Predicting Drugs Suspected of Causing Adverse Drug Reactions Using Graph Features and Attention Mechanisms
Abstract
1. Introduction
- ADR Mining: Mining potential adverse reactions from post-marketing surveillance reports of drugs, such as utilizing spontaneous reporting systems (SRS) for the passive monitoring of potential adverse drug reactions [6] or employing electronic health records (EHR) [7,8], social media platform data [9,10], and other sources for the active monitoring of adverse drug reactions [11,12,13].
- Algorithm Development: Various algorithms are developed to utilize drug structural information, target information, etc., to predict potential adverse drug reactions or to forecast drug–drug interactions (DDIs).
- The model should be capable of using generic information to assess the drugs causing ADRs in ADEs. This inference must be applicable to new drugs. For instance, by inputting patient demographics, drug SMILES encoding, and ADR information, the model should be able to infer suspected drugs.
- The model should be able to learn the relationship between the chemical structure information of existing drugs and ADRs, predicting the relationships between drugs and ADRs.
- The model should be capable of extracting the chemical structure features of drugs for tasks in drug discovery, such as predicting drug activity, toxicity, and side effects.
2. Results and Discussion
2.1. Evaluation Metrics
2.2. Identifying Suspected Drugs in Adverse Drug Reaction Events
2.2.1. Evaluation on FAERS Dataset and JADER Dataset
2.2.2. External Validation and Case Analysis
2.3. ADR Signal Detection
2.3.1. Investigation of ADRs to Mexiletine
2.3.2. Investigation of ADRs to Captopril
2.3.3. Predicting the Drug–ADR Associations for Methimazole and Propylthiouracil
2.4. Validation of Ten Tasks in the Field of Drug Discovery
3. Materials and Methods
3.1. Datasets
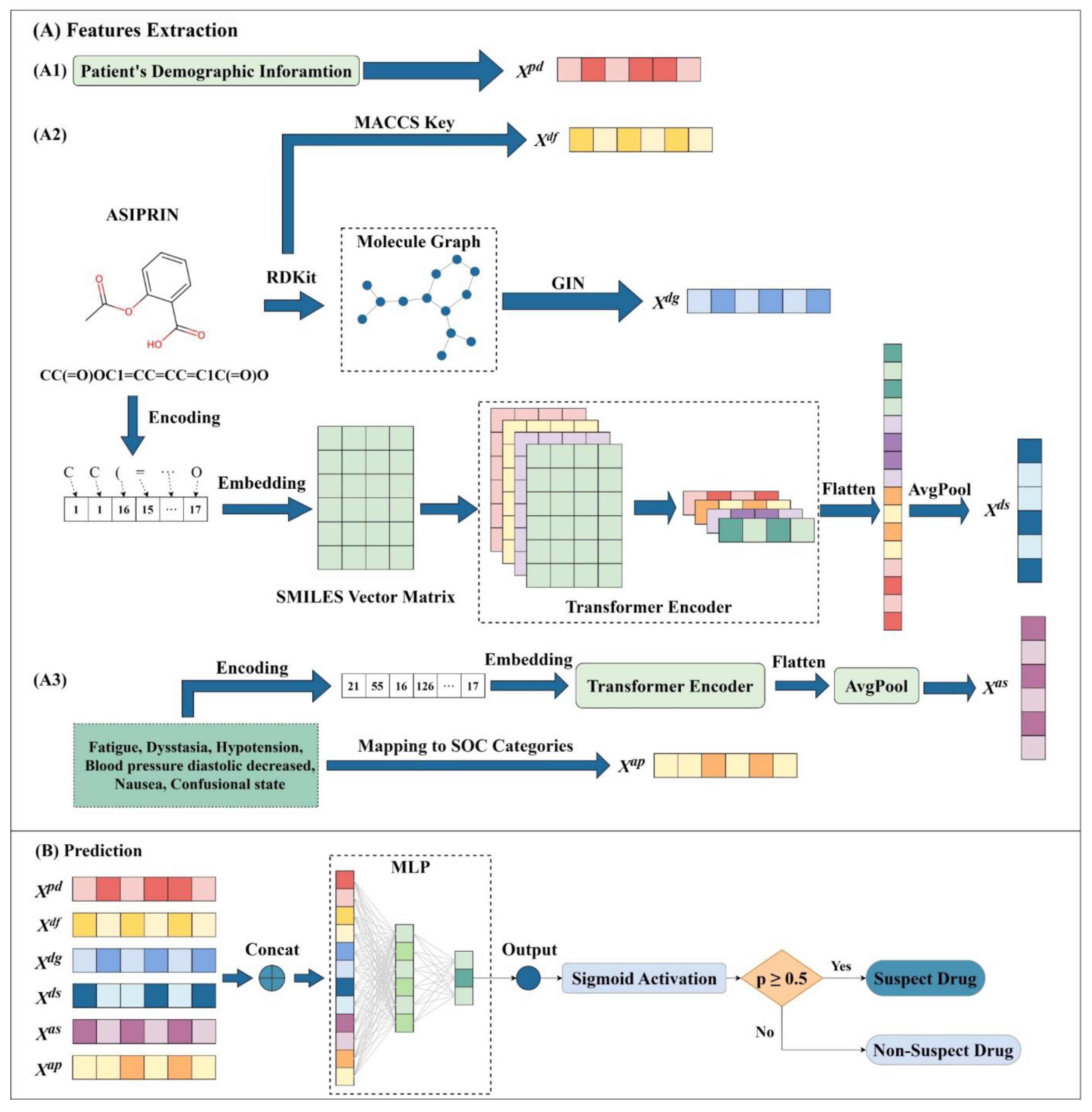
3.2. Framework of SDAJM
3.3. Extraction of Demographic Features
3.4. Extraction of Drug Features
3.4.1. Extraction of Molecular Fingerprint Features for Drugs
3.4.2. Extraction of Drug Graph Features
3.4.3. Extraction of SMILES Sequence Features
3.5. Extraction of ADR Features
3.5.1. Extraction of SOC Category Features
3.5.2. Extraction of ADR Semantic Features
3.6. Prediction
3.7. Optimization of SDAJM
3.8. Training Equipment and Time Consumption
4. Conclusions
Limitations and Future Prospects
- In the training dataset, although some drugs have known associations with the occurring ADRs, they may not necessarily be the suspect drugs causing these ADRs in the ADEs. This phenomenon may lead to confusion in the model’s understanding of the associations between the drugs and ADRs.
- While the model considers as much information as possible within a single ADE, it does not incorporate information from other drugs. This limitation is due to the current data structure. Future research should consider how to integrate additional information from other drugs and ADRs. Moreover, it would be beneficial to include data on the treatment duration and drug indications.
- The model balances drug feature extraction and ADR feature extraction using validated, ADR-related effective features. However, this results in reduced performance in drug feature extraction for other drug discovery tasks, especially in handling regression tasks. Future considerations should focus on adding more features or adopting more effective extraction methods, such as using pre-trained models, without significantly increasing the training time.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nguyen, D.A.; Nguyen, C.H.; Mamitsuka, H. A survey on adverse drug reaction studies: Data, tasks and machine learning methods. Brief. Bioinform. 2021, 22, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Rieder, M.J. Mechanisms of unpredictable adverse drug reactions. Drug Saf. 1994, 11, 196–212. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, J.; Pomeranz, B.H.; Corey, P.N. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. Jama 1998, 279, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Poudel, D.R.; Acharya, P.; Ghimire, S.; Dhital, R.; Bharati, R. Burden of hospitalizations related to adverse drug events in the USA: A retrospective analysis from large inpatient database: Adverse Drug Events Related Hospitalizations in the US. Pharmacoepidemiol. Drug Saf. 2017, 26, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Hakkarainen, K.M.; Hedna, K.; Petzold, M.; Hägg, S. Percentage of patients with preventable adverse drug reactions and preventability of adverse drug reactions--a meta-analysis. PLoS ONE 2012, 7, e33236. [Google Scholar] [CrossRef] [PubMed]
- Hoots, B.E.; Xu, L.; Kariisa, M.; Wilson, N.O.; Rudd, R.A.; Scholl, L.; Schieber, L.; Seth, P. 2018 Annual surveillance report of drug-related risks and outcomes–United States. Pediatrics 2018, 110, e53. [Google Scholar] [CrossRef]
- Noren, G.N.; Hopstadius, J.; Bate, A.; Edwards, I.R. Safety surveillance of longitudinal databases: Methodological considerations. Pharmacoepidemiol. Drug Saf. 2011, 20, 714–717. [Google Scholar] [CrossRef]
- Davis, S.E.; Zabotka, L.; Desai, R.J.; Wang, S.V.; Maro, J.C.; Coughlin, K.; Hernández-Muñoz, J.J.; Stojanovic, D.; Shah, N.H.; Smith, J.C. Use of Electronic Health Record Data for Drug Safety Signal Identification: A Scoping Review. Drug Saf. 2023, 46, 725–742. [Google Scholar] [CrossRef]
- Kass-Hout, T.A.; Alhinnawi, H. Social media in public health. Br. Med. Bull. 2013, 108, 5–24. [Google Scholar] [CrossRef]
- Sarker, A.; Ginn, R.; Nikfarjam, A.; O’Connor, K.; Smith, K.; Jayaraman, S.; Upadhaya, T.; Gonzalez, G. Utilizing social media data for pharmacovigilance: A review. J. Biomed. Inform. 2015, 54, 202–212. [Google Scholar] [CrossRef]
- Yamamoto, H.; Kayanuma, G.; Nagashima, T.; Toda, C.; Nagayasu, K.; Kaneko, S. Early Detection of Adverse Drug Reaction Signals by Association Rule Mining Using Large-Scale Administrative Claims Data. Drug Saf. 2023, 46, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Roughead, E.; Liu, L.; Pratt, N.; Li, J. Detecting potential signals of adverse drug events from prescription data. Artif. Intell. Med. 2020, 104, 101839. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.; Liu, J.; Roughead, E.; Pratt, N.; Li, J. Supervised signal detection for adverse drug reactions in medication dispensing data. Comput. Methods Programs Biomed. 2018, 161, 25–38. [Google Scholar] [CrossRef] [PubMed]
- FDA Adverse Event Reporting System (FAERS) Public Dashboard. Available online: https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard (accessed on 17 February 2024).
- European Medicines Agency. Available online: https://www.ema.europa.eu/en/homepage (accessed on 17 February 2024).
- Gallagher, R.M.; Kirkham, J.J.; Mason, J.R.; Bird, K.A.; Williamson, P.R.; Nunn, A.J.; Turner, M.A.; Smyth, R.L.; Pirmohamed, M. Development and inter-rater reliability of the Liverpool adverse drug reaction causality assessment tool. PLoS ONE 2011, 6, e28096. [Google Scholar] [CrossRef] [PubMed]
- Cocos, A.; Fiks, A.G.; Masino, A.J. Deep learning for pharmacovigilance: Recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J. Am. Med. Inf. Assoc. 2017, 24, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Cheng, M.; Yu, L.; Xiao, X. iADRGSE: A Graph-Embedding and Self-Attention Encoding for Identifying Adverse Drug Reaction in the Earlier Phase of Drug Development. Int. J. Mol. Sci. 2022, 23, 16216. [Google Scholar] [CrossRef] [PubMed]
- Tutubalina, E.; Alimova, I.; Miftahutdinov, Z.; Sakhovskiy, A.; Malykh, V.; Nikolenko, S. The Russian Drug Reaction Corpus and neural models for drug reactions and effectivesness detection in user reviews. Bioinformatics 2021, 37, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Reps, J.M.; Garibaldi, J.M.; Aickelin, U.; Soria, D.; Gibson, J.; Hubbard, R. Comparison of algorithms that detect drug side effects using electronic healthcare databases. Soft Comput. 2013, 17, 2381–2397. [Google Scholar] [CrossRef][Green Version]
- Chazard, E.; Ficheur, G.; Bernonville, S.; Luyckx, M.; Beuscart, R. Data mining to generate adverse drug events detection rules. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 823–830. [Google Scholar] [CrossRef]
- Jagannatha, A.N.; Yu, H. Bidirectional RNN for Medical Event Detection in Electronic Health Records. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; Association for Computational Linguistics: San Diego, CA, USA, 2016; pp. 473–482. [Google Scholar] [CrossRef]
- Freifeld, C.C.; Brownstein, J.S.; Menone, C.M.; Bao, W.; Filice, R.; Kass-Hout, T.; Dasgupta, N. Digital drug safety surveillance: Monitoring pharmaceutical products in twitter. Drug Saf. 2014, 37, 343–350. [Google Scholar] [CrossRef]
- Ding, P.; Zhou, X.; Zhang, X.; Wang, J.; Lei, Z. An Attentive Neural Sequence Labeling Model for Adverse Drug Reactions Mentions Extraction. IEEE Access 2018, 6, 73305–73315. [Google Scholar] [CrossRef]
- Zhang, T.; Lin, H.; Ren, Y.; Yang, L.; Xu, B.; Yang, Z.; Wang, J.; Zhang, Y. Adverse drug reaction detection via a multihop self-attention mechanism. BMC Bioinform. 2019, 20, 479. [Google Scholar] [CrossRef] [PubMed]
- Abdulrazzaq, M.M.; Ramaha, N.T.A.; Hameed, A.A.; Salman, M.; Yon, D.K.; Fitriyani, N.L.; Syafrudin, M.; Lee, S.W. Consequential Advancements of Self-Supervised Learning (SSL) in Deep Learning Contexts. Mathematics 2024, 12, 758. [Google Scholar] [CrossRef]
- Galeano, D.; Li, S.; Gerstein, M.; Paccanaro, A. Predicting the frequencies of drug side effects. Nat. Commun. 2020, 11, 4575. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zheng, K.; Li, Y.; Wang, J. A novel graph attention model for predicting frequencies of drug-side effects from multi-view data. Brief. Bioinform. 2021, 22, bbab239. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ma, T.; Yang, X.; Wang, J.; Song, B.; Zeng, X. MUFFIN: Multi-scale feature fusion for drug-drug interaction prediction. Bioinformatics 2021, 37, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Lyu, T.; Gao, J.; Tian, L.; Li, Z.; Zhang, P.; Zhang, J. MDNN: A Multimodal Deep Neural Network for Predicting Drug-Drug Interaction Events. In Proceedings of the 30th International Joint Conference on Artificial Intelligence, Montreal, QC, Canada, 19–26 August 2021; pp. 3536–3542. [Google Scholar]
- Xiong, Z.; Wang, D.; Liu, X.; Zhong, F.; Wan, X.; Li, X.; Li, Z.; Luo, X.; Chen, K.; Jiang, H.; et al. Pushing the Boundaries of Molecular Representation for Drug Discovery with the Graph Attention Mechanism. J. Med. Chem. 2020, 63, 8749–8760. [Google Scholar] [CrossRef]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016, 44, D1075–D1079. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kerndt, C.C.; Chauhan, S.; Zeltser, R. Mexiletine. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Marte, F.; Sankar, P.; Patel, P.; Cassagnol, M. Captopril. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Awosika, A.O.; Singh, G.; Correa, R. Methimazole. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Amisha, F.; Rehman, A. Propylthiouracil (PTU). In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Cooper, D.S. Antithyroid drugs. N. Engl. J. Med. 2005, 352, 905–917. [Google Scholar] [CrossRef]
- Emre, S.; Ozdemir, D.; Orhun, S.; Kalkan, G.; Sener, S. A case of severe erythema nodosum induced by methimazole. Saudi Pharm. J. 2017, 25, 813–815. [Google Scholar] [CrossRef]
- Hosoi, K.; Makino, S.; Yamano, Y.; Sasaki, M.; Takeuchi, T.; Sakane, S.; Ohsawa, N. Cryofibrinogenemia with polyarthralgia, Raynaud’s phenomenon and acral ulcer in a patient with Graves’ disease treated with methimazole. Intern. Med. 1997, 36, 439–442. [Google Scholar] [CrossRef]
- Kawachi, Y.; Nukaga, H.; Hoshino, M.; Iwata, M.; Otsuka, F. ANCA-associated vasculitis and lupus-like syndrome caused by methimazole. Clin. Exp. Dermatol. 1995, 20, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Qaisar, H.; Hossain, M.A.; Akula, M.; Cheng, J.; Patel, M.; Min, Z.; Kuzyshyn, H.; Levitt, M.; Coley, S.M.; Asif, A. Methimazole-Induced Pauci-Immune Glomerulonephritis and Anti-Phospholipid Syndrome: An Important Association to Be Aware of. J. Clin. Med. Res. 2018, 10, 786–790. [Google Scholar] [CrossRef][Green Version]
- Minkley, L.; Gohring-Frischholz, K.; Morike, K.; Lauer, U.M.; Mussig, K. Severe gastrointestinal haemorrhage after methimazole intake. Clin. Endocrinol. 2011, 74, 657–658. [Google Scholar] [CrossRef]
- Ortiz-ButrOn, R.; Blas-Valdivia, V.; Franco-Colin, M.; Pineda-Reynoso, M.; Cano-Europa, E. An increase of oxidative stress markers and the alteration of the antioxidant enzymatic system are associated with spleen damage caused by methimazole-induced hypothyroidism. Drug Chem. Toxicol. 2011, 34, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Cano-Europa, E.; Blas-Valdivia, V.; Franco-Colin, M.; Gallardo-Casas, C.A.; Ortiz-Butron, R. Methimazole-induced hypothyroidism causes cellular damage in the spleen, heart, liver, lung and kidney. Acta Histochem. 2011, 113, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Lionaki, S.; Vlachopanos, G.; Georgalis, A.; Ziakas, P.; Gakiopoulou, H.; Petra, C.; Boletis, J. Lupus nephritis and non-Hodgkin lymphoma simultaneously diagnosed in a patient on methimazole. Lupus 2013, 22, 95–98. [Google Scholar] [CrossRef]
- Arai, N.; Nemoto, K.; Oh-Ishi, S.; Nonaka, M.; Hayashihara, K.; Saito, T. Methimazole-induced ANCA-associated vasculitis with diffuse alveolar haemorrhage. Respirol. Case Rep. 2018, 6, e00315. [Google Scholar] [CrossRef]
- Akmal, A.; Kung, J. Propylthiouracil, and methimazole, and carbimazole-related hepatotoxicity. Expert. Opin. Drug Saf. 2014, 13, 1397–1406. [Google Scholar] [CrossRef]
- Kang, H.; Choi, J.D.; Jung, I.G.; Kim, D.W.; Kim, T.B.; Shin, H.K.; Kim, B.T.; Park, C.K.; Yoo, J.Y. A case of methimazole-induced acute hepatic failure in a patient with chronic hepatitis B carrier. Korean J. Intern. Med. 1990, 5, 69–73. [Google Scholar] [CrossRef]
- Khine, L.Y.; Kim, D.W.; Olajide, O.; White, C.; Shweihat, Y.; Driscoll, H. Methimazole-Induced Pleural Effusion in the Setting of Graves’ Disease. Case Rep. Endocrinol. 2019, 2019, 5748938. [Google Scholar] [CrossRef]
- Koike, K.J.; Blice, J.P.; Kylstra, J.A.; Ralston, J.S.; Self, S.E.; Ruth, N.M.; Del Priore, L.V. Frosted Branch Angiitis in Methimazole-Induced Antineutrophil Cytoplasmic Antibody-Positive Vasculitis. Retin. Cases Brief. Rep. 2018, 12, 136–139. [Google Scholar] [CrossRef]
- Wang, Y.L.; Yao, P.W.; Zhang, X.T.; Luo, Z.Z.; Wu, P.Q.; Xiao, F. Insulin Autoimmune Syndrome: 73 Cases of Clinical Analysis. Chin. Med. J. 2015, 128, 2408–2409. [Google Scholar] [CrossRef]
- Delaney, J.S. ESOL: Estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. [Google Scholar] [CrossRef]
- Mobley, D.L.; Guthrie, J.P. FreeSolv: A database of experimental and calculated hydration free energies, with input files. J. Comput.-Aided Mol. Des. 2014, 28, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, S.G.; Baumann, K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J. Chem. Inf. Model. 2009, 49, 169–184. [Google Scholar] [CrossRef] [PubMed]
- AIDS Antiviral Screen Data; NCI DTP Data. NCI Wiki. Available online: https://wiki.nci.nih.gov/display/NCIDTPdata/AIDS+Antiviral+Screen+Data (accessed on 17 February 2024).
- Subramanian, G.; Ramsundar, B.; Pande, V.; Denny, R.A. Computational Modeling of β-Secretase 1 (BACE-1) Inhibitors Using Ligand Based Approaches. J. Chem. Inf. Model. 2016, 56, 1936–1949. [Google Scholar] [CrossRef]
- Martins, I.F.; Teixeira, A.L.; Pinheiro, L.; Falcao, A.O. A Bayesian approach to in silico blood-brain barrier penetration modeling. J. Chem. Inf. Model. 2012, 52, 1686–1697. [Google Scholar] [CrossRef]
- Tox21 Data Challenge 2014. Available online: https://tripod.nih.gov/tox21/challenge (accessed on 17 February 2024).
- Artemov, G.N.; Bondarenko, S.M.; Shirokova, V.V.; Stegniy, V.N.; Sharakhov, I.V. Spatial Organization of chromosomes in malaria mosquitoes. Tsitologiia 2016, 58, 315–319. [Google Scholar]
- Gayvert, K.M.; Madhukar, N.S.; Elemento, O. A Data-Driven Approach to Predicting Successes and Failures of Clinical Trials. Cell Chem. Biol. 2016, 23, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2018, 9, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Cai, H.; Zhang, H.; Zhao, D.; Wu, J.; Wang, L. FP-GNN: A versatile deep learning architecture for enhanced molecular property prediction. Brief. Bioinform. 2022, 23, bbac408. [Google Scholar] [CrossRef] [PubMed]
- Pharmaceuticals and Medical Devices Agency. Japanese Adverse Drug Event Report Database. Available online: https://www.pmda.go.jp/safety/info-services/drugs/adr-info/suspected-adr/0005.html (accessed on 17 February 2024).
- Toropov, A.A.; Toropova, A.P.; Mukhamedzhanova, D.V.; Gutman, I. Simplified molecular input line entry system (SMILES) as an alternative for constructing quantitative structure-property relationships (QSPR). Indian J. Chem. Sect. A Inorg. Phys. Theor. Anal. 2005, 44, 1545–1552. [Google Scholar]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How Powerful are Graph Neural Networks? arXiv 2018, arXiv:1810.00826. [Google Scholar]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Representation Learning on Graphs: Methods and Applications. IEEE Data Eng. Bull. 2017, 40, 52–74. [Google Scholar]
- Hu, W.; Liu, B.; Gomes, J.; Zitnik, M.; Liang, P.; Pande, V.; Leskovec, J. Strategies for Pre-training Graph Neural Networks. arXiv 2019, arXiv:1905.12265. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015. [Google Scholar]
- Lee, C.; Yoon, J.; Schaar, M.v.d. Dynamic-DeepHit: A Deep Learning Approach for Dynamic Survival Analysis with Competing Risks Based on Longitudinal Data. IEEE Trans. Bio-Med. Eng. 2020, 67, 122–133. [Google Scholar] [CrossRef]
- Liu, M.; Sun, Z.L.; Zeng, Z.; Lam, K.M. MGF6mARice: Prediction of DNA N6-methyladenine sites in rice by exploiting molecular graph feature and residual block. Brief. Bioinform. 2022, 23, bbac082. [Google Scholar] [CrossRef] [PubMed]
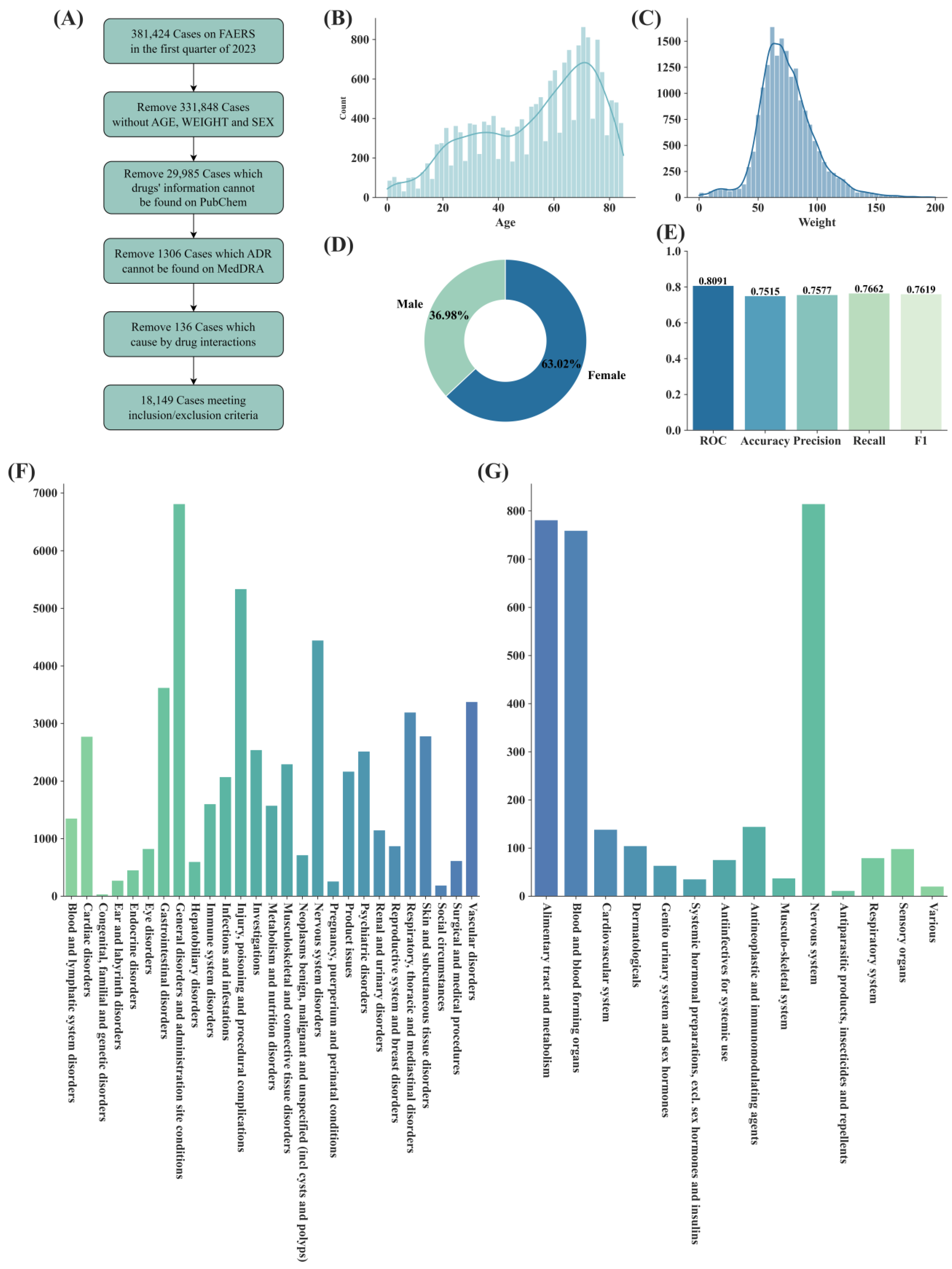
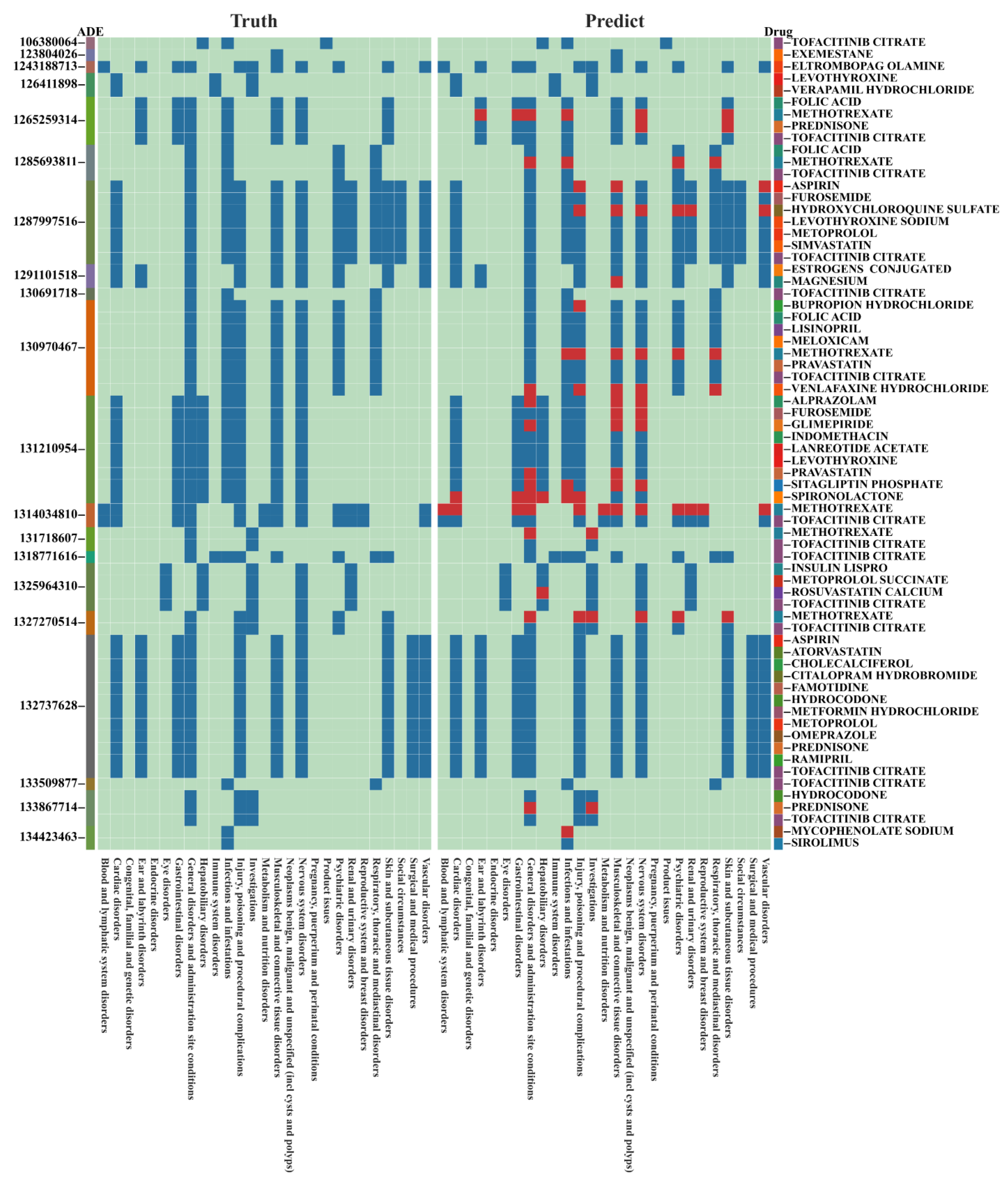
| Dataset | Split Type | Model | ROC-AUC | Accuracy | Precision | Recall | F1 |
|---|---|---|---|---|---|---|---|
| FAERS | Random | SDAJM | 0.8937 | 0.8165 | 0.8403 | 0.8406 | 0.8404 |
| Random | FPGNN-SDAJM | 0.8730 | 0.8078 | 0.8195 | 0.8445 | 0.8318 | |
| Random | ResNet | 0.8609 | 0.7860 | 0.8150 | 0.8246 | 0.8197 | |
| Random | CNN | 0.8522 | 0.7820 | 0.7908 | 0.8310 | 0.8104 | |
| Drug | SDAJM | 0.8071 | 0.7404 | 0.8185 | 0.7492 | 0.7823 | |
| Drug | FPGNN-SDAJM | 0.7875 | 0.7253 | 0.7787 | 0.7427 | 0.7603 | |
| JADER | Random | SDAJM | 0.8462 | 0.7990 | 0.7086 | 0.7325 | 0.7203 |
| Random | FPGNN-SDAJM | 0.8335 | 0.7818 | 0.7508 | 0.5877 | 0.6593 | |
| Random | ResNet | 0.8030 | 0.7593 | 0.7130 | 0.5206 | 0.6018 | |
| Random | CNN | 0.8130 | 0.7554 | 0.7747 | 0.4230 | 0.5472 |
| Primary ID: 100270603 | Age (Years): 63 | Weight (kg): 47.67 | Sex: Female |
| Drugs |
| ||
| ADRs |
| ||
| Prediction | Ibandronate sodium and alendronate sodium are suspected drugs | ||
| Evidence |
| ||
| CID | Drug | SOC | PT | Prediction | Evidence |
|---|---|---|---|---|---|
| 4178 | Mexiletine | Cardiac disorders | Ventricular extrasystoles | Yes | SIDER |
| Congestive cardiac failure | Yes | SIDER | |||
| Gastrointestinal disorders | Vomiting | Yes | SIDER | ||
| Nausea | Yes | SIDER | |||
| General disorders and administration site cond | Chest discomfort | Yes | Unconfirmed | ||
| Injury, poisoning and procedural complications | Maternal exposure during pregnancy | No | None | ||
| Investigations | Decreased ejection fraction | Yes | PMID: 17392676 | ||
| Nervous system disorders | Headache | Yes | SIDER | ||
| Intracranial hemorrhage | Yes | Unconfirmed | |||
| Cerebral hemorrhage | Yes | Unconfirmed | |||
| Pregnancy, puerperium and perinatal conditions | Subchorionic hematoma | Yes | Unconfirmed | ||
| Premature delivery | Yes | Unconfirmed |
| CID | Drug | SOC | PT | Evidence |
|---|---|---|---|---|
| 44093 | Captopril | Metabolism and nutrition disorders | Dehydration | SIDER |
| General disorders and administration site conditions | Aggravated condition | Unconfirmed | ||
| Feeling hot | Unconfirmed | |||
| Malaise | SIDER | |||
| Musculoskeletal and connective tissue disorders | Limb discomfort | Unconfirmed | ||
| Psychiatric disorders | Altered mood | Unconfirmed | ||
| Sopor | Unconfirmed | |||
| Immune system disorders | Anaphylactic shock | SIDER | ||
| Nervous system disorders | Hypokinesia | Unconfirmed |
| Drug | PT | Evidence |
|---|---|---|
| Methimazole | Exfoliative dermatitis | PMID: 15745981 |
| Erythema nodosum | PMID: 28725155 | |
| Glomerulonephritis | PMID: 30214651 | |
| Hemorrhage | PMID: 21114679 | |
| Hemoglobin | PMID: 21114679 | |
| Skin ulcer | PMID: 9213194, PMID: 8548997 | |
| Splenomegaly | PMID: 21314467, PMID: 19775732, PMID: 23263868 | |
| Vasculitis | PMID: 29760925 | |
| Hepatic failure | PMID: 19775732, PMID: 25156887, PMID: 2271514 | |
| Liver injury | PMID: 19775732, PMID: 25156887 | |
| Traumatic liver injury | PMID: 19775732, PMID: 25156887 | |
| Interstitial lung disease | Unconfirmed | |
| Rapidly progressive glomerulonephritis | PMID: 30214651 | |
| Lung infiltration | PMID: 31467736 | |
| Antineutrophil cytoplasmic antibody positivity | PMID: 27749745 | |
| Propylthiouracil | Hypoglycemic coma | Unconfirmed |
| Insulin autoimmune syndrome | PMID: 26315093 |
| Dataset | Split Type | Metric | MoleculeNet (Graph) | Chemprop (Optimized) | Attentive FP | XGBoost | FP-GNN | SDAJM |
|---|---|---|---|---|---|---|---|---|
| BACE | random | ROC | 0.898 | 0.876 | 0.889 | 0.881 | 0.883 | |
| scaffold | ROC | 0.806 (Weave) | 0.857 | 0.850 | 0.860 | 0.849 | ||
| HIV | random | ROC | 0.827 | 0.822 | 0.816 | 0.825 | 0.826 | |
| scaffold | ROC | 0.763 (GC) | 0.794 | 0.832 | 0.824 | 0.812 | ||
| MUV | random | PRC | 0.109 (Weave) | 0.053 | 0.038 | 0.068 | 0.09 | 0.093 |
| Tox21 | random | ROC | 0.829 (GC) | 0.854 | 0.852 | 0.836 | 0.815 | 0.873 |
| BBBP | random | ROC | 0.917 | 0.887 | 0.926 | 0.935 | 0.918 | |
| scaffold | ROC | 0.690 (GC) | 0.886 | 0.916 | 0.911 | |||
| ClinTox | random | ROC | 0.832 (Weave) | 0.897 | 0.904 | 0.911 | 0.840 | 0.841 |
| SIDER | random | ROC- | 0.638 (GC) | 0.658 | 0.623 | 0.642 | 0.661 | 0.779 |
| FreeSolv | random | RMSE | 1.150 (MPNN) | 1.009 | 1.091 | 1.025 | 0.905 | 1.022 |
| ESOL | random | RMSE | 0.580 (MPNN) | 0.587 | 0.587 | 0.582 | 0.675 | 0.830 |
| Lipophilicity | random | RMSE | 0.655 (GC) | 0.563 | 0.553 | 0.574 | 0.625 | 0.655 |
| Dataset | Number of Reports | Number of Drugs | Number of ADRs (PT) | Number of Suspect Drug Labels (Processed) | Number of Non-Suspect Drug Labels (Processed) |
|---|---|---|---|---|---|
| FAERS | 206,855 | 3012 | 9315 | 765,161 | 552,044 |
| JADER | 53,528 | 1407 | 3153 | 146,506 | 265,914 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Hu, Z.; Zhang, L.; Peng, B. Predicting Drugs Suspected of Causing Adverse Drug Reactions Using Graph Features and Attention Mechanisms. Pharmaceuticals 2024, 17, 822. https://doi.org/10.3390/ph17070822
Yang J, Hu Z, Zhang L, Peng B. Predicting Drugs Suspected of Causing Adverse Drug Reactions Using Graph Features and Attention Mechanisms. Pharmaceuticals. 2024; 17(7):822. https://doi.org/10.3390/ph17070822
Chicago/Turabian StyleYang, Jinxiang, Zuhai Hu, Liyuan Zhang, and Bin Peng. 2024. "Predicting Drugs Suspected of Causing Adverse Drug Reactions Using Graph Features and Attention Mechanisms" Pharmaceuticals 17, no. 7: 822. https://doi.org/10.3390/ph17070822
APA StyleYang, J., Hu, Z., Zhang, L., & Peng, B. (2024). Predicting Drugs Suspected of Causing Adverse Drug Reactions Using Graph Features and Attention Mechanisms. Pharmaceuticals, 17(7), 822. https://doi.org/10.3390/ph17070822






