A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern
Abstract
1. Introduction
2. Results
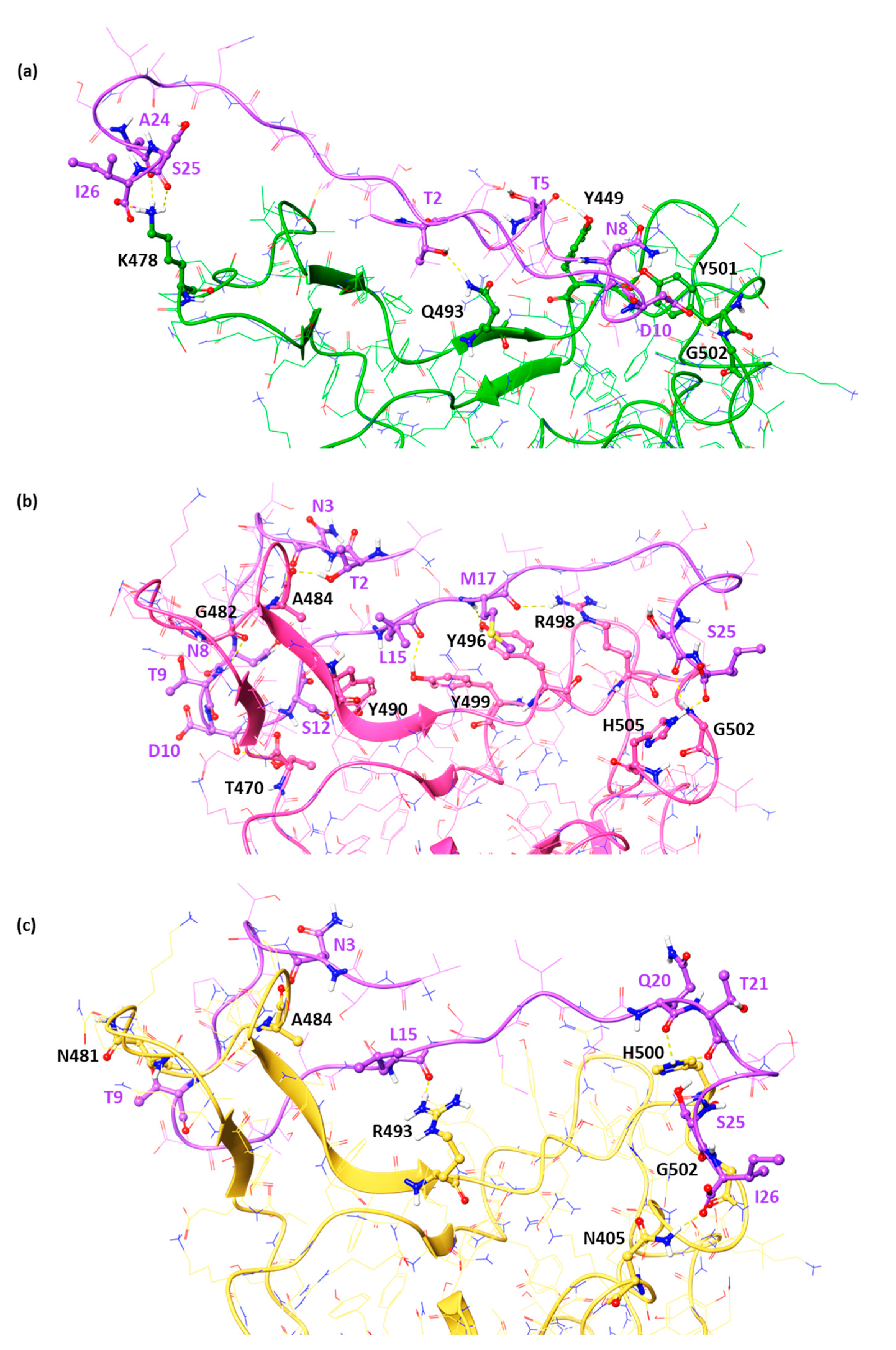
2.1. In Silico Studies of the DPP4270–295 Peptide against the RBD of Emenging Variants of Concern
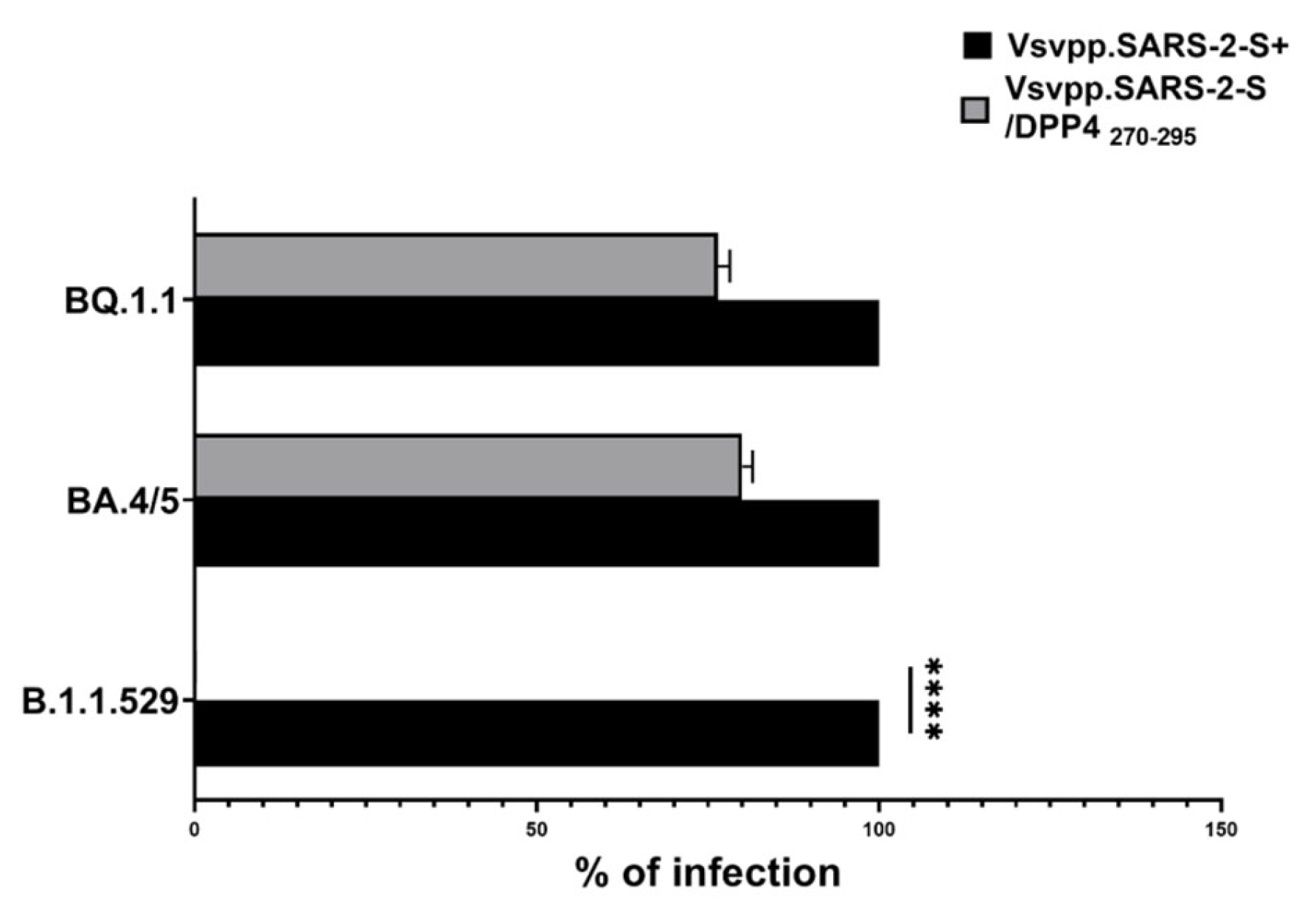
2.2. DPP4270–295 Peptide Partially Blocks SARS-CoV-2 Entry into Cells
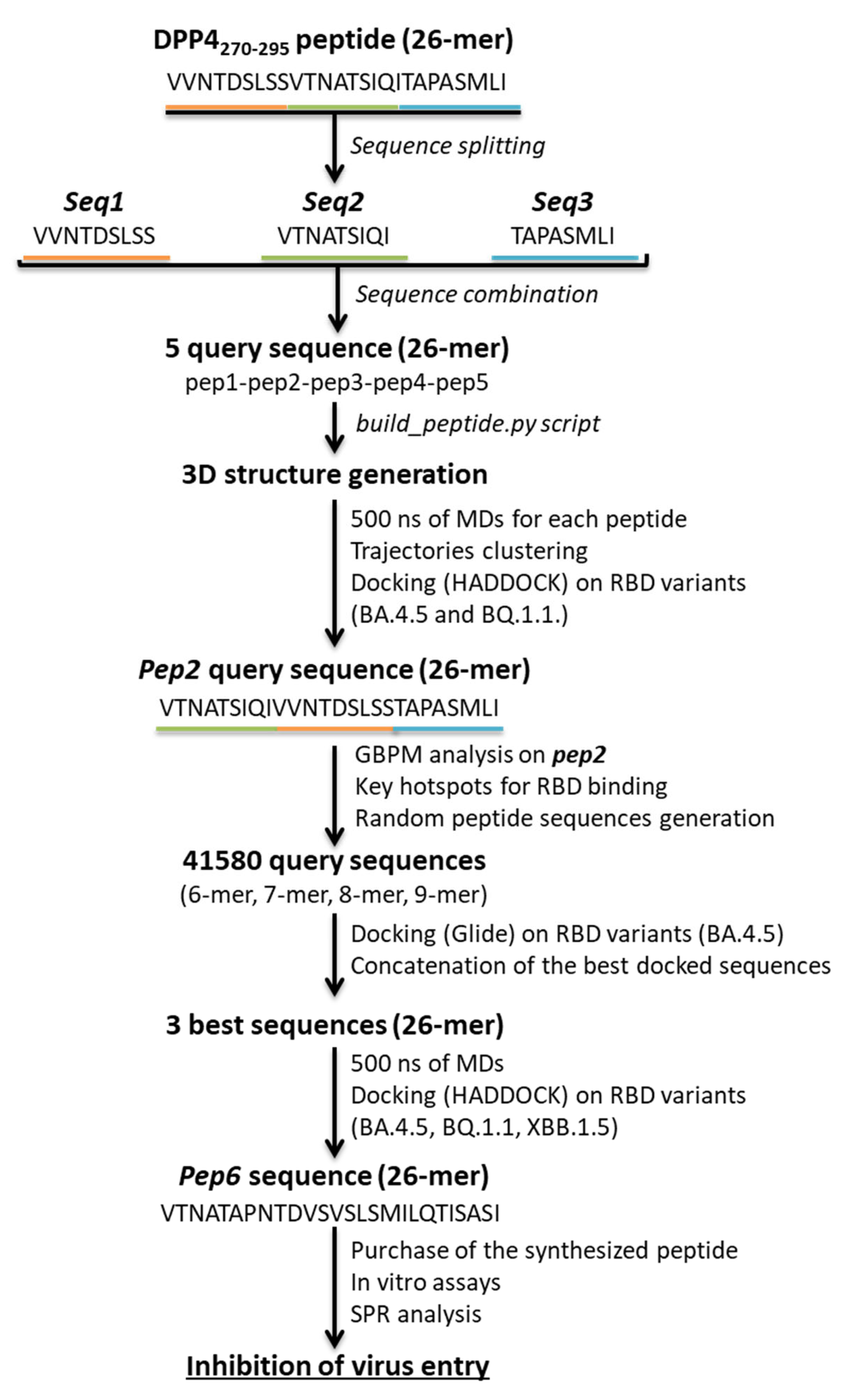
2.3. Design of Novel DPP4-Derived Binder of SARS-CoV-2 RBD Domain
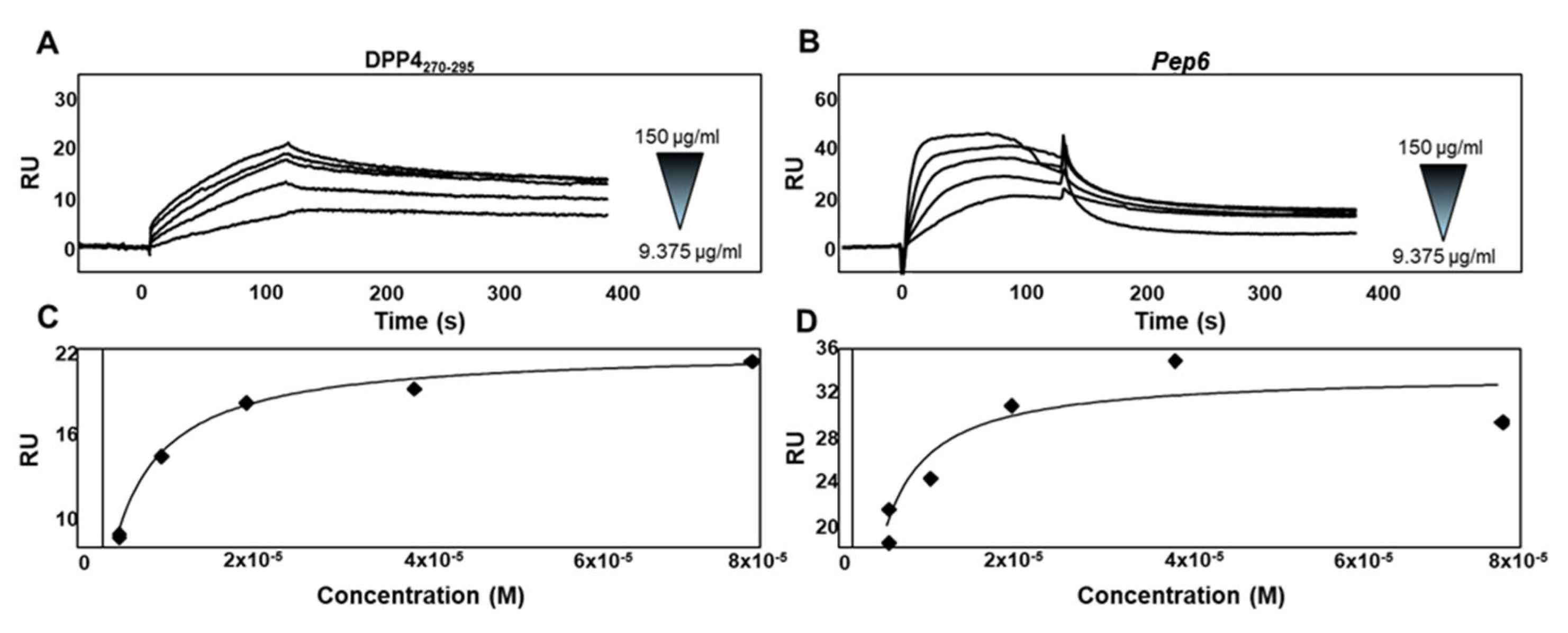
2.4. Experimental Validation of the Newly Designed Peptide pep6
3. Discussion
4. Materials and Methods
4.1. Cells
4.2. VSV_Pseudo SARS-CoV-2 Omicron B.1.1.529, BA.4/5, BQ.1.1, and XBB.1.5 Strains Spike with Luciferase Reporter
4.3. Protein Preparation of RBD Variants and Docking Simulations
4.4. Rational Peptides Design against RBD Domain
4.5. Peptide Design and Production
4.6. In Silico Analysis of the Conformations of DPP4-Derived Peptides
4.7. MTS and In Vitro Neutralization Assay of DPP4270–295 and DPP4-Derived Peptide
4.8. Surface Plasmon Resonance (SPR) Assay
4.9. Gene Expression Analysis
4.10. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. Available online: www.who.int/news-room/fact-sheets/detail/coronavirus-disease-(covid-19) (accessed on 5 May 2023).
- Atella, V.; Scandizzo, P.L. Chapter 1—The origins of infections: Some basic concepts. In The COVID-19 Disruption and the Global Health Challenge; Atella, V., Scandizzo, P.L., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 3–20. [Google Scholar]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Atella, V.; Scandizzo, P.L. Chapter 11—How to manage the risk of new pandemics. In The COVID-19 Disruption and the Global Health Challenge; Atella, V., Scandizzo, P.L., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 409–438. [Google Scholar]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Zé-Zé, L.; Duarte, S.; Vieira, L.; Guiomar, R.; Gomes, J.P.; Gordo, I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health 2022, 10, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Romeo, I.; Prandi, I.G.; Giombini, E.; Gruber, C.E.M.; Pietrucci, D.; Borocci, S.; Abid, N.; Fava, A.; Beccari, A.R.; Chillemi, G. The spike mutants website: A worldwide used resource against SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 13082. [Google Scholar] [CrossRef]
- Malet, I.; Ambrosio, F.A.; Subra, F.; Herrmann, B.; Leh, H.; Bouger, M.-C.; Artese, A.; Katlama, C.; Talarico, C.; Romeo, I. Pathway involving the N155H mutation in HIV-1 integrase leads to dolutegravir resistance. J. Antimicrob. Chemother. 2018, 73, 1158–1166. [Google Scholar] [CrossRef]
- Lupia, A.; Moraca, F.; Bagetta, D.; Maruca, A.; Ambrosio, F.A.; Rocca, R.; Catalano, R.; Romeo, I.; Talarico, C.; Ortuso, F. Computer-based techniques for lead identification and optimization II: Advanced search methods. Phys. Sci. Rev. 2019, 5, 20180114. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiao, B.; Qu, L.; Yang, D.; Liu, R. The development of COVID-19 treatment. Front. Immunol. 2023, 14, 1125246. [Google Scholar] [CrossRef]
- Todaro, B.; Ottalagana, E.; Luin, S.; Santi, M. Targeting peptides: The new generation of targeted drug delivery systems. Pharmaceutics 2023, 15, 1648. [Google Scholar] [CrossRef]
- Lui, G.; Guaraldi, G. Drug treatment of COVID-19 infection. Curr. Opin. Pulm. Med. 2023, 29, 174. [Google Scholar] [CrossRef]
- Konno, Y.; Kimura, I.; Uriu, K.; Fukushi, M.; Irie, T.; Koyanagi, Y.; Sauter, D.; Gifford, R.J.; Nakagawa, S.; Sato, K. SARS-CoV-2 orf3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020, 32, 108185. [Google Scholar] [CrossRef]
- Cox, M.; Peacock, T.P.; Harvey, W.T.; Hughes, J.; Wright, D.W.; Willett, B.J.; Thomson, E.; Gupta, R.K.; Peacock, S.J.; Robertson, D.L. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2023, 21, 112–124. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The major cell entry receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Vankadari, N.; Wilce, J.A. Emerging covid-19 coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020, 9, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Murdocca, M.; Citro, G.; Romeo, I.; Lupia, A.; Miersch, S.; Amadio, B.; Bonomo, A.; Rossi, A.; Sidhu, S.S.; Pandolfi, P.P. Peptide platform as a powerful tool in the fight against COVID-19. Viruses 2021, 13, 1667. [Google Scholar] [CrossRef] [PubMed]
- Ortuso, F.; Langer, T.; Alcaro, S. Gbpm: Grid-based pharmacophore model: Concept and application studies to protein–protein recognition. Bioinformatics 2006, 22, 1449–1455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fenton, C.; Keam, S.J. Emerging small molecule antivirals may fit neatly into COVID-19 treatment. Drugs Ther. Perspect. 2022, 38, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Mattiuzzi, C.; Henry, B.M. Updated picture of SARS-CoV-2 variants and mutations. Diagnosis 2021, 9, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. The Coronavirus is Mutating—Does It Matter? Nature Publishing Group: London, UK, 2020. [Google Scholar]
- Solerte, S.B.; Di Sabatino, A.; Galli, M.; Fiorina, P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol. 2020, 57, 779–783. [Google Scholar] [CrossRef]
- Sebastián-Martín, A.; Sánchez, B.G.; Mora-Rodríguez, J.M.; Bort, A.; Díaz-Laviada, I. Role of dipeptidyl peptidase-4 (DPP4) on COVID-19 physiopathology. Biomedicines 2022, 10, 2026. [Google Scholar] [CrossRef] [PubMed]
- Moroy, G.; Tuffery, P. Peptide-based strategies against SARS-CoV-2 attack: An updated in silico perspective. Front. Drug Discov. 2022, 2, 899477. [Google Scholar] [CrossRef]
- Rawat, K.; Kumari, P.; Saha, L. COVID-19 vaccine: A recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharmacol. 2021, 892, 173751. [Google Scholar] [CrossRef] [PubMed]
- Saini, J.; Kaur, P.; Malik, N.; Lakhawat, S.S.; Sharma, P.K. Antimicrobial peptides: A promising tool to combat multidrug resistance in SARS-CoV-2 era. Microbiol. Res. 2022, 265, 127206. [Google Scholar] [CrossRef]
- Lega, S.; Naviglio, S.; Volpi, S.; Tommasini, A. Recent insight into SARS-CoV-2 immunopathology and rationale for potential treatment and preventive strategies in COVID-19. Vaccines 2020, 8, 224. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, F.; Deb, D.; Srivastava, A.P.; Liò, P.; Liso, A. From infection to immunity: Understanding the response to SARS-CoV-2 through in-silico modeling. Front. Immunol. 2021, 12, 646972. [Google Scholar] [CrossRef] [PubMed]
- Holenya, P.; Lange, P.J.; Reimer, U.; Woltersdorf, W.; Panterodt, T.; Glas, M.; Wasner, M.; Eckey, M.; Drosch, M.; Hollidt, J.M. Peptide microarray-based analysis of antibody responses to SARS-CoV-2 identifies unique epitopes with potential for diagnostic test development. Eur. J. Immunol. 2021, 51, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.H.; Downard, K.M. Mass spectrometry analytical responses to the SARS-CoV-2 coronavirus in review. TrAC Trends Anal. Chem. 2021, 142, 116328. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic strategies for COVID-19: Progress and lessons learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef]
- VanPatten, S.; He, M.; Altiti, A.; Cheng, K.F.; Ghanem, M.H.; Al-Abed, Y. Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics. Future Med. Chem. 2020, 12, 1647–1656. [Google Scholar] [CrossRef]
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.-M.; Loh, H.-S.; Le, C.F. The potential of antiviral peptides as COVID-19 therapeutics. Front. Pharmacol. 2020, 11, 575444. [Google Scholar] [CrossRef] [PubMed]
- Zarkesh, K.; Akbarian, M.; Tayebi, L.; Uversky, V.N.; Rubio-Casillas, A.; Redwan, E.M. Harnessing antiviral peptides as means for SARS-CoV-2 control. COVID 2023, 3, 975–986. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Krut’, V.G.; Chuvpilo, S.A.; Astrakhantseva, I.V.; Kozlovskaya, L.I.; Efimov, G.A.; Kruglov, A.A.; Drutskaya, M.S.; Nedospasov, S.A. Will peptides help to stop COVID-19? Biochemistry 2022, 87, 590–604. [Google Scholar] [CrossRef]
- Gil, C.; Ginex, T.; Maestro, I.; Nozal, V.; Barrado-Gil, L.; Cuesta-Geijo, M.Á.; Urquiza, J.; Ramírez, D.; Alonso, C.; Campillo, N.E. COVID-19: Drug targets and potential treatments. J. Med. Chem. 2020, 63, 12359–12386. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-1: Maestro; Schrödinger LLC.: New York, NY, USA, 2018.
- Yin, W.; Xu, Y.; Xu, P.; Cao, X.; Wu, C.; Gu, C.; He, X.; Wang, X.; Huang, S.; Yuan, Q. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 2022, 375, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J. Virological characteristics of the SARS-CoV-2 omicron BA. 2 subvariants, including BA. 4 and BA. 5. Cell 2022, 185, 3992–4007.e3916. [Google Scholar] [CrossRef] [PubMed]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput.-Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef]
- Schrödinger Release 2018−1: Epik; Schrödinger LLC.: New York, NY, USA, 2018.
- Schrödinger Release 2018−1: Impact; Schrödinger, LLC.: New York, NY, USA, 2018.
- Schrödinger Release 2018−1: Prime; Schrödinger, LLC.: New York, NY, USA, 2018.
- Tubert-Brohman, I.; Sherman, W.; Repasky, M.; Beuming, T. Improved docking of polypeptides with Glide. J. Chem. Inf. Model. 2013, 53, 1689–1699. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Spitalieri, P.; Centofanti, F.; Murdocca, M.; Scioli, M.G.; Latini, A.; Di Cesare, S.; Citro, G.; Rossi, A.; Orlandi, A.; Miersch, S. Two different therapeutic approaches for SARS-CoV-2 in hipscs-derived lung organoids. Cells 2022, 11, 1235. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 84. [Google Scholar]
- Van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; van Dijk, M.; De Vries, S.; Bonvin, A. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Romeo, I.; Brizzi, A.; Pessina, F.; Ambrosio, F.A.; Aiello, F.; Belardo, C.; Carullo, G.; Costa, G.; De Petrocellis, L.; Frosini, M. In silico-guided rational drug design and synthesis of novel 4-(thiophen-2-yl) butanamides as potent and selective TRPV1 agonists. J. Med. Chem. 2023, 66, 6994–7015. [Google Scholar] [CrossRef] [PubMed]
- Bivacqua, R.; Romeo, I.; Barreca, M.; Barraja, P.; Alcaro, S.; Montalbano, A. HSV-1 glycoprotein d and its surface receptors: Evaluation of protein–protein interaction and targeting by triazole-based compounds through in silico approaches. Int. J. Mol. Sci. 2023, 24, 7092. [Google Scholar] [CrossRef]
- Struelens, M.J.; Ludden, C.; Werner, G.; Sintchenko, V.; Jokelainen, P.; Ip, M. Real-time genomic surveillance for enhanced control of infectious diseases and antimicrobial resistance. Front. Sci. 2024, 2, 1298248. [Google Scholar] [CrossRef]


| HADDOCK Score * | Cluster Size | RMSD ** | VdW Energy | Electrostatic Energy | Desolvation Energy | BSA *** | Z-Score **** | |
|---|---|---|---|---|---|---|---|---|
| RBD B.1.1.529 | −80.6 ± 2.8 | 39 | 3.2 ± 0.1 | −53.6 ± 2.6 | −87.1 ± 11.3 | −17.1 ± 2.1 | 1577.5 ± 35.4 | −1.9 |
| RBD BA.4/5 | −74.4 ± 2.9 | 50 | 3.6 ± 0.1 | −63.0 ± 2.0 | −44.2 ± 7.4 | −17.0 ± 3.4 | 1586.7 ± 76.0 | −2.5 |
| RBD BQ.1.1 | −77.7 ± 2.9 | 20 | 3.8 ± 0.1 | −55.3 ± 3.0 | −95.1 ± 21.0 | −9.3 ± 2.0 | 1718.3 ± 94.2 | −1.6 |
| DPP4270–295–RBD BA4.5 | DPP4270–295–RBD BQ.1.1 | |||
|---|---|---|---|---|
| Residue | Total Score | Quartile | Total Score | Quartile |
| T4 | −3.31 | Q3 | - | - |
| D5 | −17.17 | Q1 | −6.79 | Q2 |
| L7 | - | - | −4.92 | Q4 |
| S9 | −8.55 | Q1 | - | - |
| V10 | - | - | −10.98 | Q1 |
| T11 | −1.93 | Q4 | −20.06 | Q1 |
| N12 | −0.94 | Q4 | - | - |
| A13 | −5.20 | Q2 | - | - |
| T14 | −8.39 | Q2 | - | - |
| I16 | −4.82 | Q2 | - | - |
| Q17 | −34.50 | Q1 | −12.11 | Q1 |
| I18 | - | . | −5.43 | Q3 |
| A22 | −1.73 | Q4 | −3.20 | Q4 |
| S23 | −2.86 | Q3 | −5.46 | Q2 |
| M24 | - | - | −5.16 | Q3 |
| Pep2–RBD BA4.5 | Pep2–RBD BQ.1.1 | |||
| Residue | Total Score | Quartile | Total Score | Quartile |
| V1 | −18.55 | Q1 | - | - |
| T2 | −0.76 | Q4 | - | - |
| Q8 | −21.99 | Q1 | - | - |
| I9 | −5.78 | Q3 | - | - |
| N12 | −5.80 | Q2 | - | - |
| T13 | −17.52 | Q2 | - | - |
| D14 | - | - | −22.57 | Q1 |
| S15 | - | - | −14.58 | Q1 |
| L16 | −0.52 | Q4 | - | - |
| S17 | - | - | −7.23 | Q3 |
| S18 | - | - | −8.47 | Q3 |
| T19 | - | - | −10.99 | Q2 |
| A22 | - | - | −0.24 | Q4 |
| L25 | - | - | −1.76 | Q4 |
| I26 | - | - | −12.22 | Q2 |
| HADDOCK Score * | Cluster Size | RMSD ** | VdW Energy | Electrostatic Energy | Desolvation Energy | BSA *** | Z-Score **** | |
|---|---|---|---|---|---|---|---|---|
| RBD BA.4/5 | −97.1 ± 2.4 | 56 | 0.4 ± 0.2 | −56.3 ± 4.1 | −208.6 ± 35.1 | −10.2 ± 1.7 | 1522.2 ± 41.9 | −2.4 |
| RBD BQ.1.1 | −91.3 ± 1.1 | 31 | 0.4 ± 0.3 | −61.6 ± 2.9 | −102.2 ± 23.0 | −16.8 ± 2.1 | 1669.1 ± 36.0 | −2.6 |
| RBD XBB.1.5 | −91.7 ± 2.7 | 37 | 0.7 ± 0.4 | −61.6 ± 6.2 | −126.2 ± 14.9 | −11.4 ± 5.3 | 1702.5 ± 37.4 | −2.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murdocca, M.; Romeo, I.; Citro, G.; Latini, A.; Centofanti, F.; Bugatti, A.; Caccuri, F.; Caruso, A.; Ortuso, F.; Alcaro, S.; et al. A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern. Pharmaceuticals 2024, 17, 891. https://doi.org/10.3390/ph17070891
Murdocca M, Romeo I, Citro G, Latini A, Centofanti F, Bugatti A, Caccuri F, Caruso A, Ortuso F, Alcaro S, et al. A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern. Pharmaceuticals. 2024; 17(7):891. https://doi.org/10.3390/ph17070891
Chicago/Turabian StyleMurdocca, Michela, Isabella Romeo, Gennaro Citro, Andrea Latini, Federica Centofanti, Antonella Bugatti, Francesca Caccuri, Arnaldo Caruso, Francesco Ortuso, Stefano Alcaro, and et al. 2024. "A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern" Pharmaceuticals 17, no. 7: 891. https://doi.org/10.3390/ph17070891
APA StyleMurdocca, M., Romeo, I., Citro, G., Latini, A., Centofanti, F., Bugatti, A., Caccuri, F., Caruso, A., Ortuso, F., Alcaro, S., Sangiuolo, F., & Novelli, G. (2024). A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern. Pharmaceuticals, 17(7), 891. https://doi.org/10.3390/ph17070891










