Exploring the Underlying Mechanisms of Qingxing Granules Treating H1N1 Influenza Based on Network Pharmacology and Experimental Validation
Abstract
1. Introduction
2. Results
2.1. Component Target Prediction of Qingxing Granules
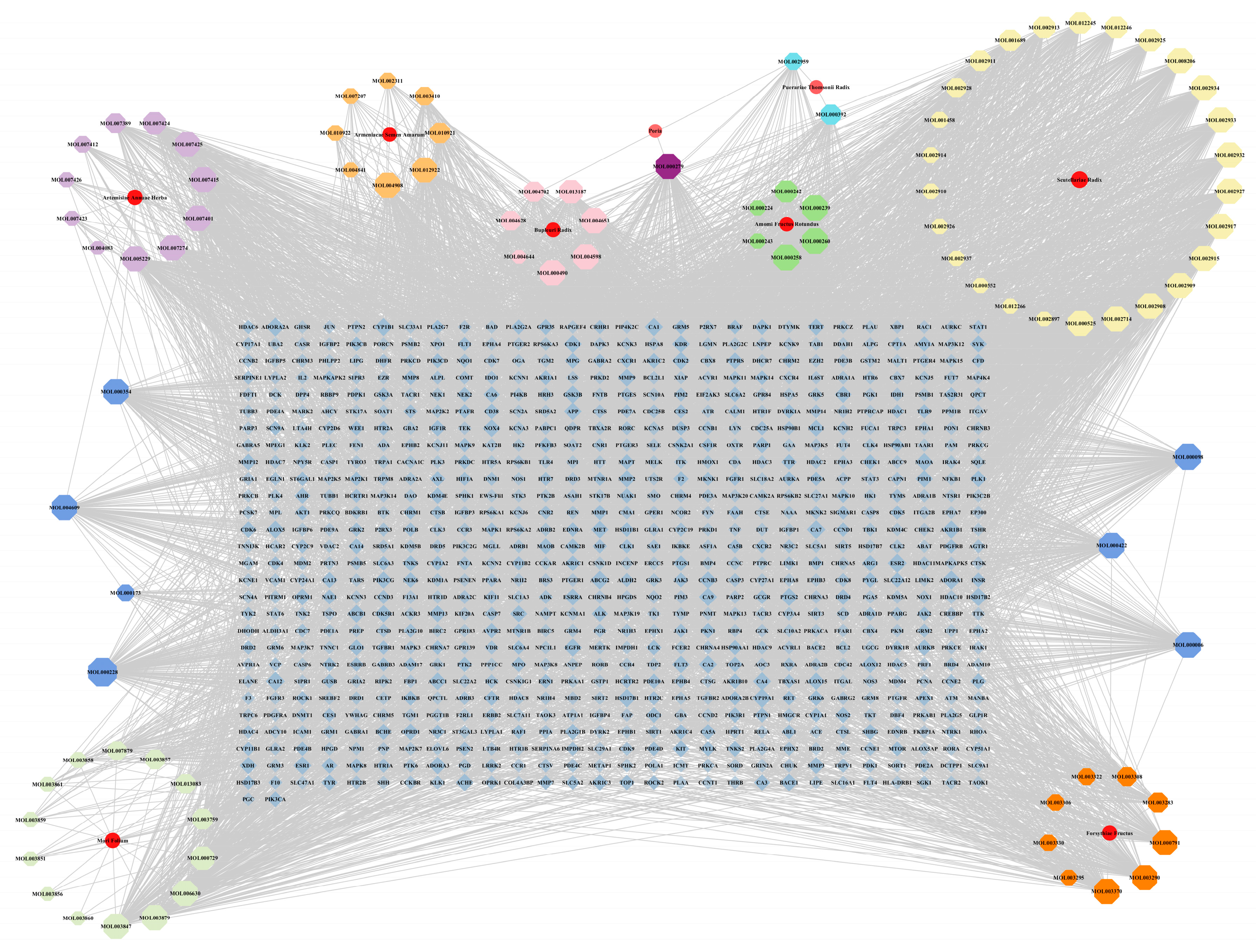
2.2. “Medicinal Material-Effective Component-Target” Network Map of Qingxing Granules
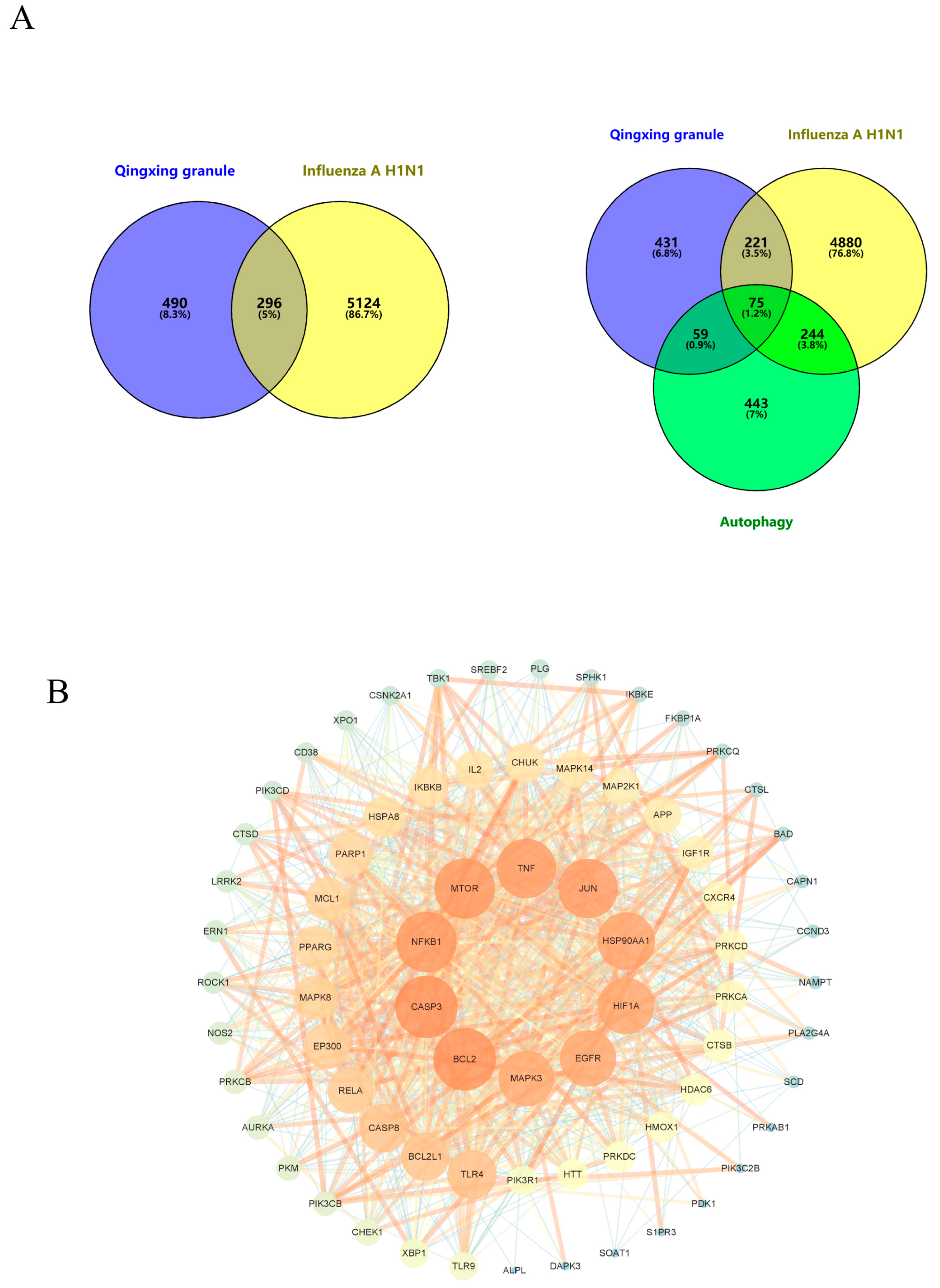
2.3. Prediction of Influenza A H1N1 Targets
2.4. Prediction of Autophagy-Related Genes
2.5. Prediction of Qingxing Granules PPI Network
2.6. GO Enrichment Analysis
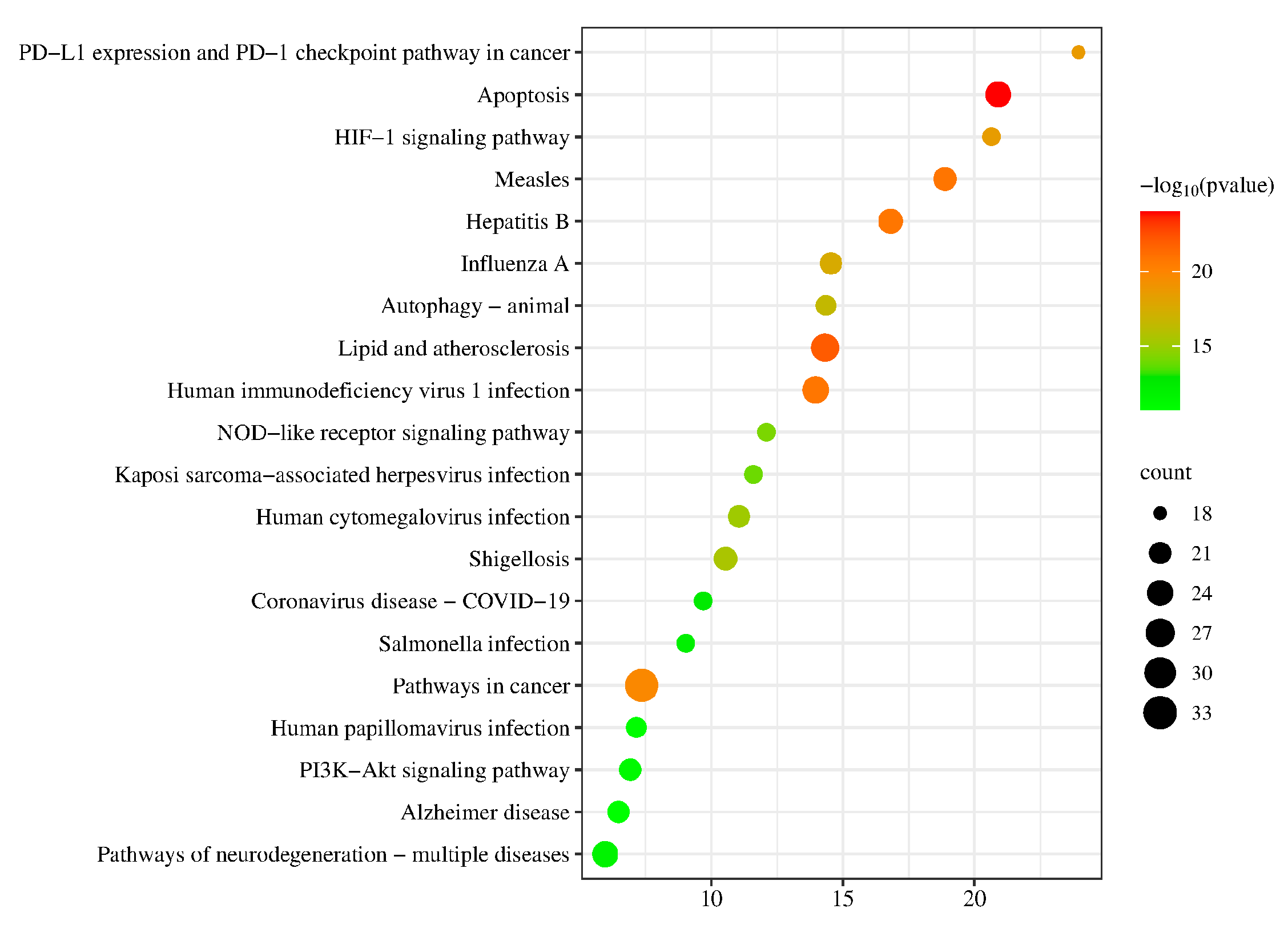
2.7. KEGG Pathway Enrichment Analysis
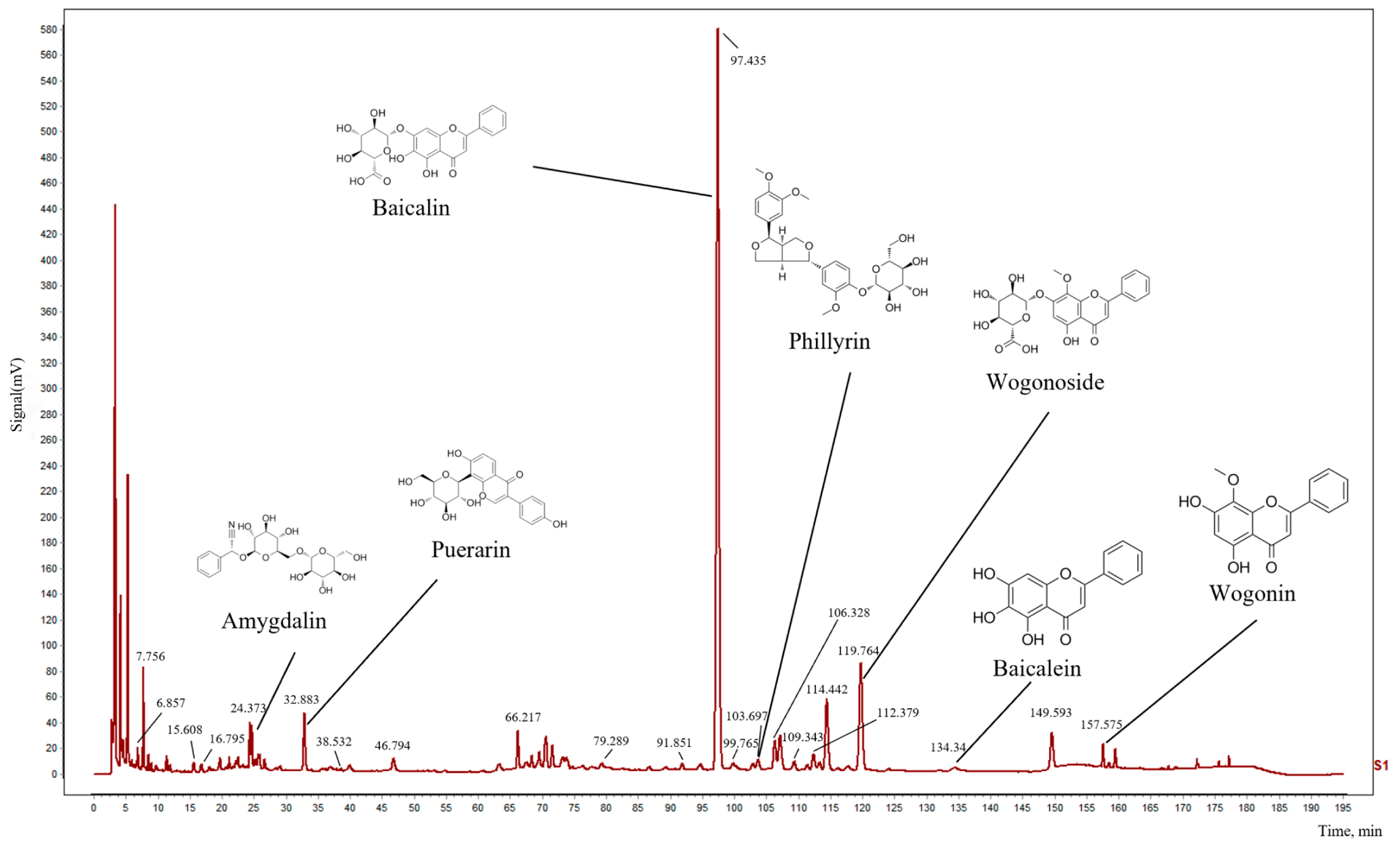
2.8. Determination of the Main Chemical Components in QX
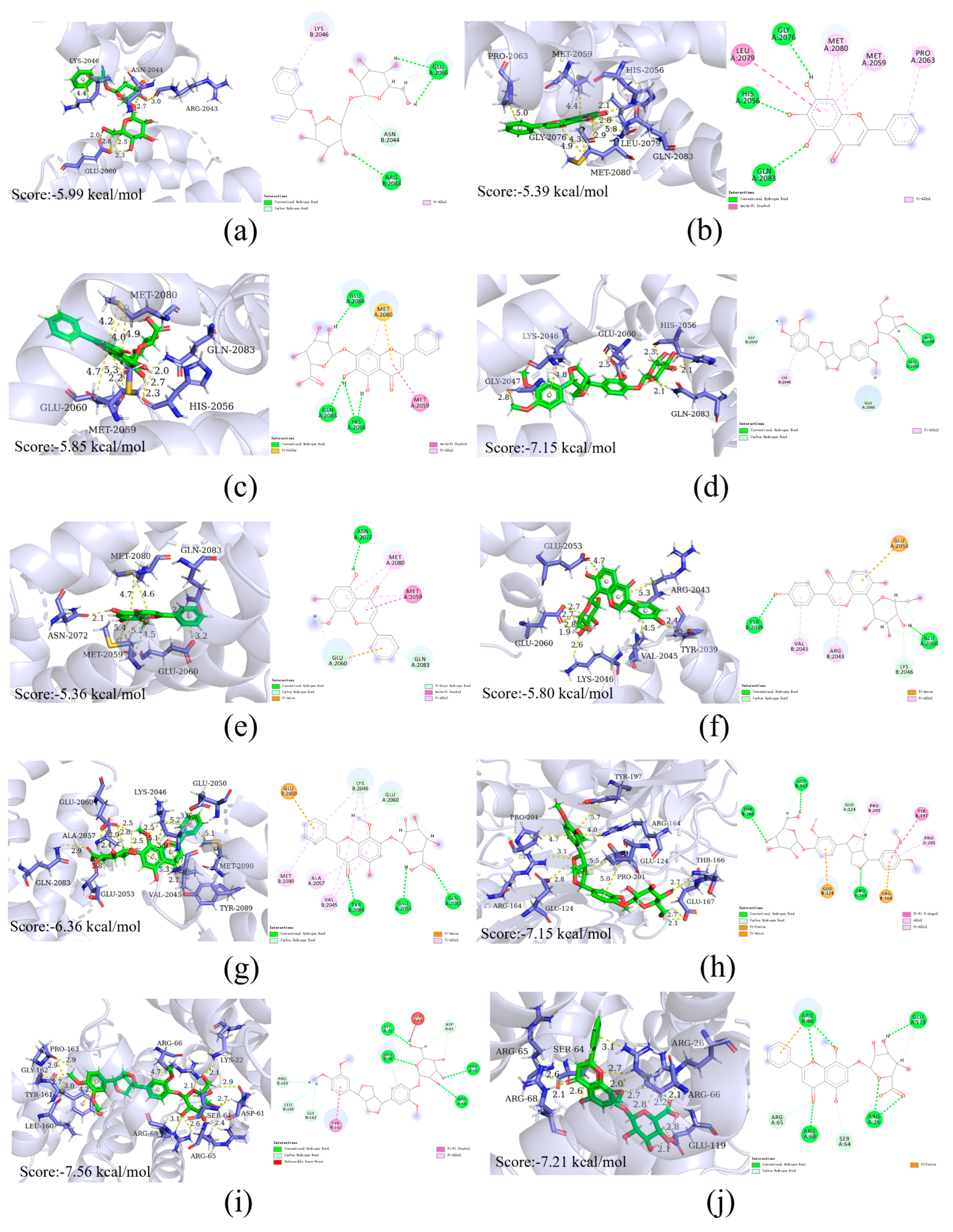
2.9. Molecular Docking
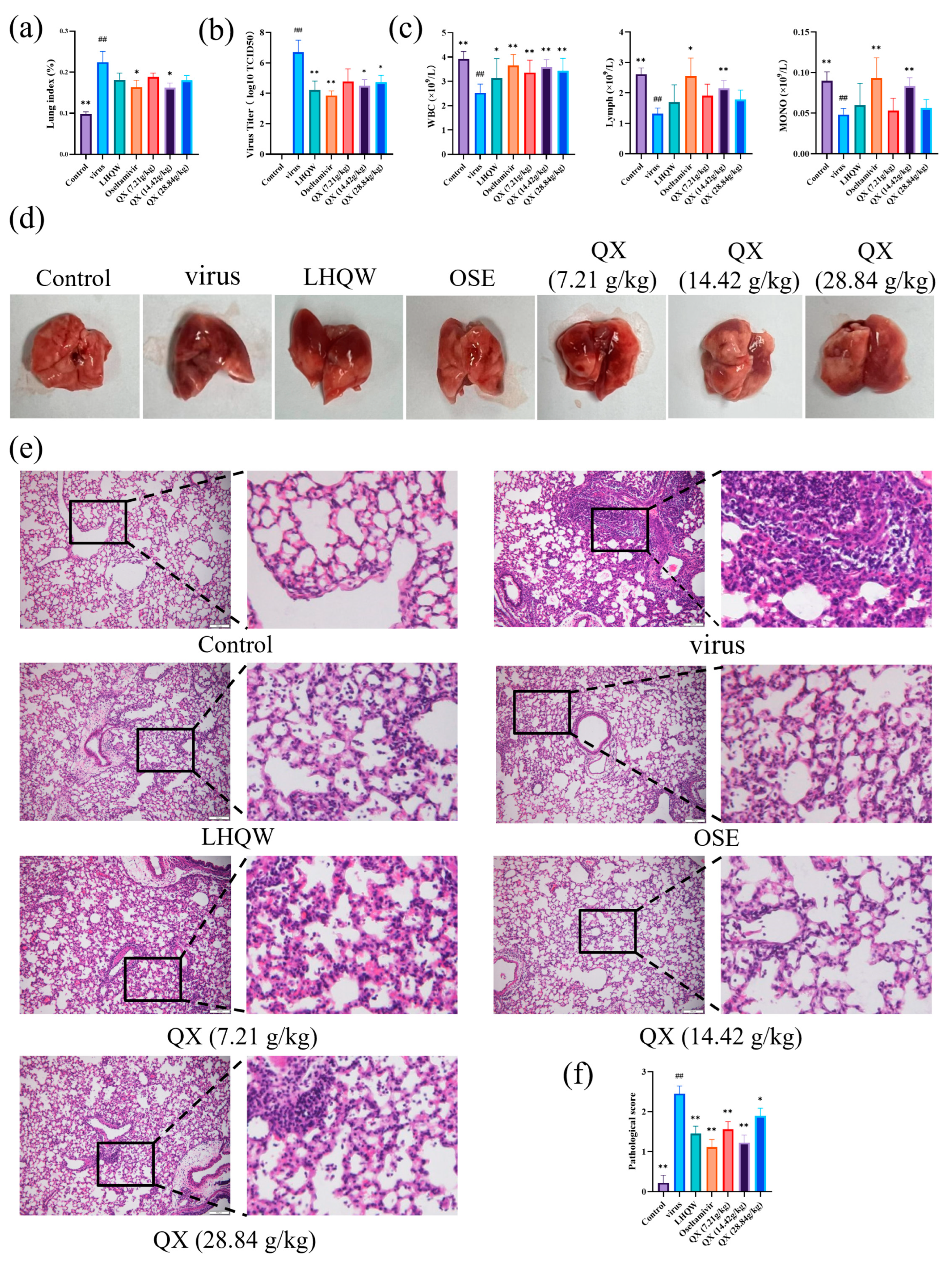
2.10. Improved Effects of QX on Mice Infected with H1N1
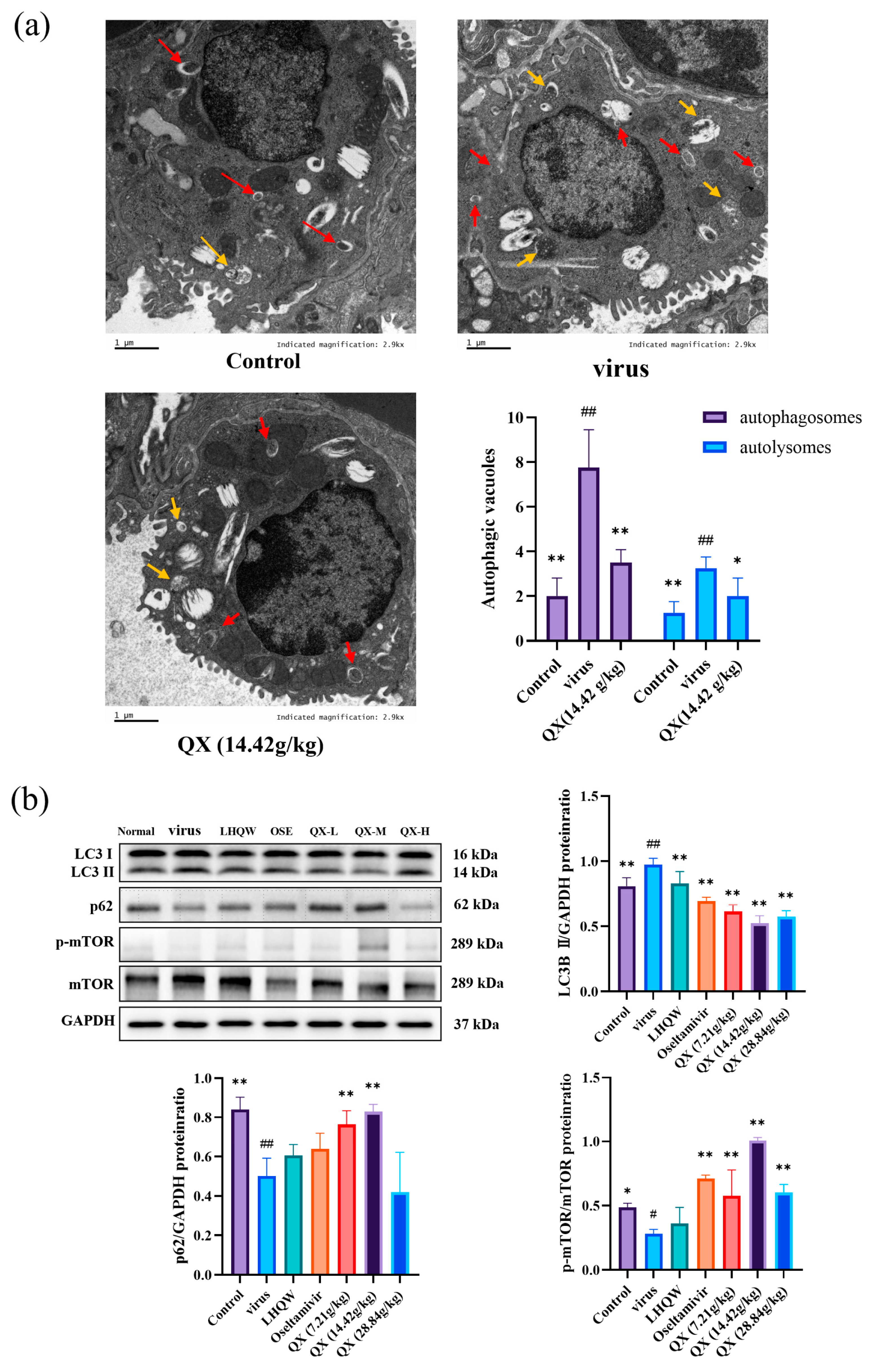
2.11. Transmission Electron Microscopy
2.12. Western Blot
3. Discussion
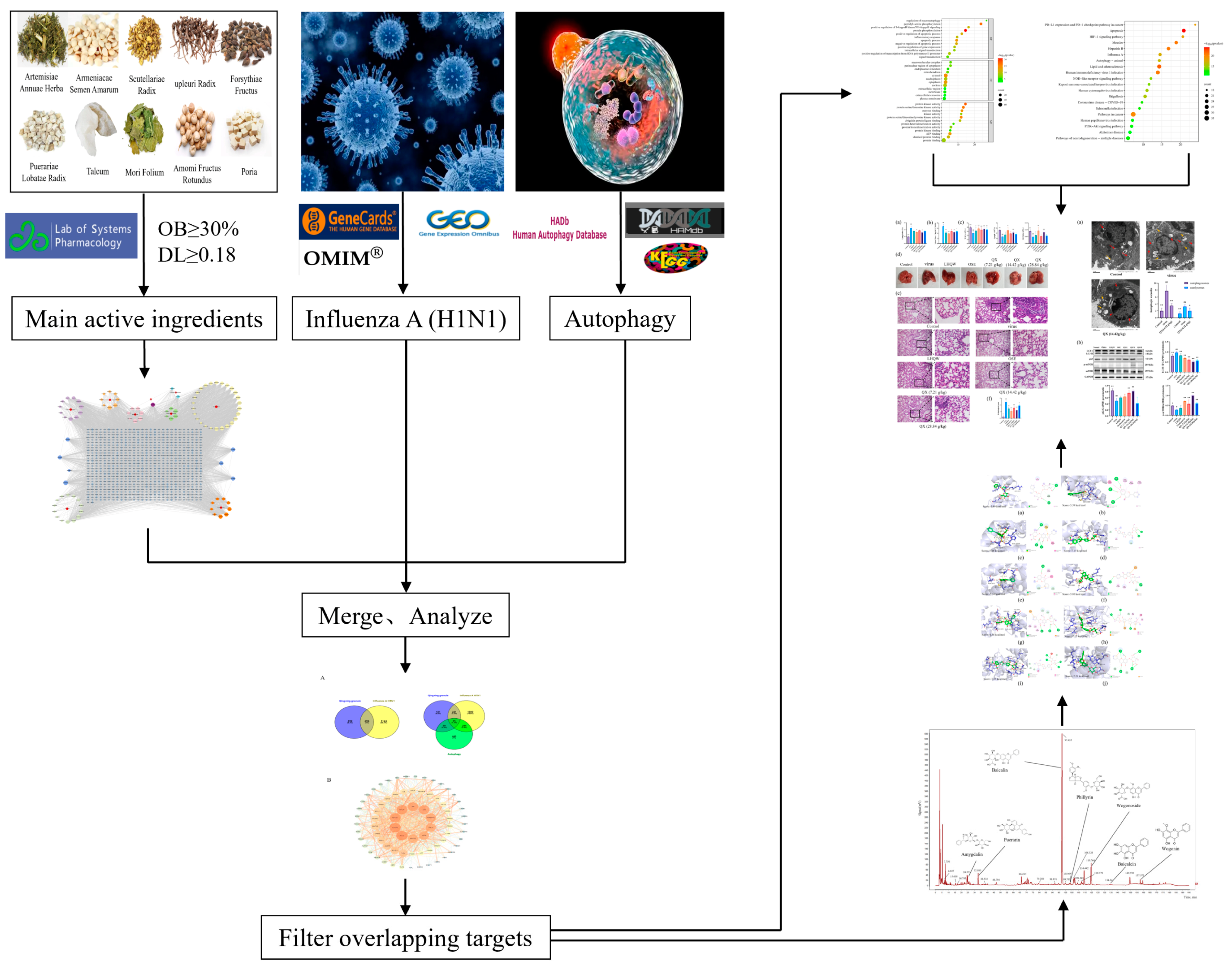
4. Materials and Methods
4.1. Screening of Active Biological Compounds and Related Targets in Qingxing Granules
4.2. Network Construction of “Medicinal Material-Active Component-Target” of Qingxing Granules
4.3. Collection of Targets Related to Influenza A H1N1
4.4. Collection of Autophagy-Related Genes
4.5. Construction and Analysis of the Protein–Protein Interaction (PPI) Network
4.6. GO Enrichment Analysis and KEGG Pathway Enrichment Analysis
4.7. Medicinal Herbs
4.8. Preparation Process of Qingxing Granules
4.9. HPLC Analysis of Qingxing Granules
4.10. Molecular Docking
4.11. Animals and Viruses
4.12. Animal Model
4.13. Routine Blood Test
4.14. Hematoxylin and Eosin (HE) Staining
4.15. Transmission Electron Microscopy
4.16. Western Blot Analysis
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Influenza (Seasonal). 2023. Volume 2023. Available online: https://www.who.int/ (accessed on 19 December 2023).
- Park, J.E.; Ryu, Y. Transmissibility and severity of influenza virus by subtype. Infect. Genet. Evol. 2018, 65, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Hagau, N.; Slavcovici, A.; Gonganau, D.N.; Oltean, S.; Dirzu, D.S.; Brezoszki, E.S.; Maxim, M.; Ciuce, C.; Mlesnite, M.; Gavrus, R.L.; et al. Clinical aspects and cytokine response in severe H1N1 influenza A virus infection. Crit. Care 2010, 14, R203. [Google Scholar] [CrossRef]
- Huo, C.; Tang, Y.; Li, X.; Han, D.; Gu, Q.; Su, R.; Liu, Y.; Reiter, R.J.; Liu, G.; Hu, Y.; et al. Melatonin alleviates lung injury in H1N1-infected mice by mast cell inactivation and cytokine storm suppression. PLoS Pathog. 2023, 19, e1011406. [Google Scholar] [CrossRef]
- Antipov, E.A.; Pokryshevskaya, E.B. The effects of adverse drug reactions on patients’ satisfaction: Evidence from publicly available data on Tamiflu (oseltamivir). Int. J. Med. Inf. 2019, 125, 30–36. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Hui, D.S.; Zambon, M.; Wentworth, D.E.; Monto, A.S. Influenza. Lancet 2022, 400, 693–706. [Google Scholar] [CrossRef]
- Zang, F.; Chen, Y.; Lin, Z.; Cai, Z.; Yu, L.; Xu, F.; Wang, J.; Zhu, W.; Lu, H. Autophagy is involved in regulating the immune response of dendritic cells to influenza A (H1N1) pdm09 infection. Immunology 2016, 148, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, X.; Liu, D.; Fan, Z.; Hu, X.; Yan, J.; Wang, M.; Gao, G.F. Autophagy is involved in influenza A virus replication. Autophagy. Autophagy 2009, 5, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Lőrincz, P.; Juhász, G. Autophagosome-Lysosome Fusion. J. Mol. Biol. 2020, 432, 2462–2482. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.-S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.-I.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Lee, Y.K.; Lee, J.A. Role of the mammalian ATG8/LC3 family in autophagy: Differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 2016, 49, 424–430. [Google Scholar] [CrossRef]
- Gannagé, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Rämer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, B.; Ghavami, S.; Rahim, M.N.; Klonisch, T.; Halayko, A.J.; Coombs, K.M. Autophagy activation is required for influenza A virus-induced apoptosis and replication. Biochim. Biophys. Acta. Mol. Cell Res. 2018, 1865, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Zhirnov, O.P.; Klenk, H.D. Influenza A virus proteins NS1 and hemagglutinin along with M2 are involved in stimulation of autophagy in infected cells. J. Virol. 2013, 87, 13107–13114. [Google Scholar] [CrossRef] [PubMed]
- Nayak, M.K.; Agrawal, A.S.; Bose, S.; Naskar, S.; Bhowmick, R.; Chakrabarti, S.; Sarkar, S.; Chawla-Sarkar, M. Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 2014, 69, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Zhang, J.; Wu, B.; Li, Z.; Wu, J.; Bie, M.J. Therapeutic Effect of Artesunate on Influenza A Viral Pneumonia. Sichuan Da Xue Xue Bao Yi Xue Ban 2022, 53, 1055–1060. [Google Scholar] [CrossRef]
- Chen, J.; Duan, M.; Zhao, Y.; Ling, F.; Xiao, K.; Li, Q.; Li, B.; Lu, C.; Qi, W.; Zeng, Z.; et al. Saikosaponin A inhibits influenza A virus replication and lung immunopathology. Oncotarget 2015, 6, 42541–42556. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y. Study of the Effects and Mechanisms of Qingxing Granules in Treating Dengue Fever Based on In Vivo and In Vitro Models. Master’s Thesis, Guangzhou University of Chinese Medicine, Guangzhou, China, 2023. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef] [PubMed]
- Abdelwhab, E.M.; Veits, J.; Mettenleiter, T.C. Biological fitness and natural selection of amantadine resistant variants of avian influenza H5N1 viruses. Virus Res. 2017, 228, 109–113. [Google Scholar] [CrossRef]
- Mitrasinovic, P.M. Advances in the structure-based design of the influenza A neuraminidase inhibitors. Curr. Drug Targets 2010, 11, 315–326. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Z.; Lin, C.; Ren, H.; Li, Y.; Zhang, Y.; Qu, Y.; Li, H.; Ma, S.; Xia, H.; et al. A/(H1N1) pdm09 NS1 promotes viral replication by enhancing autophagy through hijacking the IAV negative regulatory factor LRPPRC. Autophagy 2023, 19, 1533–1550. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, B.; Cho, S.Y.; Oh, K.S.; Kim, S.H.; Kim, Y.O.; Jeong, E.H.; Nguyen, T.T.; Kim, S.H.; Kim, I.S.; Kwon, J.; et al. Effectiveness of Periodic Treatment of Quercetin against Influenza A Virus H1N1 through Modulation of Protein Expression. J. Agric. Food Chem. 2016, 64, 4416–4425. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, S.; Du, L.; Jiang, S. A new role of neuraminidase (NA) in the influenza virus life cycle: Implication for developing NA inhibitors with novel mechanism of action. Rev. Med. Virol. 2016, 26, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.-L.; Wang, H.-D.; Lee, S.M.; Wang, Y.-T.; Du, G.-H. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg. Med. Chem. 2008, 16, 7141–7147. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Abdal Dayem, A.; Choi, H.Y.; Kim, Y.B.; Cho, S.G. Antiviral effect of methylated flavonol isorhamnetin against influenza. PLoS ONE 2015, 10, e0121610. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, S.; Liu, D.; Wei, H.; Wang, X.; Yan, F. Exploration of the potential mechanism of Pushen capsule in the treatment of vascular dementia based on network pharmacology and experimental verification. J. Ethnopharmacol. 2022, 298, 115632. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Martin, J.F.; Martin-Loeches, I.; Rello, J.; Antón, A.; Almansa, R.; Xu, L.; Lopez-Campos, G.; Pumarola, T.; Ran, L.; Ramirez, P.; et al. Host adaptive immunity deficiency in severe pandemic influenza. Crit. Care 2010, 14, R167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, B.; Jin, H.; Liu, Z.; Li, Y. Differential expression of autophagy related genes in tongue cancer and its influence on prognosis based on TCGA and HADb database. Oral Biomed. 2020, 11, 232–236. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; Dong, J.; Zhang, L.; Ouyang, D.; Cheng, Y.; Chen, A.F.; Lu, A.P.; Cao, D.S. HAMdb: A database of human autophagy modulators with specific pathway and disease information. J. Cheminform. 2018, 10, 34. [Google Scholar] [CrossRef]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Huang, W.; Zhao, J.; Yang, Z. Liu Shen Wan inhibits influenza a virus and excessive virus-induced inflammatory response via suppression of TLR4/NF-κB signaling pathway in vitro and in vivo. J. Ethnopharmacol. 2020, 252, 112584. [Google Scholar] [CrossRef] [PubMed]
- Sang, H. Study on Anti-Influenza A Virus Effect of Cepharanthine in Vitro and Vivo; Southern Medical University: Guangzhou, China, 2023. [Google Scholar] [CrossRef]
- Matute-Bello, G.; Winn, R.K.; Jonas, M.; Chi, E.Y.; Martin, T.R.; Liles, W.C. Fas (CD95) induces alveolar epithelial cell apoptosis in vivo: Implications for acute pulmonary inflammation. Am. J. Pathol. 2001, 158, 153–161. [Google Scholar] [CrossRef] [PubMed]

| Mol ID | Compound Name | Molecular Function | 2D Structure | Degree | BC | CC | ASPL |
|---|---|---|---|---|---|---|---|
| MOL000228 | (2R)-7-hydroxy-5-methoxy-2-phenylchroman-4-one | C16H14O4 | 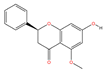 | 131 | 0.0628 | 0.3839 | 2.6045 |
| MOL000098 | Quercetin | C15H10O7 | 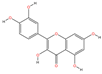 | 108 | 0.0093 | 0.3767 | 2.6542 |
| MOL000258 | Dehydrodiisoeugenol | C19H20O4 | 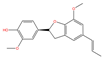 | 108 | 0.0513 | 0.3751 | 2.6655 |
| MOL000006 | Luteolin | C15H10O6 | 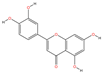 | 106 | 0.0087 | 0.3748 | 2.6677 |
| MOL000260 | 5-[(2R,3R)-7-methoxy-3-methyl-5-[(E)-prop-1-enyl]-2,3-dihydrobenzofuran-2-yl]-1,3-benzodioxole | C20H20O4 |  | 105 | 0.0777 | 0.3729 | 2.6813 |
| MOL000239 | Jaranol | C17H14O6 | 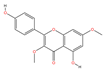 | 104 | 0.0106 | 0.3745 | 2.6700 |
| MOL000422 | Kaempferol | C15H10O6 | 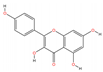 | 104 | 0.0078 | 0.3754 | 2.6632 |
| MOL000354 | Isorhamnetin | C16H12O7 | 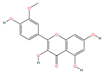 | 102 | 0.0069 | 0.3732 | 2.6790 |
| MOL004609 | Areapillin | C18H16O8 |  | 102 | 0.0068 | 0.3729 | 2.6813 |
| MOL000490 | Petunidin | C16H13O7+ | 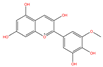 | 101 | 0.0087 | 0.3735 | 2.6768 |
| Number | Target Name | Degree | BC | CC | ASPL |
|---|---|---|---|---|---|
| 1 | BCL2 | 56 | 0.04368725 | 0.81818182 | 1.22222222 |
| 2 | CASP3 | 56 | 0.04635699 | 0.81818182 | 1.22222222 |
| 3 | NFKB1 | 54 | 0.03216106 | 0.8 | 1.25 |
| 4 | MTOR | 54 | 0.052009 | 0.8 | 1.25 |
| 5 | JUN | 53 | 0.03213428 | 0.79120879 | 1.26388889 |
| 6 | TNF | 53 | 0.03952339 | 0.79120879 | 1.26388889 |
| 7 | HSP90AA1 | 51 | 0.03260088 | 0.77419355 | 1.29166667 |
| 8 | EGFR | 50 | 0.03866376 | 0.76595745 | 1.30555556 |
| 9 | HIF1A | 50 | 0.03640685 | 0.76595745 | 1.30555556 |
| 10 | MAPK3 | 49 | 0.05609154 | 0.75789474 | 1.31944444 |
| Marker Ingredients | Retention Time (min) | Peak Area | Relative Retention Time | Relative Peak Area |
|---|---|---|---|---|
| Amygdalin | 24.373 | 412935 | 0.2501 | 0.0251 |
| Puerarin | 32.883 | 869521 | 0.3375 | 0.0529 |
| Baicalin | 97.435 | 16427378 | 1.0000 | 1.0000 |
| Phillyrin | 103.697 | 201714 | 1.0643 | 0.0123 |
| Wogonoside | 119.764 | 2860469 | 1.2292 | 0.1741 |
| Baicalein | 134.34 | 188517 | 1.3788 | 0.0115 |
| Wogonin | 157.575 | 252499 | 1.6172 | 0.0154 |
| Compound | Affinity kcal/mol | Protein | BCL2 | CASP3 | NFKB1 | MTOR |
|---|---|---|---|---|---|---|
| (2R)-7-hydroxy-5-methoxy-2-phenylchroman-4-one | −5.17 | −6.26 | −4.77 | −5.63 | ||
| Quercetin | −5.58 | −5.59 | −5.30 | −5.61 | ||
| Dehydrodiisoeugenol | −6.01 | −6.06 | −6.06 | −5.38 | ||
| Amygdalin | −6.97 | −6.56 | −7.06 | −5.99 | ||
| Puerarin | −6.69 | −6.50 | −5.85 | −5.80 | ||
| Baicalin | −6.85 | −6.52 | −6.75 | −5.85 | ||
| Phillyrin | −7.56 | −7.15 | −7.09 | −7.15 | ||
| Wogonoside | −7.21 | −6.94 | −6.49 | −6.36 | ||
| Baicalein | −5.13 | −5.55 | / | −5.39 | ||
| Wogonin | −5.33 | −5.64 | −5.01 | −5.36 | ||
| Number | Herb Name Chinese Spelling | English Name | Latin Name | Plant Part | Weight Percentage |
|---|---|---|---|---|---|
| 1 | Qing Hao | Artemisiae Annuae Herba | Artemisia annua L. | Aboveground | 10.42% |
| 2 | Ku Xingren | Armeniacae Semen Amarum | Prunus armeniaca L. var. ansu Maxim. | Seed | 10.42% |
| 3 | Chai Hu | Bupleuri Radix | Bupleurum chinense DC. | Root | 10.42% |
| 4 | Huang Qin | Scutellariae Radix | Scutellaria baicalensis Georgi | Root | 10.42% |
| 5 | Lian Qiao | Forsythiae Fructus | Forsythia suspensa (Thunb.) Vahl | Fructus | 10.42% |
| 6 | Ge Gen | Puerariae Lobatae Radix | Pueraria lobata (Willd.) Ohwi | Root | 10.42% |
| 7 | Hua Shi | Talcum | Talcum | Ore | 10.42% |
| 8 | Sang Ye | Mori Folium | Morus alba L. | Leaf | 10.42% |
| 9 | Dou Kou | Amomi Fructus Rotundus | Amomum kravanh Pierre ex Gagnep. | Fructus | 6.25% |
| 10 | Fu Ling | Poria | Poria Cocos (Schw.) Wolf | Sclerotium | 10.42% |
| Time (min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|
| 0~10 | 5% | 95% |
| 10~15 | 5~10% | 95~90% |
| 15~55 | 10~12% | 90~88% |
| 55~60 | 12~16% | 88~84% |
| 60~80 | 16~18% | 84~82% |
| 80~100 | 18~22% | 82~78% |
| 100~140 | 22~26% | 78~74% |
| 140~160 | 26~56% | 74~44% |
| 160~170 | 56~90% | 44~10% |
| 170~195 | 90~5% | 10~95% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, H.; Zhang, L.; Sun, H.; Zheng, S.; Zhang, H.; Yuan, S.; Zhou, J.; Fang, Z.; Song, J.; Mei, M.; et al. Exploring the Underlying Mechanisms of Qingxing Granules Treating H1N1 Influenza Based on Network Pharmacology and Experimental Validation. Pharmaceuticals 2024, 17, 731. https://doi.org/10.3390/ph17060731
Du H, Zhang L, Sun H, Zheng S, Zhang H, Yuan S, Zhou J, Fang Z, Song J, Mei M, et al. Exploring the Underlying Mechanisms of Qingxing Granules Treating H1N1 Influenza Based on Network Pharmacology and Experimental Validation. Pharmaceuticals. 2024; 17(6):731. https://doi.org/10.3390/ph17060731
Chicago/Turabian StyleDu, Hujun, Lianying Zhang, Haoxiang Sun, Shaoqin Zheng, Hongying Zhang, Shijia Yuan, Jiuyao Zhou, Zihao Fang, Jianping Song, Manxue Mei, and et al. 2024. "Exploring the Underlying Mechanisms of Qingxing Granules Treating H1N1 Influenza Based on Network Pharmacology and Experimental Validation" Pharmaceuticals 17, no. 6: 731. https://doi.org/10.3390/ph17060731
APA StyleDu, H., Zhang, L., Sun, H., Zheng, S., Zhang, H., Yuan, S., Zhou, J., Fang, Z., Song, J., Mei, M., & Deng, C. (2024). Exploring the Underlying Mechanisms of Qingxing Granules Treating H1N1 Influenza Based on Network Pharmacology and Experimental Validation. Pharmaceuticals, 17(6), 731. https://doi.org/10.3390/ph17060731






