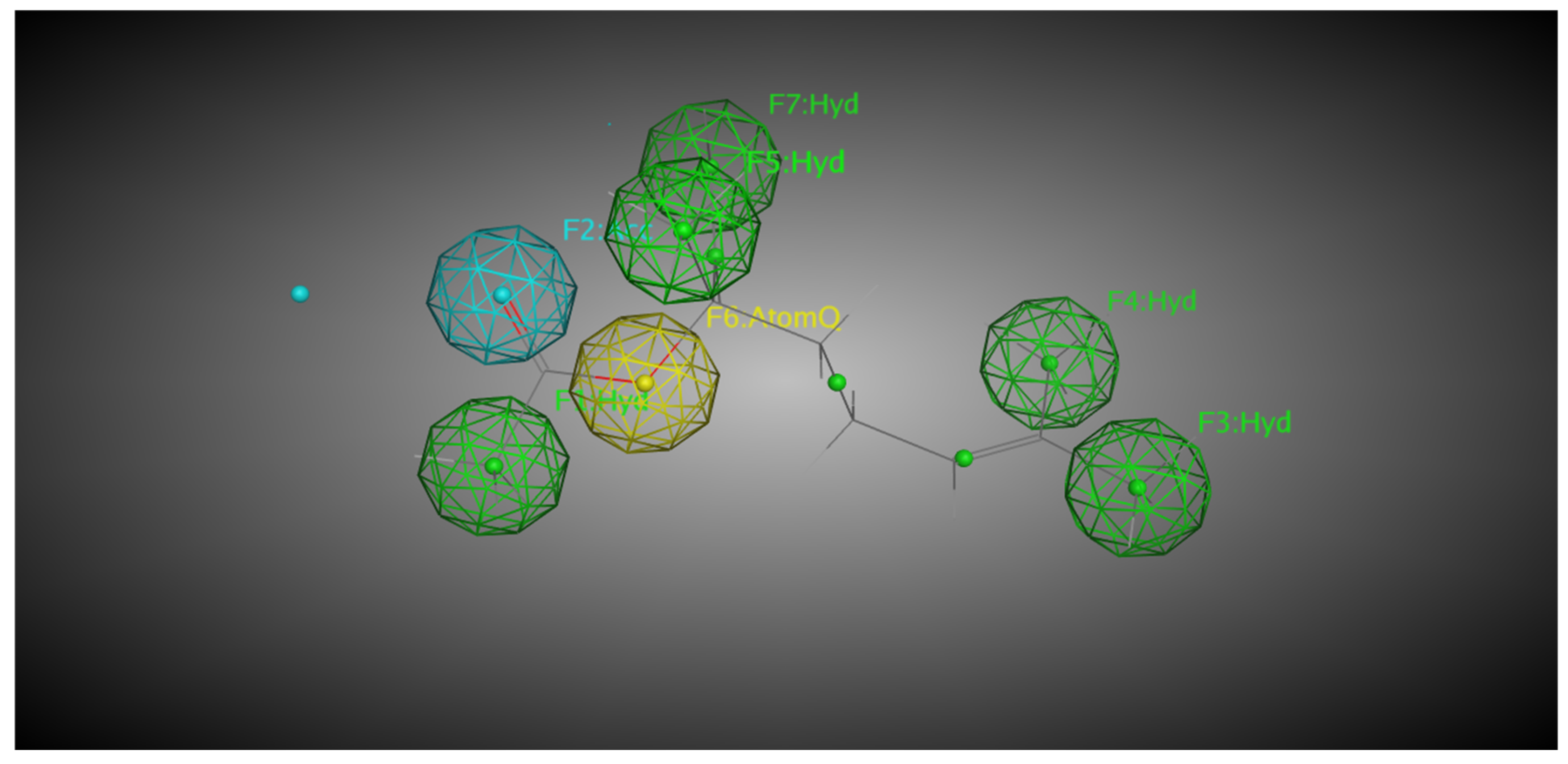
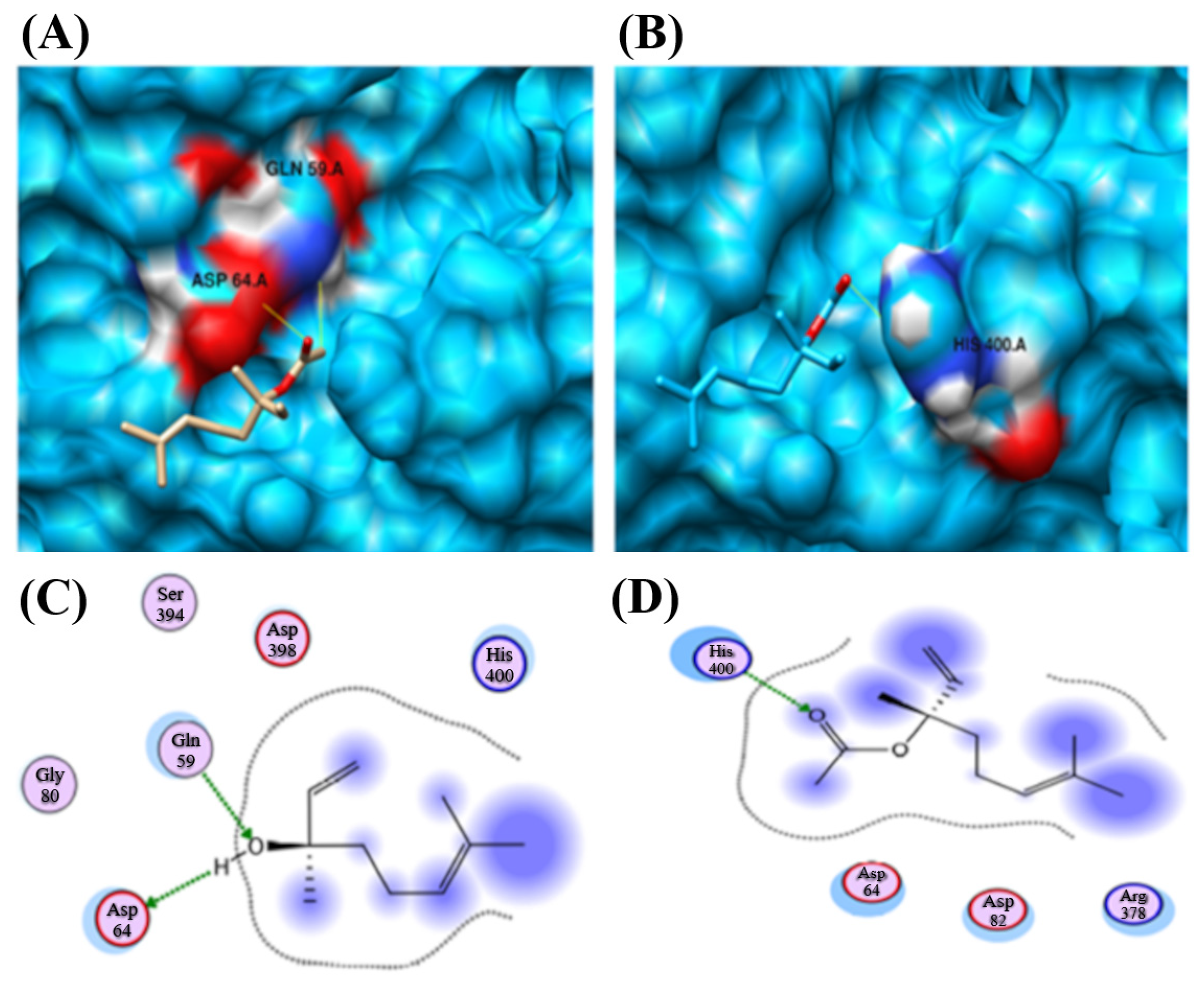


| Protein | Compound Ligand Name | Autodock Vina Binding Affinity | Binding Residues/Number of Hydrogen Bonds |
|---|---|---|---|
| CYP2B11 | Linalool | −4.096 | LYS 122, ARG 126/2 |
| CYP2B11 | Linalyl Acetate | −4.643 | ASN 397, LYS 479/3 |
| CYP2C21 | Linalool | −4.487 | ASN 474, SER 51/2 |
| CYP2C21 | Linalyl Acetate | −4.998 | ASN 474, SER 51/2 |
| CYP2D15 | Linalool | −4.144 | SER 62, GLN 65, ASN 85 and ASN 401/4 |
| CYP2D15 | Linalyl Acetate | −4.199 | ARG 197 and TYR 200/3 |
| Protein | Compound Ligand Name | Docking Scores | GBVI/WSA 1 | Binding Residues/Number of Hydrogen Bonds |
|---|---|---|---|---|
| CYP2B11 | Linalool | −7.624 | −15.8441 | GLN 59 and ASP 64/2 |
| CYP2B11 | Linalyl Acetate | −6.917 | −13.7862 | HIS 400/1 |
| CYP2C21 | Linalool | −7.612 | −13.345 | ASN 204, SER 208/2 |
| CYP2C21 | Linalyl Acetate | −6.983 | −12.722 | No interaction |
| CYP2D15 | Linalool | −6.533 | −13.6314 | HIS 112/1 |
| CYP2D15 | Linalyl Acetate | −6.711 | −13.2242 | ASN 255/1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares-Santos, R.R.; Jaiswal, A.K.; Ferreira, R.C.M.; Azevedo, V.A.d.C.; Aburjaile, F.F.; Soto-Blanco, B. Interactions of Linalool and Linalyl Acetate with Selected Dog Cytochrome P450 (CYP) Proteins Identified by In Silico Drug Discovery Followed by Molecular Docking Analysis. Pharmaceuticals 2025, 18, 1499. https://doi.org/10.3390/ph18101499
Soares-Santos RR, Jaiswal AK, Ferreira RCM, Azevedo VAdC, Aburjaile FF, Soto-Blanco B. Interactions of Linalool and Linalyl Acetate with Selected Dog Cytochrome P450 (CYP) Proteins Identified by In Silico Drug Discovery Followed by Molecular Docking Analysis. Pharmaceuticals. 2025; 18(10):1499. https://doi.org/10.3390/ph18101499
Chicago/Turabian StyleSoares-Santos, Raquel Rodrigues, Arun Kumar Jaiswal, Renata Cristina Mendes Ferreira, Vasco Ariston de Carvalho Azevedo, Flávia Figueira Aburjaile, and Benito Soto-Blanco. 2025. "Interactions of Linalool and Linalyl Acetate with Selected Dog Cytochrome P450 (CYP) Proteins Identified by In Silico Drug Discovery Followed by Molecular Docking Analysis" Pharmaceuticals 18, no. 10: 1499. https://doi.org/10.3390/ph18101499
APA StyleSoares-Santos, R. R., Jaiswal, A. K., Ferreira, R. C. M., Azevedo, V. A. d. C., Aburjaile, F. F., & Soto-Blanco, B. (2025). Interactions of Linalool and Linalyl Acetate with Selected Dog Cytochrome P450 (CYP) Proteins Identified by In Silico Drug Discovery Followed by Molecular Docking Analysis. Pharmaceuticals, 18(10), 1499. https://doi.org/10.3390/ph18101499









