Identification of South African Plant-Based Bioactive Compounds as Potential Inhibitors against the SARS-CoV-2 Receptor
Abstract
1. Introduction
2. Results
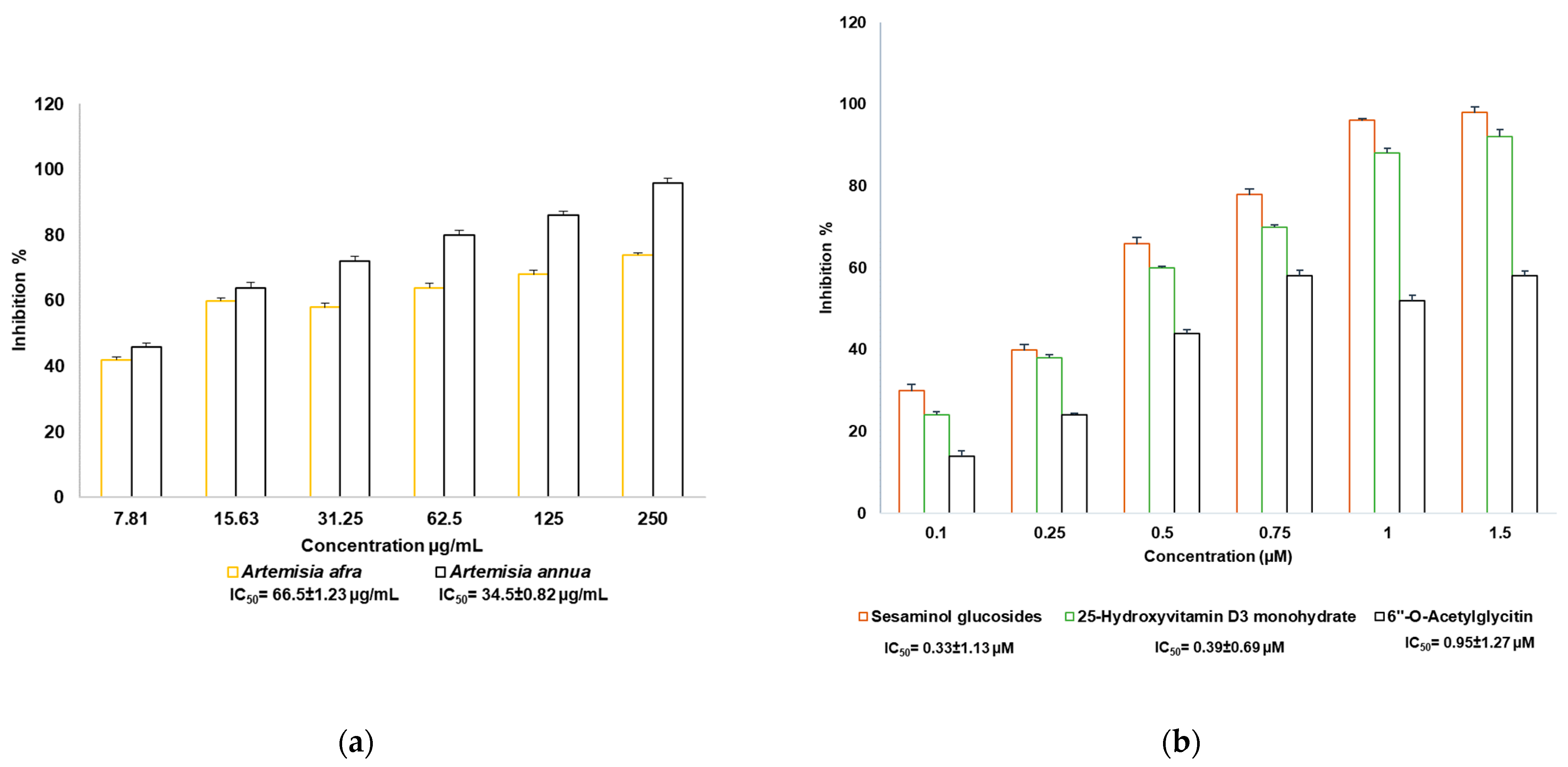
2.1. Resazurin Cell Viability Assay
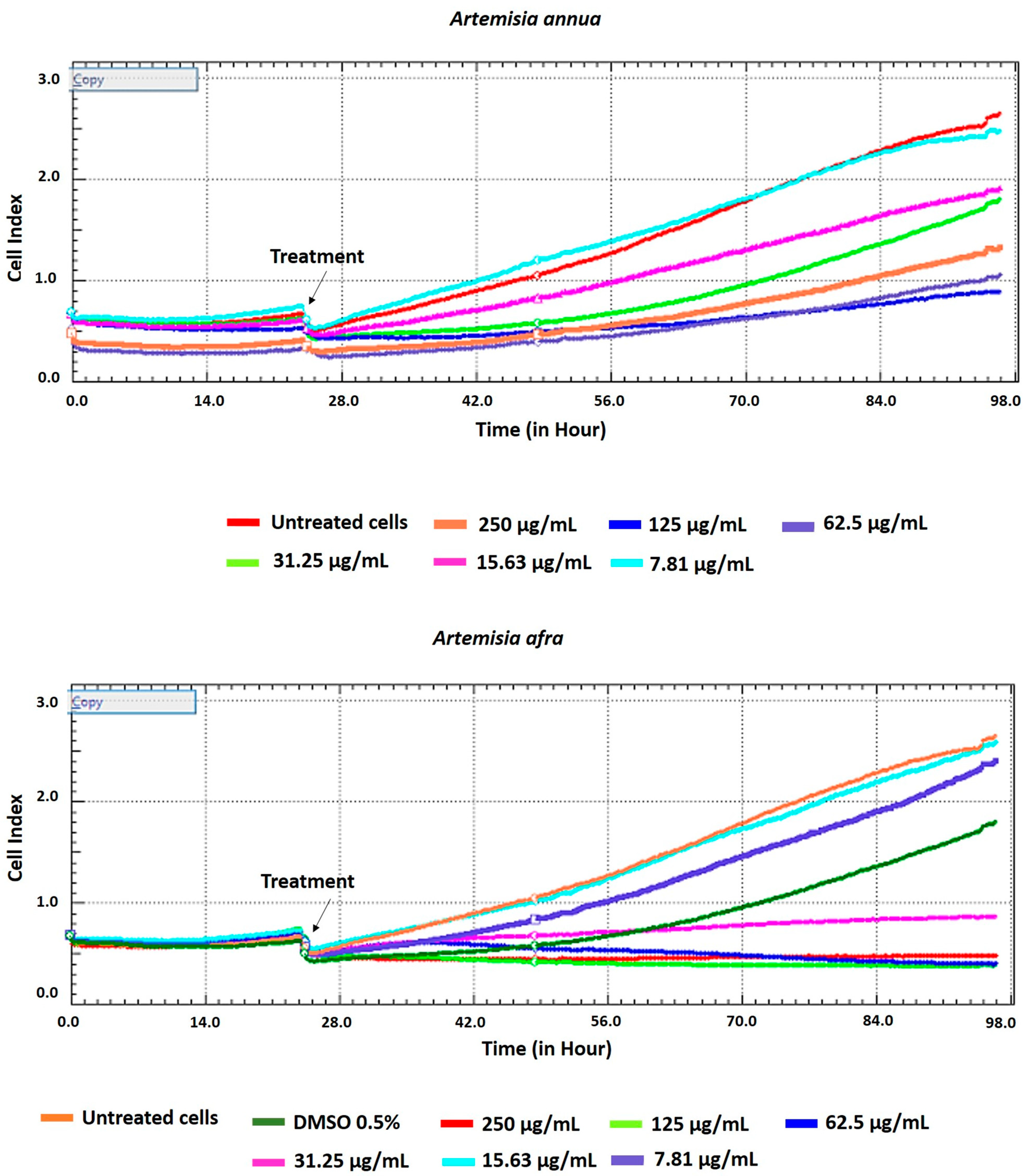
2.2. Cell Viability Assay Using xCELLigence Real-Time Cell Analyzer
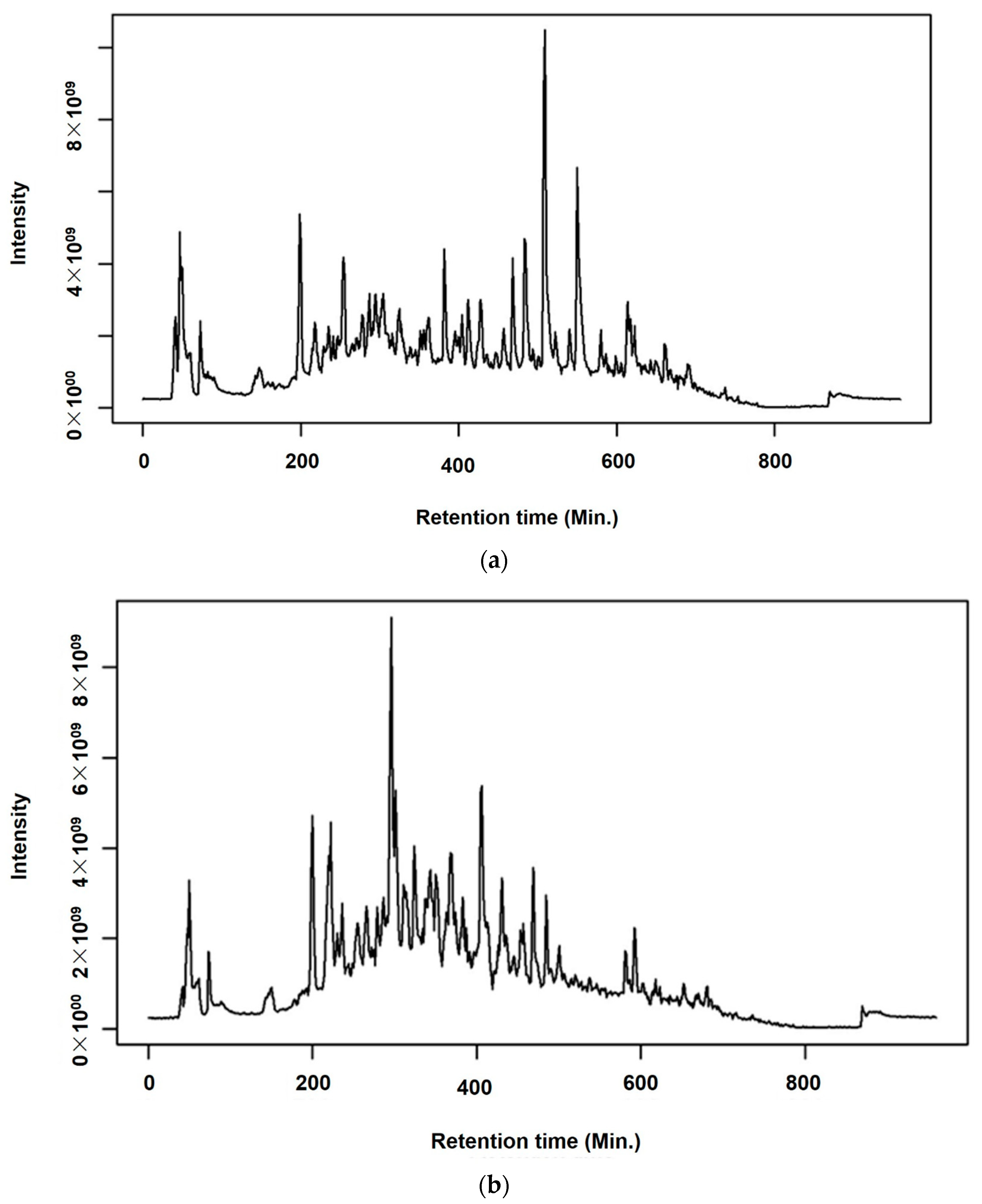
2.3. Chemical Profiling of A. annua and A. afra Leaves
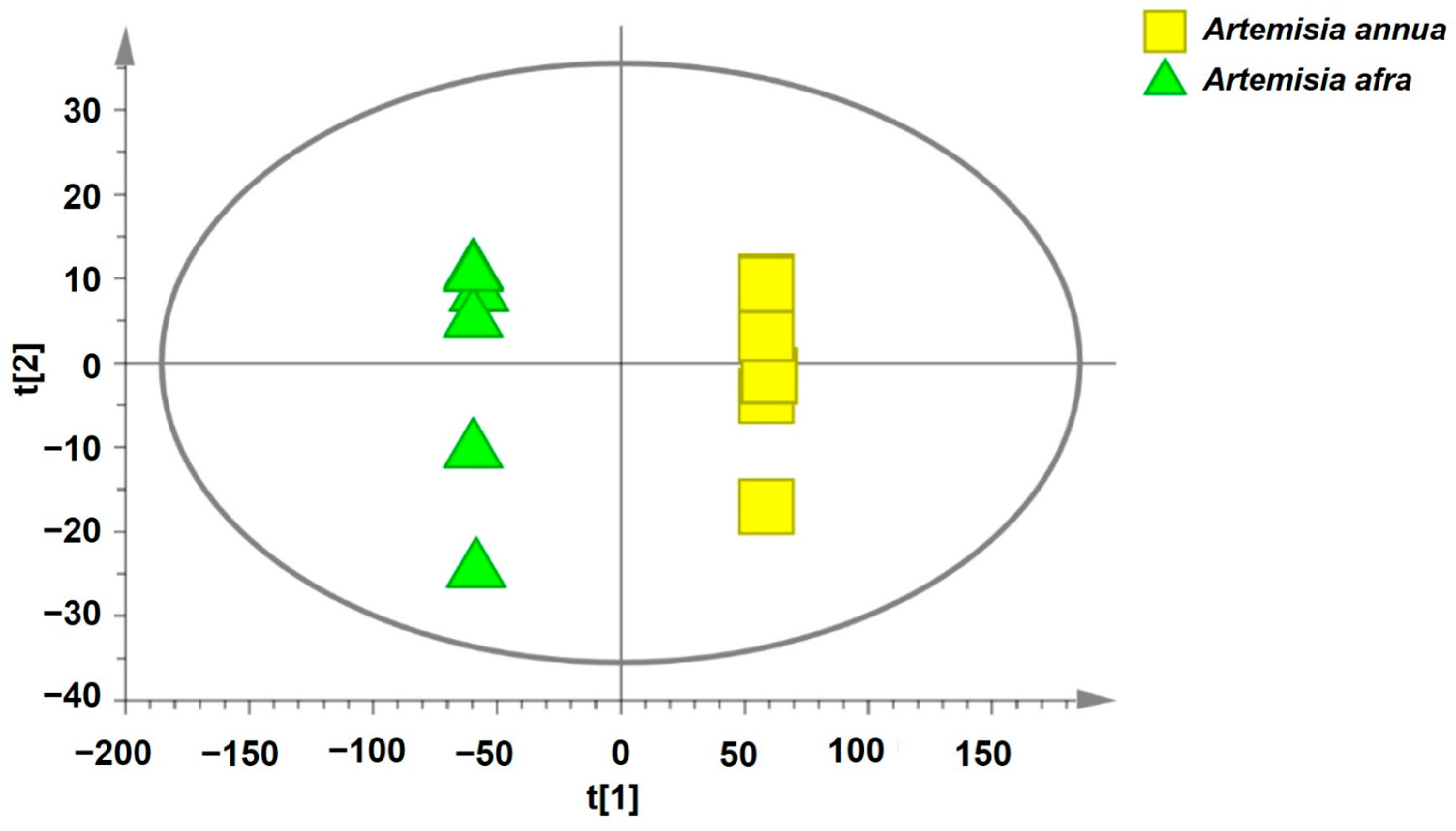
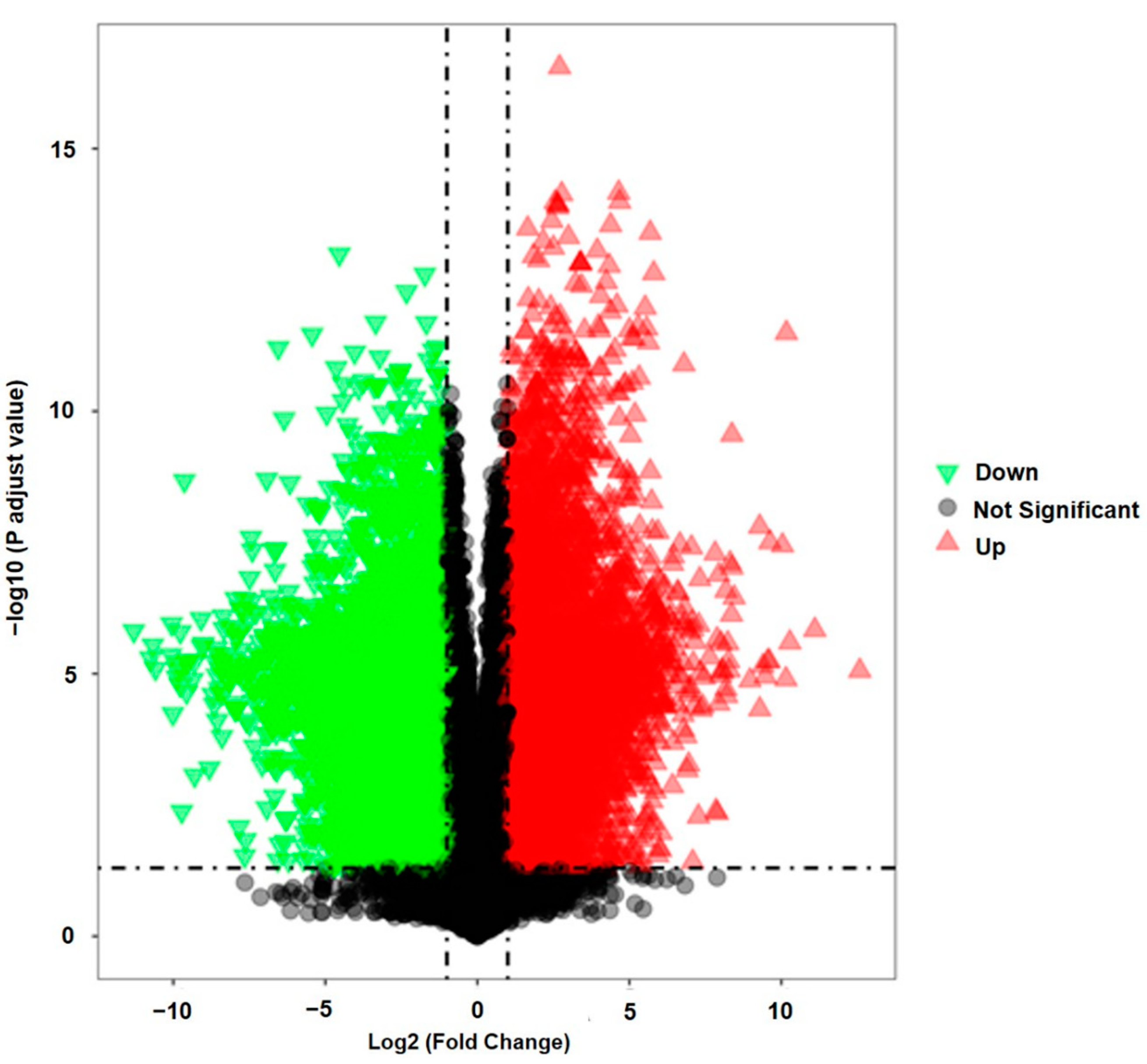
2.4. Determination of the Bioactive Compounds Utilizing Supervised Pattern Recognition Analysis
2.5. Binding Site Prediction of the SARS-CoV-2 Spike S Glycoprotein (6LZG)
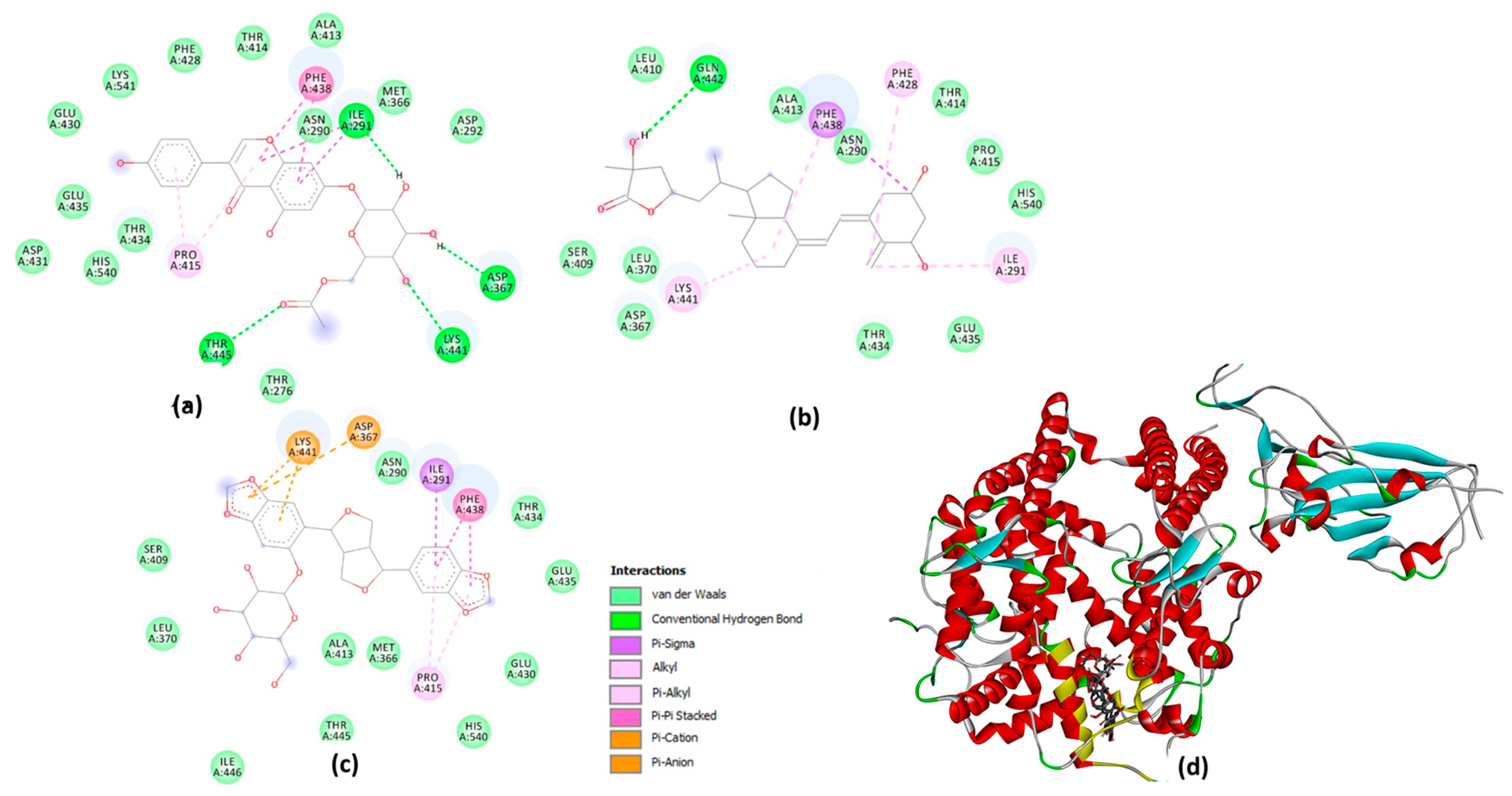
2.6. Molecular Docking Studies on the Plant-Based Bioactive Compounds
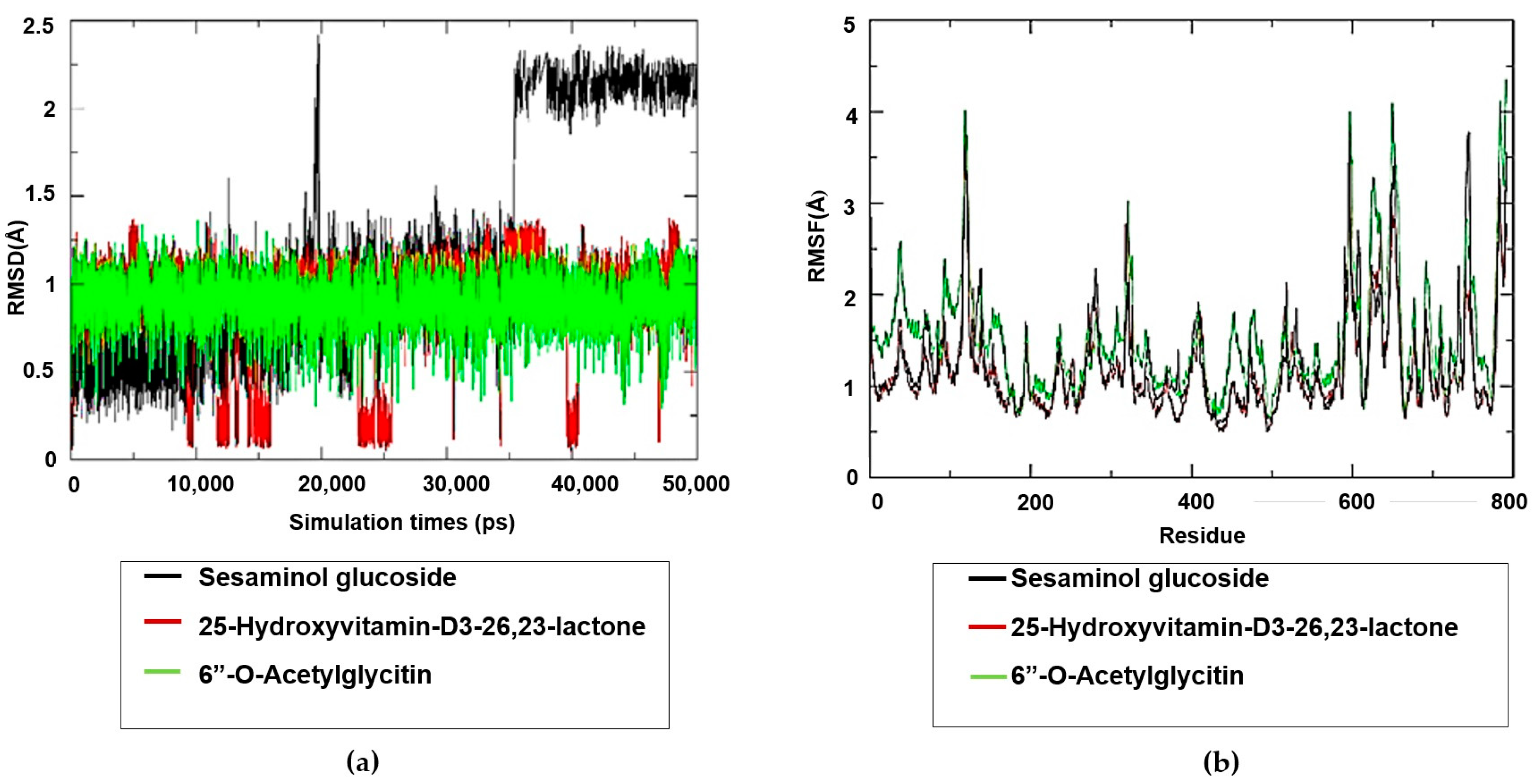
2.7. Molecular Dynamics Simulation Studies on the Plant-Based Bioactive Compounds
2.8. SARS-CoV-2 Enzyme In Vitro Inhibition Assay
3. Discussion
4. Materials and Methods
4.1. Plant Collection and Identification
4.2. Preparation of Plant Extracts
4.3. Resazurin Cell Viability Assay
4.4. Cell Viability Assay Using xCELLigence Real-Time Cell Analyzer
4.5. Metabolomics: Establishment of the Metabolites’ Bioactive Compounds
4.5.1. Instruments and Reagents
4.5.2. Sample Preparation
4.5.3. Analysis of the Prepared Plant Samples Using UPLC-MS/MS
4.6. Binding Site Prediction
4.7. Molecular Docking Studies on the Bioactive Compounds
4.8. Molecular Dynamics Simulation
4.9. SARS-CoV-2 Enzyme In Vitro Inhibition Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, F.; Chen, L.; Chen, H.; Yan, Z.; Liu, Y. Discovery of the key active compounds in Citri Reticulatae Pericarpium (Citrus reticulata “Chachi”) and their therapeutic potential for the treatment of COVID-19 based on comparative metabolomics and network pharmacology. Front. Pharmacol. 2022, 13, 1048926. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Wang, S.; Liang, P.; Wang, Y.; Zhang, X.; Jia, Q.; Fu, J.; Han, S.; He, L. Screening and evaluation of anti-SARS-CoV-2 components from Ephedra sinica by ACE2/CMC-HPLC-IT-TOF-MS approach. Anal. Bioanal. Chem. 2021, 413, 2995–3004. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Samprathi, M.; Jayashree, M. Biomarkers in COVID-19: An Up-To-Date Review. Front. Pediatr. 2021, 8, 607647. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Angeletti, S.; Minieri, M.; Giovannetti, M.; Benvenuto, D.; Pascarella, S.; Sagnelli, C.; Bianchi, M.; Bernardini, S.; Ciccozzi, M. COVID-19 Outbreak: An Overview. Chemotherapy 2019, 64, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Rasaily, S.; Haldipur, A.C.; Srividya, N. Indian red rice phenolic metabolites as potential natural inhibitors of SARS-CoV-2 main protease: A metabolomic and in silico study. Ann. Phytomed. 2021, 1, S40–S50. [Google Scholar] [CrossRef]
- Liu, J.; Xie, W.; Wang, Y.; Xiong, Y.; Chen, S.; Han, J.; Wu, Q. A comparative overview of COVID-19, MERS and SARS: Review article. Int. J. Surg. 2020, 81, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.; Tripathi, A.; Kumar, N.; Ganguly, N.K. A Comprehensive Overview on COVID-19: Future Perspectives. Front. Cell. Infect. Microbiol. 2021, 11, 744903. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Oronsky, B.; Larson, C.; Hammond, T.C.; Oronsky, A.; Kesari, S.; Lybeck, M.; Reid, T.R. A Review of Persistent Post-COVID Syndrome (PPCS). Clin. Rev. Allergy Immunol. 2023, 64, 66–74. [Google Scholar] [CrossRef]
- Raveendran, A.V.; Jayadevan, R.; Sashidharan, S. Long COVID: An overview. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 869–875. [Google Scholar] [CrossRef]
- Akindele, A.J.; Sowemimo, A.; Agunbiade, F.O.; Sofidiya, M.O.; Awodele, O.; Ade-Ademilua, O.; Orabueze, I.; Ishola, I.O.; Ayolabi, C.I.; Salu, O.B.; et al. Bioprospecting for anti-COVID-19 interventions from African medicinal plants: A review. Nat. Prod. Commun. 2022, 17, 1934578X221096968. [Google Scholar] [CrossRef]
- Invernizzi, L.; Moyo, P.; Cassel, J.; Isaacs, F.J.; Salvino, J.M.; Montaner, L.J.; Tietjen, I.; Maharaj, V. Use of hyphenated analytical techniques to identify the bioactive constituents of Gunnera perpensa L., a South African medicinal plant, which potently inhibit SARS-CoV-2 spike glycoprotein-host ACE2 binding. Anal. Bioanal. Chem. 2022, 414, 3971–3985. [Google Scholar] [CrossRef] [PubMed]
- Dwarka, D.; Agoni, C.; Mellem, J.J.; Soliman, M.E.; Baijnath, H. Identification of potential SARS-CoV-2 inhibitors from South African medicinal plant extracts using molecular modelling approaches. S. Afr. J. Bot. 2020, 133, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Richman, D.D. COVID-19 vaccines: Implementation, limitations and opportunities. Glob. Health Med. 2021, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, A.; Jailani, A.A.K.; Mandal, B. Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness. Vaccines 2023, 11, 1347. [Google Scholar] [CrossRef] [PubMed]
- Gillim-Ross, L.; Subbarao, K. Emerging respiratory viruses: Challenges and vaccine strategies. Clin. Microbiol. Rev. 2006, 19, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Berlanga, B.; Pniewski, T. Plant-Based Vaccines in Combat against Coronavirus Diseases. Vaccines 2022, 10, 138. [Google Scholar] [CrossRef]
- Mehrbod, P.; Abdalla, M.A.; Njoya, E.M.; Ahmed, A.S.; Fotouhi, F.; Farahmand, B.; Gado, D.A.; Tabatabaian, M.; Fasanmi, O.G.; Eloff, J.N.; et al. South African medicinal plant extracts active against influenza A virus. BMC Complement. Altern. Med. 2018, 18, 112. [Google Scholar] [CrossRef]
- World Health Organization. WHO Monograph on Good Agricultural and Collection Practices (GACP) for Artemisia annua L. 2006. Available online: https://iris.who.int/bitstream/handle/10665/43509/9241594438_eng.pdf?sequence=1 (accessed on 11 January 2024).
- Kapepula, P.M.; Kabengele, J.K.; Kingombe, M.; Van Bambeke, F.; Tulkens, P.M.; Kishabongo, A.S.; Decloedt, E.; Zumla, A.; Tiberi, S.; Suleman, F.; et al. Artemisia spp. Derivatives for COVID-19 Treatment: Anecdotal Use, Political Hype, Treatment Potential, Challenges, and Road Map to Randomized Clinical Trials. Am. J. Trop. Med. Hyg. 2020, 103, 960–964. [Google Scholar] [CrossRef]
- Bilia, A.R.; Santomauro, F.; Sacco, C.; Bergonzi, M.C.; Donato, R. Essential Oil of Artemisia annua L.: An Extraordinary Component with Numerous Antimicrobial Properties. Evid.-Based Complement. Altern. Med. 2014, 2014, 159819. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.F.; Luthria, D.L.; Sasaki, T.; Heyerick, A. Flavonoids from Artemisia annua L. as antioxidants and their potential synergism with artemisinin against malaria and cancer. Molecules 2010, 15, 3135–3170. [Google Scholar] [CrossRef] [PubMed]
- Jackson, W.P.U. Origins and Meanings of Names of South African Plant Genera; University of Cape Town UCT Ecolab: Cape Town, South Africa, 1990; ISBN 0799212849. [Google Scholar]
- Van Wyk, B.E.; Van Oudtshoorn, B.; Gerike, N. Medicinal Plants of South Africa; Briza Publications: Pretoria, South Africa, 1997; 142p. [Google Scholar]
- Liu, N.Q.; Van der Kooy, F.; Verpoorte, R. Artemisia afra: A potential flagship for African medicinal plants. S. Afr. J. Bot. 2009, 75, 185–195. [Google Scholar] [CrossRef]
- Motshudi, M.C.; Olaokun, O.O.; Mkolo, N.M. Evaluation of GC × GC-TOF-MS untargeted metabolomics, cytotoxicity and antimicrobial activity of leaf extracts of Artemisia afra (Jacq.) purchased from three local vendors. J. King Saud Univ. Sci. 2021, 33, 101422. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Supports Scientifically-Proven Traditional Medicine. 2020. Available online: https://www.afro.who.int/news/who-supports-scientifically-proven-traditional-medicine (accessed on 11 January 2024).
- Göbel, S.; Kortum, F.; Chavez, K.J.; Jordan, I.; Sandig, V.; Reichl, U.; Altomonte, J.; Genzel, Y. Cell-line screening and process development for a fusogenic oncolytic virus in small-scale suspension cultures. Appl. Microbiol. Biotechnol. 2022, 106, 4945–4961. [Google Scholar] [CrossRef] [PubMed]
- Dolskiy, A.A.; Grishchenko, I.V.; Yudkin, D.V. Cell Cultures for Virology: Usability, Advantages, and Prospects. Int. J. Mol. Sci. 2020, 21, 7978. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273. [Google Scholar] [CrossRef]
- Modrof, J.; Kerschbaum, A.; Farcet, M.R.; Niemeyer, D.; Corman, V.M.; Kreil, T.R. SARS-CoV-2 and the safety margins of cell-based biological medicinal products. Biologicals 2020, 68, 122–124. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Schrödinger Release 2020-3: Protein Preparation Wizard; Schrödinger: New York, NY, USA, 2020.
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Natesh, J.; Mondal, P.; Penta, D.; Abdul Salam, A.A.; Meeran, S.M. Culinary spice bioactives as potential therapeutics against SARS-CoV-2: Computational investigation. Comput. Biol. Med. 2021, 128, 104102. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S. Current perspectives on the beneficial effects of soybean isoflavones and their metabolites on plants. Food Sci. Biotechnol. 2022, 31, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Boland, R.; Skliar, M.; Curino, A.; Milanesi, L. Vitamin D compounds in plants. Plant Sci. 2003, 164, 357–369. [Google Scholar] [CrossRef]
- Aghaee, E.; Ghodrati, M.; Ghasemi, J.B. In silico exploration of novel protease inhibitors against coronavirus 2019 (COVID-19). Inform. Med. Unlocked 2021, 23, 100516. [Google Scholar] [CrossRef] [PubMed]
- Bepari, A.K.; Reza, H.M. Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation. PeerJ 2021, 9, e11261. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Rehman, M.T.; AlAjmi, M.F.; Hussain, A. Natural Compounds as Inhibitors of SARS-CoV-2 Main Protease (3CLpro): A Molecular Docking and Simulation Approach to Combat COVID-19. Curr. Pharm. Des. 2021, 27, 3577–3589. [Google Scholar] [CrossRef]
- Ogunyemi, O.M.; Gyebi, G.A.; Ibrahim, I.M.; Olaiya, C.O.; Ocheje, J.O.; Fabusiwa, M.M.; Adebayo, J.O. Dietary stigmastane-type saponins as promising dual-target directed inhibitors of SARS-CoV-2 proteases: A structure-based screening. RSC Adv. 2021, 11, 33380–33398. [Google Scholar] [CrossRef]
- Ahmed, N.Z.; John Davis, G.D.; Khan, A.A.; Prabhakar, L.; Ram Paratap, M.; Afnaan, Z.; Devi Sri, M.; Anwar, N. Arq Ajīb—A wonder Unani formulation for inhibiting SARS-CoV-2 spike glycoprotein and main protease—An in silico approach. J. Complement. Integr. Med. 2021, 20, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Ekowati, J.; Tejo, B.A.; Maulana, S.; Kusuma, W.A.; Fatriani, R.; Ramadhanti, N.S.; Norhayati, N.; Nofianti, K.A.; Sulistyowaty, M.I.; Zubair, M.S.; et al. Potential Utilization of Phenolic Acid Compounds as Anti-Inflammatory Agents through TNF-α Convertase Inhibition Mechanisms: A Network Pharmacology, Docking, and Molecular Dynamics Approach. ACS Omega 2023, 8, 46851–46868. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.S.; Maulana, S.; Widodo, A.; Pitopang, R.; Arba, M.; Hariono, M. GC-MS, LC-MS/MS, Docking and Molecular Dynamics Approaches to Identify Potential SARS-CoV-2 3-Chymotrypsin-Like Protease Inhibitors from Zingiber officinale Roscoe. Molecules 2021, 26, 5230. [Google Scholar] [CrossRef] [PubMed]
- Mani, J.S.; Johnson, J.B.; Steel, J.C.; Broszczak, D.A.; Neilsen, P.M.; Walsh, K.B.; Naiker, M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020, 284, 197989. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Réniam, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. COVID-19 Vaccine Coverage, World Data. 2023. Available online: https://data.who.int/dashboards/covid19/vaccines?n=c (accessed on 10 April 2024).
- Ma, L.L.; Liu, H.M.; Liu, X.M.; Yuan, X.Y.; Xu, C.; Wang, F.; Lin, J.Z.; Xu, R.C.; Zhang, D.K. Screening S protein—ACE2 blockers from natural products: Strategies and advances in the discovery of potential inhibitors of COVID-19. Eur. J. Med. Chem. 2021, 226, 113857. [Google Scholar] [CrossRef] [PubMed]
- Arokiyaraj, S.; Stalin, A.; Kannan, B.S.; Shin, H. Geranii herba as a Potential Inhibitor of SARS-CoV-2 main 3CLpro, spike RBD, and regulation of unfolded protein response: An in-silico approach. Antibiotics 2020, 9, 863. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Bretz, G.P.M.; Campos, J.R.; Veloso, A.A.; Gomes, K.B.; Fernandes, A.B. Impact of COVID-19 pandemic on serum 25-hydroxyvitamin D levels in Brazilian patients. J. Lab. Precis. Med. 2023, 8, 31. [Google Scholar] [CrossRef]
- Bae, J.H.; Choe, H.J.; Holick, M.F.; Lim, S. Association of vitamin D status with COVID-19 and its severity: Vitamin D and COVID-19: A narrative review. Rev. Endocr. Metab. Disord. 2022, 3, 579–599. [Google Scholar] [CrossRef]
- Prinsloo, S.; Rialet, P.; Carlos, C.B. A cell viability assay to determine the cytotoxic Effects of Water contaminated by microbes. S. Afr. J. Sci. 2013, 109, 1–4. [Google Scholar] [CrossRef]
- Roche Diagnostics GmbH. Introduction of the RTCA DP Instrument. RTCA DP Instrument Operator’s Manual A; ACEA Biosciences Inc.: San Diego, CA, USA, 2008; Available online: http://www.cytometrie-imagerie-saint-antoine.org/media/4046/RTCA_DP_System_Operator_Manual_v3.pdf (accessed on 5 February 2024).
- Fabre, N.; Rustan, I.; de Hoffmann, E.; Quetin-Leclercq, J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2020-1: Desmond Molecular Dynamics System; Schrödinger: New York, NY, USA, 2020.
- Schrödinger Release 2020-1: Maestro Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2020.

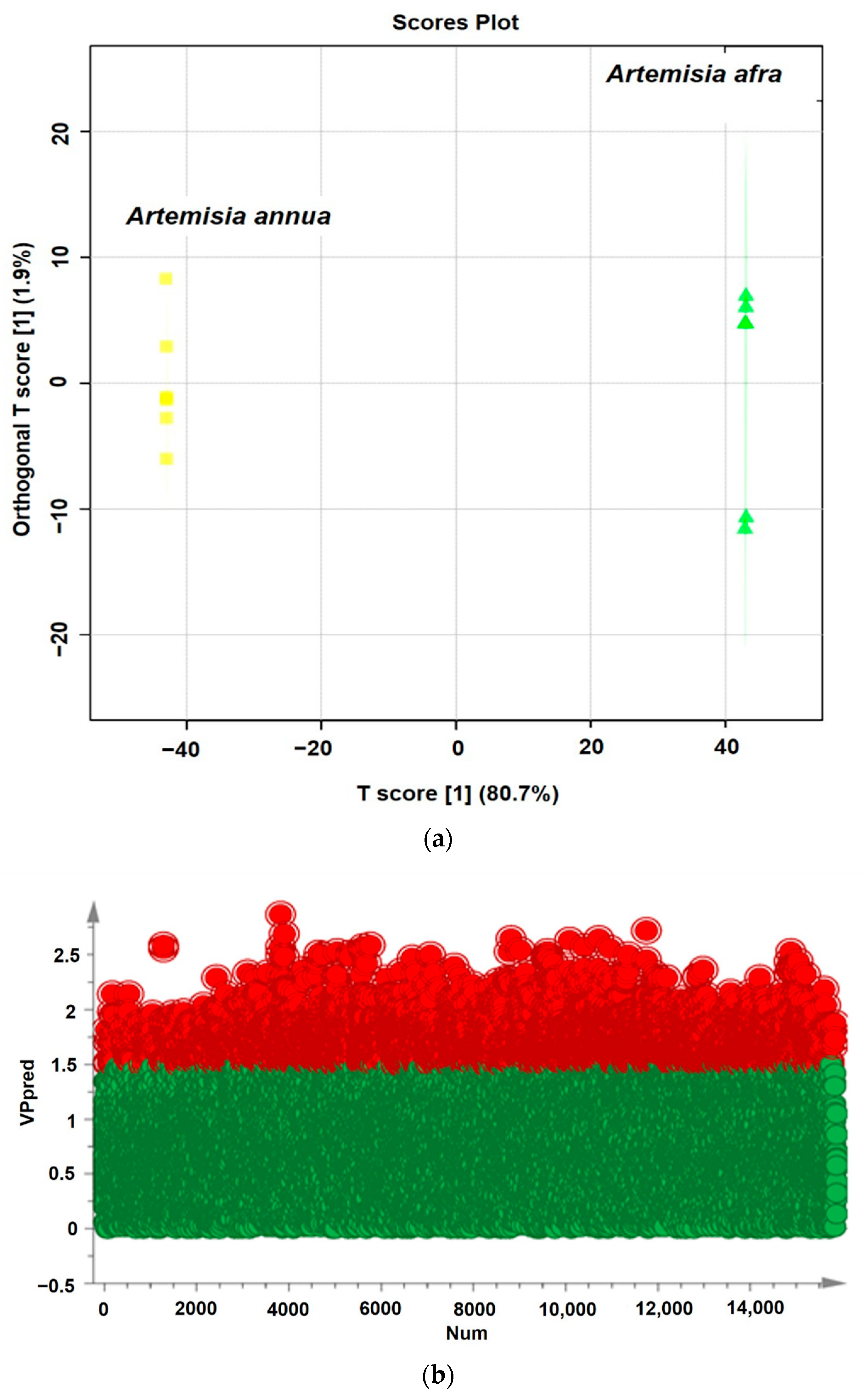



| Model Type | Number of Model Dimensions | N (a) | R2X (cum) (b) | R2Y (cum) (b) | Q2 (cum) (c) |
|---|---|---|---|---|---|
| Parameters | |||||
| PCA-X | 2 | 12 | 0.799 | - | 0.697 |
| PLS-DA | 2 | 12 | 0.793 | 1 | 0.999 |
| OPLS-DA | 1 + 1 + 0 | 12 | 0.793 | 1 | 0.999 |
| Blank | Positive Control | Test Inhibitor | |
|---|---|---|---|
| 1× Immuno Buffer 1 | 40 μL | 20 μL | 20 μL |
| Inhibitor buffer (no inhibitor) | 10 μL | 10 μL | - |
| ACE2-Biotin (1 ng/μL) | 20 μL | 20 μL | |
| Test Inhibitor | - | - | 10 μL |
| Total | 50 μL | 50 μL | 50 μL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mkolo, N.M.; Naidoo, C.M.; Kadye, R.; Obi, C.L.; Iweriebor, B.C.; Olaokun, O.O.; Prinsloo, E.; Zubair, M.S. Identification of South African Plant-Based Bioactive Compounds as Potential Inhibitors against the SARS-CoV-2 Receptor. Pharmaceuticals 2024, 17, 821. https://doi.org/10.3390/ph17070821
Mkolo NM, Naidoo CM, Kadye R, Obi CL, Iweriebor BC, Olaokun OO, Prinsloo E, Zubair MS. Identification of South African Plant-Based Bioactive Compounds as Potential Inhibitors against the SARS-CoV-2 Receptor. Pharmaceuticals. 2024; 17(7):821. https://doi.org/10.3390/ph17070821
Chicago/Turabian StyleMkolo, Nqobile Monate, Clarissa Marcelle Naidoo, Rose Kadye, Chikwelu Lawrence Obi, Benson Chucks Iweriebor, Oyinlola Oluwunmi Olaokun, Earl Prinsloo, and Muhammad Sulaiman Zubair. 2024. "Identification of South African Plant-Based Bioactive Compounds as Potential Inhibitors against the SARS-CoV-2 Receptor" Pharmaceuticals 17, no. 7: 821. https://doi.org/10.3390/ph17070821
APA StyleMkolo, N. M., Naidoo, C. M., Kadye, R., Obi, C. L., Iweriebor, B. C., Olaokun, O. O., Prinsloo, E., & Zubair, M. S. (2024). Identification of South African Plant-Based Bioactive Compounds as Potential Inhibitors against the SARS-CoV-2 Receptor. Pharmaceuticals, 17(7), 821. https://doi.org/10.3390/ph17070821







