BRCA1 and Its Vulnerable C-Terminal BRCT Domain: Structure, Function, Genetic Mutations and Links to Diagnosis and Treatment of Breast and Ovarian Cancer
Abstract
1. Introduction
2. Structure and Function of BRCA1-BRCT Domain
3. Association of BRCA1-BRCT Domains Mutations and Breast Cancer
4. Impact of BRCT Domain Mutations on Breast Cancer Prognosis
5. Association of BRCA1-BRCT Domain Mutations and Ovarian Cancer
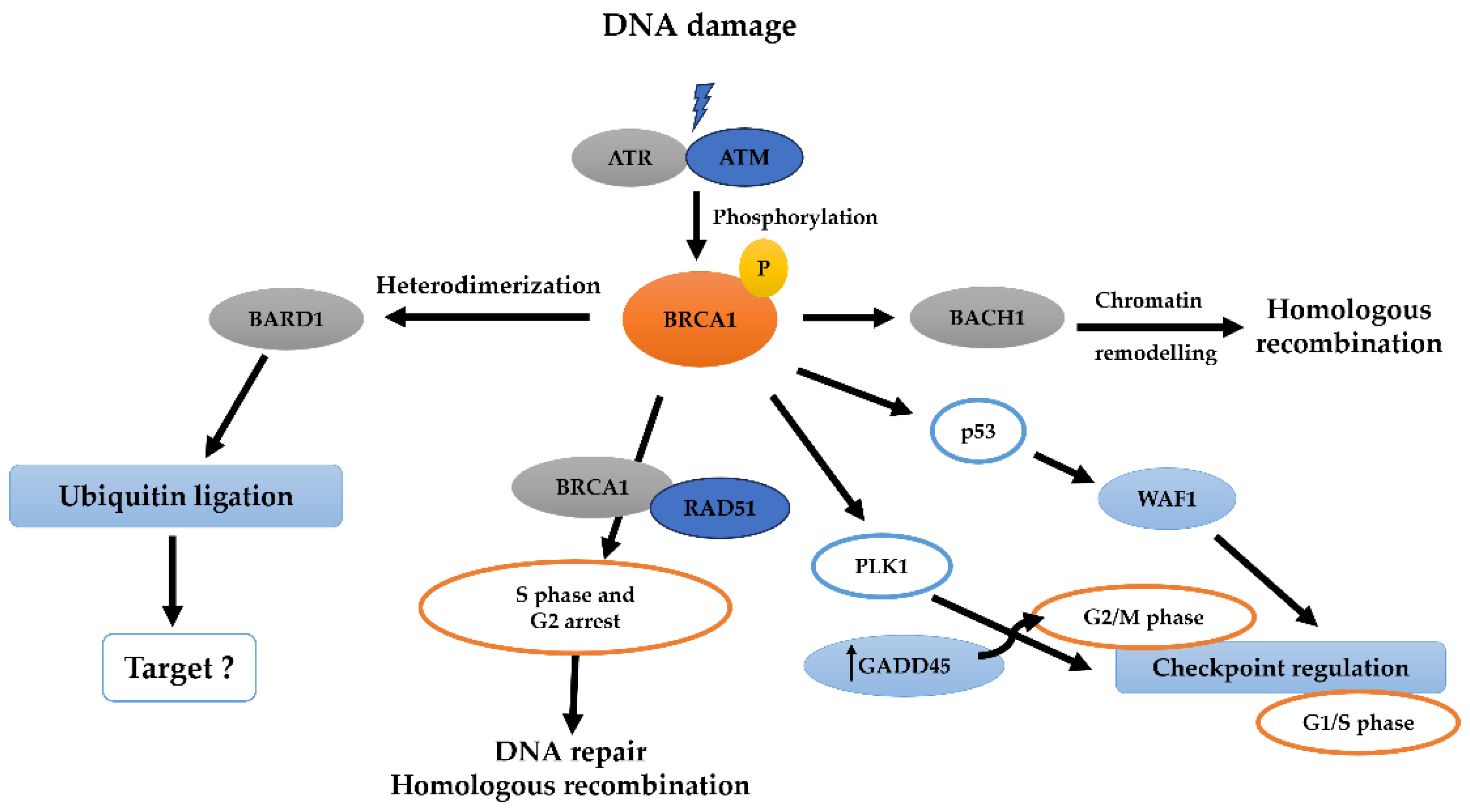
6. BRCA1-BRCT and Signaling Pathways in Breast and Ovarian Cancer
7. Genetic Testing and Screening for BRCA1 Mutations
8. Treatment Strategies for BRCA1 Mutation Carrier Patients
8.1. Surgery
8.2. Chemotherapy
8.3. Hormone Therapy
8.4. Targeted Therapy, like PARPS Inhibitors
9. Discussion
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- National Cancer Institute. BRCA Gene Mutations: Cancer Risk and Genetic Testing; National Cancer Institute: Washington, DC, USA, 2020. [Google Scholar]
- Grzelak, D. Treatment Options for Germline BRCA-Mutated Metastatic Pancreatic Adenocarcinoma. J. Adv. Pract. Oncol. 2021, 12, 488. [Google Scholar]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef]
- Gorodetska, I.; Kozeretska, I.; Dubrovska, A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer 2019, 10, 2109–2127. [Google Scholar] [CrossRef]
- Wang, B. BRCA1 tumor suppressor network: Focusing on its tail. Cell Biosci. 2012, 2, 6. [Google Scholar] [CrossRef]
- Clark, S.L.; Rodriguez, A.M.; Snyder, R.R.; Hankins, G.D.; Boehning, D. Structure-function of the tumor suppressor BRCA1. Comput. Struct. Biotechnol. J. 2012, 1, e201204005. [Google Scholar] [CrossRef]
- Li, Q.; Engebrecht, J. BRCA1 and BRCA2 Tumor Suppressor Function in Meiosis. Front. Cell Dev. Biol. 2021, 9, 668309. [Google Scholar] [CrossRef]
- Krivokuca, A.; Mihajlovic, M.; Susnjar, S.; Spasojevic, I.B.; Minic, I.; Popovic, L.; Brankovic-Magic, M. Mutational profile of hereditary breast and ovarian cancer—Establishing genetic testing guidelines in a developing country. Curr. Probl. Cancer 2022, 46, 100767. [Google Scholar] [CrossRef]
- Lee, M.S.; Green, R.; Marsillac, S.M.; Coquelle, N.; Williams, R.S.; Yeung, T.; Foo, D.; Hau, D.D.; Hui, B.; Monteiro, A.N.; et al. Comprehensive Analysis of Missense Variations in the BRCT Domain of BRCA1 by Structural and Functional Assays. Cancer Res. 2010, 70, 4880–4890. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Meza, J.E.; King, M.-C.; Klevit, R.E. BRCA1 RING Domain Cancer-predisposing Mutations. J. Biol. Chem. 2001, 276, 41399–41406. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-F.; Miyake, T.; Ye, Q.; Li, R. Characterization of a Novel Trans-Activation Domain of BRCA1 That Functions in Concert with the BRCA1 C-terminal (BRCT) Domain. J. Biol. Chem. 2000, 275, 40910–40915. [Google Scholar] [CrossRef] [PubMed]
- Pulver, E.M.; Mukherjee, C.; van de Kamp, G.; Roobol, S.J.; Rother, M.B.; van der Gulden, H.; de Bruijn, R.; Lattanzio, M.V.; van der Burg, E.; Drenth, A.P.; et al. A BRCA1 Coiled-Coil Domain Variant Disrupting PALB2 Interaction Promotes the Development of Mammary Tumors and Confers a Targetable Defect in Homologous Recombination Repair. Cancer Res. 2021, 81, 6171–6182. [Google Scholar] [CrossRef]
- Wu, Q.; Jubb, H.; Blundell, T.L. Phosphopeptide interactions with BRCA1 BRCT domains: More than just a motif. Prog. Biophys. Mol. Biol. 2015, 117, 143–148. [Google Scholar] [CrossRef]
- Zhang, X.; Moréra, S.; Bates, P.A.; Whitehead, P.C.; Coffer, A.I.; Hainbucher, K.; Nash, R.A.; Sternberg, M.J.; Lindahl, T.; Freemont, P.S. Structure of an XRCC1 BRCT domain: A new protein-protein interaction module. EMBO J. 1998, 17, 6404–6411. [Google Scholar] [CrossRef]
- Peña-Guerrero, J.; Fernández-Rubio, C.; García-Sosa, A.T.; Nguewa, P.A. BRCT Domains: Structure, Functions, and Implications in Disease—New Therapeutic Targets for Innovative Drug Discovery against Infections. Pharmaceutics 2023, 15, 1839. [Google Scholar] [CrossRef]
- Shiozaki, E.N.; Gu, L.; Yan, N.; Shi, Y. Structure of the BRCT Repeats of BRCA1 Bound to a BACH1 Phosphopeptide: Implications for Signaling. Mol. Cell 2004, 14, 405–412. [Google Scholar] [CrossRef]
- BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 2004, 11, 519–525. [CrossRef]
- Ouchi, T.; Monteiro, A.N.A.; August, A.; Aaronson, S.A.; Hanafusa, H. BRCA1 regulates p53-dependent gene expression. Proc. Natl. Acad. Sci. USA 1998, 95, 2302–2306. [Google Scholar] [CrossRef] [PubMed]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.C.Y.; Glover, J.M. BRCT domains. Cell Cycle 2011, 10, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.D.; Ryu, W.-J.; Han, H.J.; Kim, T.Y.; Kim, M.H.; Sohn, J. Molecular Characterization of BRCA1 c.5339T>C Missense Mutation in DNA Damage Response of Triple-Negative Breast Cancer. Cancers 2022, 14, 2405. [Google Scholar] [CrossRef] [PubMed]
- Petitalot, A.; Dardillac, E.; Jacquet, E.; Nhiri, N.; Guirouilh-Barbat, J.; Julien, P.; Bouazzaoui, I.; Bonte, D.; Feunteun, J.; Schnell, J.A.; et al. Combining Homologous Recombination and Phosphopeptide-binding Data to Predict the Impact of BRCA1 BRCT Variants on Cancer Risk. Mol. Cancer Res. 2019, 17, 54–69. [Google Scholar] [CrossRef]
- Drikos, I.; Boutou, E.; Kastritis, P.L.; Vorgias, C.E. BRCA1-BRCT Mutations Alter the Subcellular Localization of BRCA1 In Vitro. Anticancer Res. 2021, 41, 2953–2962. [Google Scholar] [CrossRef]
- Di Masi, A.; Gullotta, F.; Cappadonna, V.; Leboffe, L.; Ascenzi, P. Cancer predisposing mutations in BRCT domains. IUBMB Life 2011, 63, 503–512. [Google Scholar] [CrossRef]
- Laitman, Y.; Feng, B.-J.; Zamir, I.M.; Weitzel, J.N.; Duncan, P.; Port, D.; Thirthagiri, E.; Teo, S.-H.; Evans, G.; Latif, A.; et al. Haplotype analysis of the 185delAG BRCA1 mutation in ethnically diverse populations. Eur. J. Hum. Genet. 2012, 21, 212–216. [Google Scholar] [CrossRef]
- Hamel, N.; Feng, B.-J.; Foretova, L.; Stoppa-Lyonnet, D.; A Narod, S.; Imyanitov, E.; Sinilnikova, O.; Tihomirova, L.; Lubinski, J.; Gronwald, J.; et al. On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur. J. Hum. Genet. 2010, 19, 300–306. [Google Scholar] [CrossRef]
- NCI Dictionary of Cancer Terms—NCI 2011. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/ (accessed on 24 October 2023).
- Badgujar, D.C.; Sawant, U.; Mahadik, H.; Gadewal, N.; Varma, A.K. Pathogenicity of Mutations Discovered in BRCA1 BRCT Domains is Characterized by Destabilizing the Hydrophobic Interactions. J. Cancer Sci. Ther. 2012, 4, 386–393. [Google Scholar] [CrossRef]
- Joo, W.S.; Jeffrey, P.D.; Cantor, S.B.; Finnin, M.S.; Livingston, D.M.; Pavletich, N.P. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002, 16, 583–593. [Google Scholar] [CrossRef]
- Williams, R.S.; Chasman, D.I.; Hau, D.D.; Hui, B.; Lau, A.Y.; Glover, J.N.M. Detection of Protein Folding Defects Caused by BRCA1-BRCT Truncation and Missense Mutations. J. Biol. Chem. 2003, 278, 53007–53016. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Wang, S.M. Classification of VUS and unclassified variants in BRCA1 BRCT repeats by molecular dynamics simulation. Comput. Struct. Biotechnol. J. 2020, 18, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R. Hereditary breast and ovarian cancer (HBOC): Review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer 2020, 28, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, C.; Goff, B.; Burstein, H.J. Cancer Risks and Management of BRCA1/2 Carriers without Cancer. Available online: https://www.uptodate.com/contents/cancer-risks-and-management-of-brca1-2-carriers-without-cancer (accessed on 10 January 2024).
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; Van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Xu, Y.; Zhou, Y.D.; Yao, R.; Wu, H.W.; Zhu, Q.L.; Wang, C.J.; Mao, F.; Lin, Y.; Shen, S.J.; et al. The prognostic comparison among unilateral, bilateral, synchronous bilateral, and metachronous bilateral breast cancer: A meta-analysis of studies from recent decade (2008–2018). Cancer Med. 2019, 8, 2908–2918. [Google Scholar] [CrossRef]
- Copson, E.R.; Maishman, T.C.; Tapper, W.J.; I Cutress, R.; Greville-Heygate, S.; Altman, D.G.; Eccles, B.; Gerty, S.; Durcan, L.T.; Jones, L.; et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): A prospective cohort study. Lancet Oncol. 2018, 19, 169–180. [Google Scholar] [CrossRef]
- Sukumar, J.; Gast, K.; Quiroga, D.; Lustberg, M.; Williams, N. Triple-negative breast cancer: Promising prognostic biomarkers currently in development. Expert Rev. Anticancer Ther. 2021, 21, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Boyle, P. Triple-negative breast cancer: Epidemiological considerations and recommendations. Ann. Oncol. 2012, 23 (Suppl. S6), vi7–vi12. [Google Scholar] [CrossRef]
- Baretta, Z.; Mocellin, S.; Goldin, E.; Olopade, O.I.; Huo, D. Effect of BRCA germline mutations on breast cancer prognosis. Medicine 2016, 95, e4975. [Google Scholar] [CrossRef]
- Creeden, J.F.; Nanavaty, N.S.; Einloth, K.R.; Gillman, C.E.; Stanbery, L.; Hamouda, D.M.; Dworkin, L.; Nemunaitis, J. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer 2021, 21, 1154. [Google Scholar] [CrossRef]
- Gorski, J.W.; Ueland, F.R.; Kolesar, J.M. CCNE1 Amplification as a Predictive Biomarker of Chemotherapy Resistance in Epithelial Ovarian Cancer. Diagnostics 2020, 10, 279. [Google Scholar] [CrossRef]
- Bruchim, I.; Fishman, A.; Friedman, E.; Goldberg, I.; Chetrit, A.; Barshack, I.; Dekel, E.; Hirsh-Yechezkel, G.; Modan, B.; Kopolovic, J. Analyses of p53 expression pattern and BRCA mutations in patients with double primary breast and ovarian cancer. Int. J. Gynecol. Cancer 2004, 14, 251–258. [Google Scholar] [CrossRef]
- Zheng, M.; Hu, Y.; Gou, R.; Liu, O.; Nie, X.; Li, X.; Liu, Q.; Hao, Y.; Liu, J.; Lin, B. Identification of immune-enhanced molecular subtype associated with BRCA1 mutations, immune checkpoints and clinical outcome in ovarian carcinoma. J. Cell. Mol. Med. 2020, 24, 2819–2831. [Google Scholar] [CrossRef]
- Wei, Y.; Ou, T.; Lu, Y.; Wu, G.; Long, Y.; Pan, X.; Yao, D. Classification of ovarian cancer associated with BRCA1 mutations, immune checkpoints, and tumor microenvironment based on immunogenomic profiling. PeerJ 2020, 8, e10414. [Google Scholar] [CrossRef]
- Clarke, B.; Tinker, A.V.; Lee, C.H.; Subramanian, S.; van de Rijn, M.; Turbin, D.; Kalloger, S.; Han, G.; Ceballos, K.; Cadungog, M.G.; et al. Intraepithelial T cells and prognosis in ovarian carcinoma: Novel associations with stage, tumor type, and BRCA1 loss. Mod. Pathol. 2009, 22, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2021, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, T.; Fisher, P.; Ganesan, S.; Efstratiadis, A. Tumorigenesis in mice carrying a truncating Brca1 mutation. Minerva Anestesiol. 2001, 15, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Pao, G.M.; Chen, H.-W.; Verma, I.M.; Hunter, T. Enhancement of BRCA1 E3 Ubiquitin Ligase Activity through Direct Interaction with the BARD1 Protein. J. Biol. Chem. 2003, 278, 5255–5263. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.-X. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006, 34, 1416–1426. [Google Scholar] [CrossRef]
- Christou, C.M.; Kyriacou, K. BRCA1 and Its Network of Interacting Partners. Biology 2013, 2, 40–63. [Google Scholar] [CrossRef]
- Xiang, T.; Ohashi, A.; Huang, Y.; Pandita, T.K.; Ludwig, T.; Powell, S.N.; Yang, Q. Negative Regulation of AKT Activation by BRCA1. Cancer Res. 2008, 68, 10040–10044. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Nadhan, R.; Latha, N.R.; Krishnan, N.; Warrier, A.V.; Srinivas, P. Deregulated estrogen receptor signaling and DNA damage response in breast tumorigenesis. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1875, 188482. [Google Scholar] [CrossRef]
- Hilton, H.N.; Graham, J.D.; Clarke, C.L. Minireview: Progesterone Regulation of Proliferation in the Normal Human Breast and in Breast Cancer: A Tale of Two Scenarios? Mol. Endocrinol. 2015, 29, 1230–1342. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5414684/ (accessed on 18 October 2023). [CrossRef]
- Kim, O.; Park, E.Y.; Kwon, S.Y.; Shin, S.; Emerson, R.E.; Shin, Y.-H.; DeMayo, F.J.; Lydon, J.P.; Coffey, D.M.; Hawkins, S.M.; et al. Targeting progesterone signaling prevents metastatic ovarian cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 31993–32004. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.J.; Olopade, O.I. Disparities in Genetic Testing: Thinking Outside the BRCA Box. J. Clin. Oncol. 2006, 24, 2197–2203. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Lynch, H.T.; Neuhausen, S.L.; Narod, S.A.; Veer, L.V.; Garber, J.E.; Evans, G.; Isaacs, C.; Daly, M.B.; Matloff, E.; et al. Prophylactic Oophorectomy in Carriers of BRCA1 or BRCA2 Mutations. N. Engl. J. Med. 2002, 346, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Eisen, A.; Lubinski, J.; Klijn, J.; Moller, P.; Lynch, H.T.; Offit, K.; Weber, B.; Rebbeck, T.; Neuhausen, S.L.; Ghadirian, P.; et al. Breast Cancer Risk Following Bilateral Oophorectomy in BRCA1 and BRCA2 Mutation Carriers: An International Case-Control Study. J. Clin. Oncol. 2005, 23, 7491–7496. [Google Scholar] [CrossRef]
- Armstrong, D.K.N.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef]
- Hodgson, A.; Turashvili, G. Pathology of Hereditary Breast and Ovarian Cancer. Front. Oncol. 2020, 10, 531790. [Google Scholar] [CrossRef] [PubMed]
- Melki, R.; Melloul, M.; Aissaoui, S.; EL Harroudi, T.; Boukhatem, N. Increased prevalence of the founder BRCA1 c.5309G>T and recurrent BRCA2 c.1310_1313delAAGA mutations in breast cancer families from Northerstern region of Morocco: Evidence of geographical specificity and high relevance for genetic counseling. BMC Cancer 2023, 23, 339. [Google Scholar] [CrossRef]
- Frey, M.K.; Finch, A.; Kulkarni, A.; Akbari, M.R.; Chapman-Davis, E. Genetic Testing for All: Overcoming Disparities in Ovarian Cancer Genetic Testing. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 471–482. [Google Scholar] [CrossRef]
- de Sabando, A.R.; Lafuente, E.U.; García-Amigot, F.; Sánchez, A.A.; Garofalo, L.M.; Moreno, S.; Ardanaz, E.; Ramos-Arroyo, M.A. Genetic and clinical characterization of BRCA-associated hereditary breast and ovarian cancer in Navarra (Spain). BMC Cancer 2019, 19, 1145. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.; Lynch, H.T.; Neuhausen, S.L.; Veer, L.V.; Garber, J.E.; Evans, G.R.; Narod, S.A.; Isaacs, C.; Matloff, E.; et al. Bilateral Prophylactic Mastectomy Reduces Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers: The PROSE Study Group. J. Clin. Oncol. 2004, 22, 1055–1062. [Google Scholar] [CrossRef]
- Conte, C.; Pelligra, S.; Sarpietro, G.; Montana, G.D.; Della Corte, L.; Bifulco, G.; Martinelli, C.; Ercoli, A.; Palumbo, M.; Cianci, S. Hereditary Women’s Cancer: Management and Risk-Reducing Surgery. Medicina 2023, 59, 300. [Google Scholar] [CrossRef]
- Godet, I.; Gilkes, D.M. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr. Cancer Sci. Ther. 2017, 4. [Google Scholar] [CrossRef]
- McGuire, K.P.; Arthur, D.W.; Mamounas, E.P. Updates on Management of Hereditary Breast Cancer: New Data on PARP Inhibitors Change Recommendations Regarding the Multidisciplinary Care of Breast Cancer Patients with BRCA Mutations. Ann. Surg. Oncol. 2022, 29, 6504–6507. [Google Scholar] [CrossRef]
- Peddie, N.; Agnew, S.; Crawford, M.; Dixon, D.; MacPherson, I.; Fleming, L. The impact of medication side effects on adherence and persistence to hormone therapy in breast cancer survivors: A qualitative systematic review and thematic synthesis. Breast 2021, 58, 147–159. [Google Scholar] [CrossRef]
- Yar, M.S.; Haider, K.; Gohel, V.; Siddiqui, N.A.; Kamal, A. Synthetic lethality on drug discovery: An update on cancer therapy. Expert Opin. Drug Discov. 2020, 15, 823–832. [Google Scholar] [CrossRef]
- Dong, R.; Ding, T.; Li, Z. Update on poly(ADP-ribose) polymerase inhibitors resistance in ovarian cancer. Front. Pharmacol. 2023, 14, 1164395. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhang, Y.; Meng, H.; Miao, H.; Jiang, Y.; Zhang, L.; Cheng, W. Bractoppin, a BRCA1 carboxy-terminal domain (BRCT) inhibitor, suppresses tumor progression in ovarian borderline tumor organoids. Biochem. Biophys. Res. Commun. 2023, 638, 76–83. [Google Scholar] [CrossRef]
- Bhardwaj, B.K.; Thankachan, S.; Magesh, P.; Venkatesh, T.; Tsutsumi, R.; Suresh, P.S. Current Update on Nanotechnology-Based Approaches in Ovarian Cancer Therapy. Reprod. Sci. 2022, 30, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 2022, 23, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.; Pino, M.A.; Karchin, R.; Beddor, J.; Godinho-Netto, M.; Mesquita, R.D.; Rodarte, R.S.; Vaz, D.C.; Monteiro, V.A.; Manoukian, S.; et al. Analysis of a set of missense, frameshift, and in-frame deletion variants of BRCA1. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2009, 660, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Danishuddin; Khan, S.; Kim, J.J. From cancer big data to treatment: Artificial intelligence in cancer research. J. Gene Med. 2023, 26, e3629. [Google Scholar]



| Mutation | Gene Location | Variant Significance | Effect on BRCT | Reference |
|---|---|---|---|---|
| Lys1702 Thr | c.5105A>C | Unknown | Destabilize the interaction with Ser group of BACH1/CtIP proteins. | [28] |
| Arg1699Pro | c.5096G>C | Likely pathogenic and uncertain significance | Affect residues in the surface cleft of BRCT domain structure. Disrupt interaction with pSer990 in BACH1 protein: a 20-fold lower binding affinity. | [16] |
| Val 1696 Leu | c.5086G>C | Uncertain significance | Affect the interaction between BRCT domain BACH1 protein (50% reductions in BACH1 binding in rat). | [29] |
| Ala1708Glu Met1775Arg | c.5123C>A c.5324T>G | Pathogenic | Ablate the double-strand break repair and transcription function of BRCA1. Destabilize the BRCT fold. Inhibit BRCT interactions with histone deacetylases BACH1, and the transcriptional co-repressor CtIP. | [30] |
| Gly1763Val Leu1786Pro | 5407 G>T 5476 T>C | Pathogenic Pathogenic | These mutations are far away from the interaction site between BRCA1 and its phosphor-ligands, which may at least partially explain why these two mutants have no deleterious effect on BRCA1 function. | [15] |
| Tyr 1853 Asn | c.5558A>G | Pathogenic/Likely pathogenic | Deleterious impact on protein structure and function. Impact phosphopeptide binding | [9] |
| Criteria | Details |
|---|---|
| Young | Cancer diagnosis occurring at a younger age than usual. Breast cancer diagnosed in individuals under the age of 50. |
| Rare | Ovarian cancer, breast cancer in men, and pancreatic cancer among blood relatives. |
| Bilateral | Cancer affecting both organs in a pair, such as both breasts. |
| Multiple |
|
| Familial mutation | An identified genetic mutation in a family member. |
| Ethnic predisposition | Jewish ancestry of Ashkenazi origin. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, T.; Alzneika, S.; Riguene, E.; Al-maraghi, S.; Alabdulrazzak, A.; Al-Khal, N.; Fetais, S.; Thanassoulas, A.; AlFarsi, H.; Nomikos, M. BRCA1 and Its Vulnerable C-Terminal BRCT Domain: Structure, Function, Genetic Mutations and Links to Diagnosis and Treatment of Breast and Ovarian Cancer. Pharmaceuticals 2024, 17, 333. https://doi.org/10.3390/ph17030333
Ismail T, Alzneika S, Riguene E, Al-maraghi S, Alabdulrazzak A, Al-Khal N, Fetais S, Thanassoulas A, AlFarsi H, Nomikos M. BRCA1 and Its Vulnerable C-Terminal BRCT Domain: Structure, Function, Genetic Mutations and Links to Diagnosis and Treatment of Breast and Ovarian Cancer. Pharmaceuticals. 2024; 17(3):333. https://doi.org/10.3390/ph17030333
Chicago/Turabian StyleIsmail, Tala, Safa Alzneika, Emna Riguene, Salwa Al-maraghi, Aya Alabdulrazzak, Noof Al-Khal, Sara Fetais, Angelos Thanassoulas, Halema AlFarsi, and Michail Nomikos. 2024. "BRCA1 and Its Vulnerable C-Terminal BRCT Domain: Structure, Function, Genetic Mutations and Links to Diagnosis and Treatment of Breast and Ovarian Cancer" Pharmaceuticals 17, no. 3: 333. https://doi.org/10.3390/ph17030333
APA StyleIsmail, T., Alzneika, S., Riguene, E., Al-maraghi, S., Alabdulrazzak, A., Al-Khal, N., Fetais, S., Thanassoulas, A., AlFarsi, H., & Nomikos, M. (2024). BRCA1 and Its Vulnerable C-Terminal BRCT Domain: Structure, Function, Genetic Mutations and Links to Diagnosis and Treatment of Breast and Ovarian Cancer. Pharmaceuticals, 17(3), 333. https://doi.org/10.3390/ph17030333








