Immunomodulatory Peptides as Vaccine Adjuvants and Antimicrobial Agents
Abstract
1. Introduction
2. Results
2.1. Retrieval of Cryptic Immunomodulatory Peptides from AMPs in Arthropods
2.2. Identification of Immunomodulatory Peptides with Antimicrobial and Anticancer Properties
2.3. Docking Bifunctional Immunomodulatory Peptides with TLRs
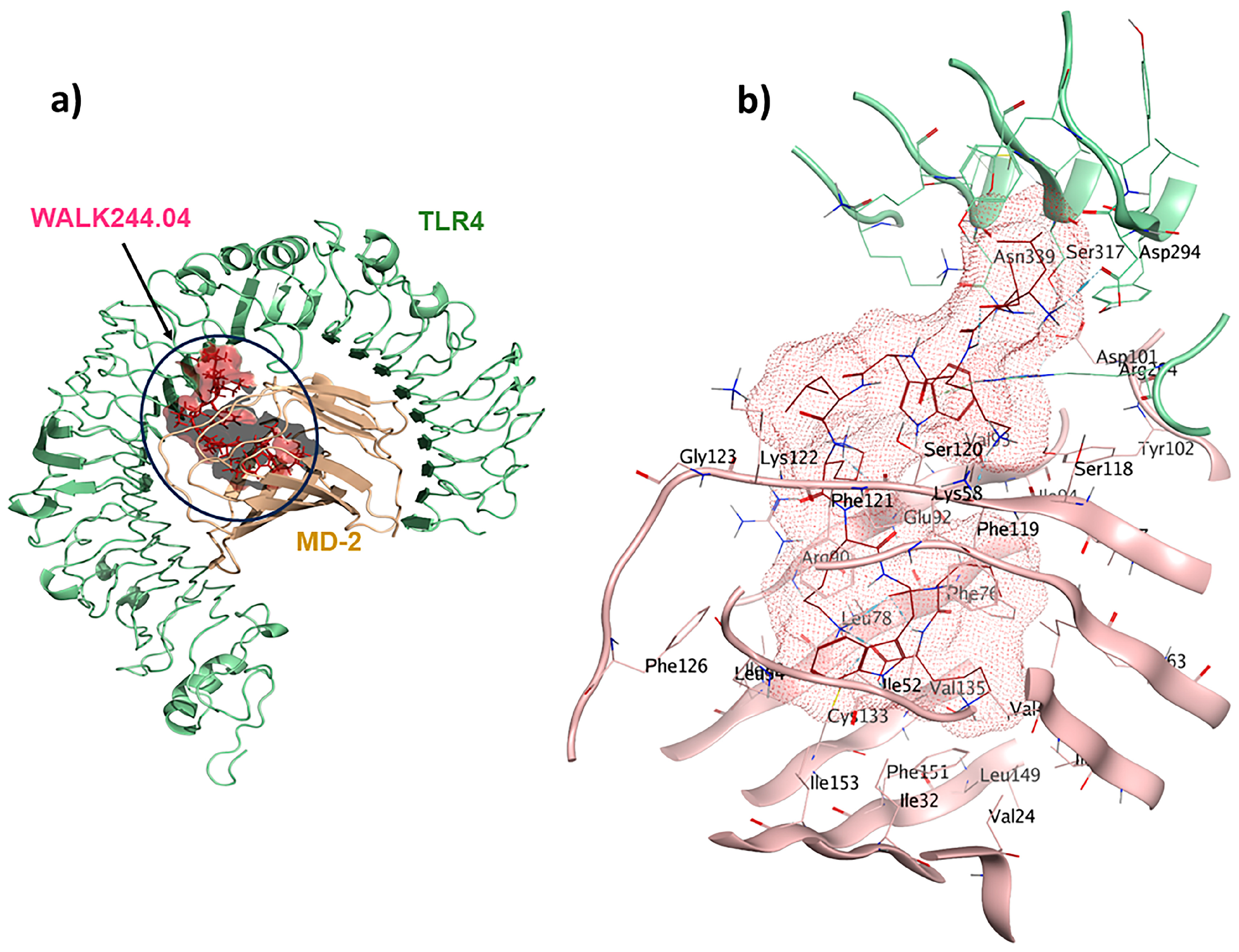
2.4. TLR4/MD2-WALK244.04 Complex as the Positive Control
2.5. TLR1/TLR2-SPalf2-453 as an Antiviral Immunomodulator
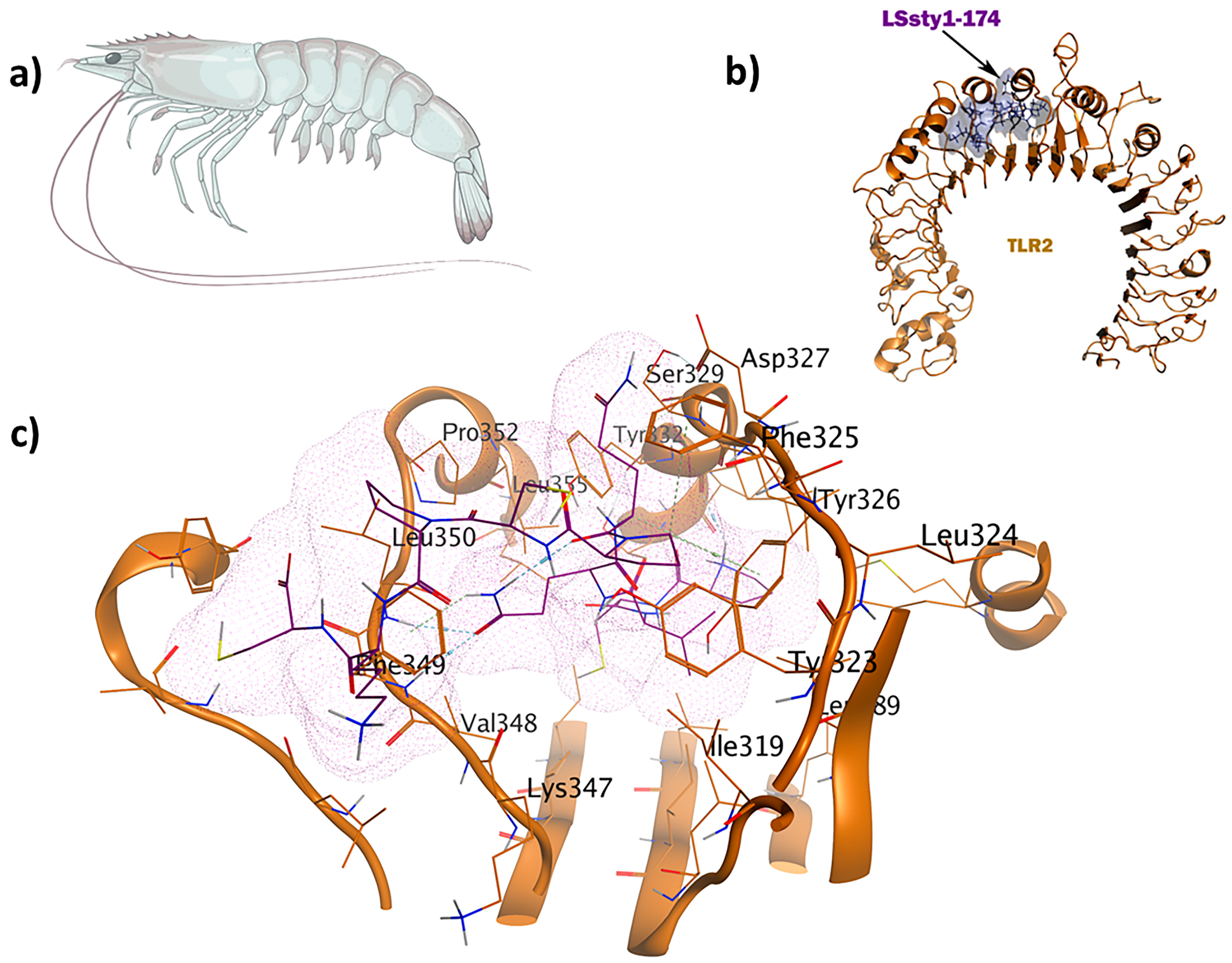
2.6. Antitubercular and Antifungal Immunomodulators
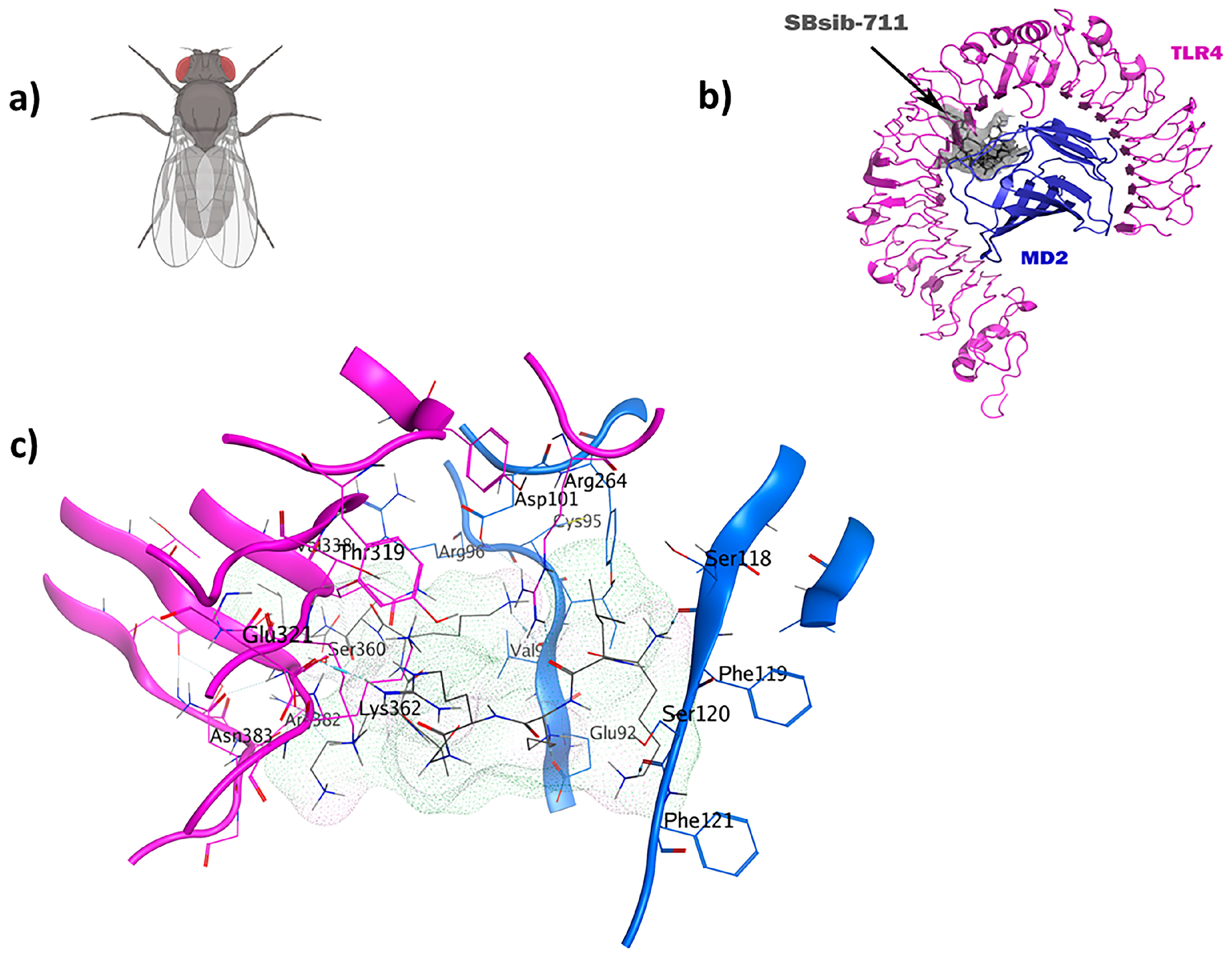
2.7. TLR4/MD2-SBsib-711 as an Anticancer Immunomodulator
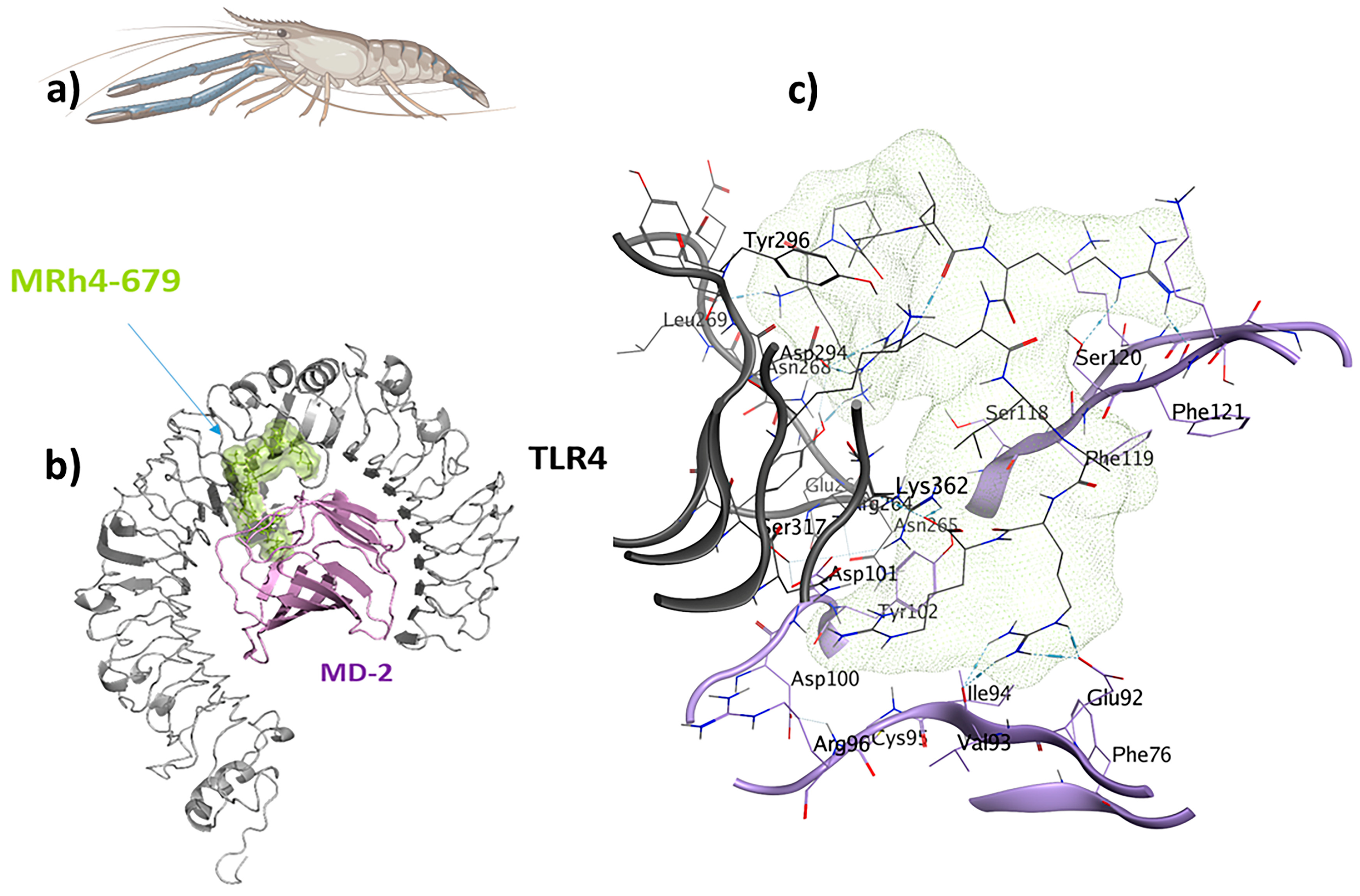
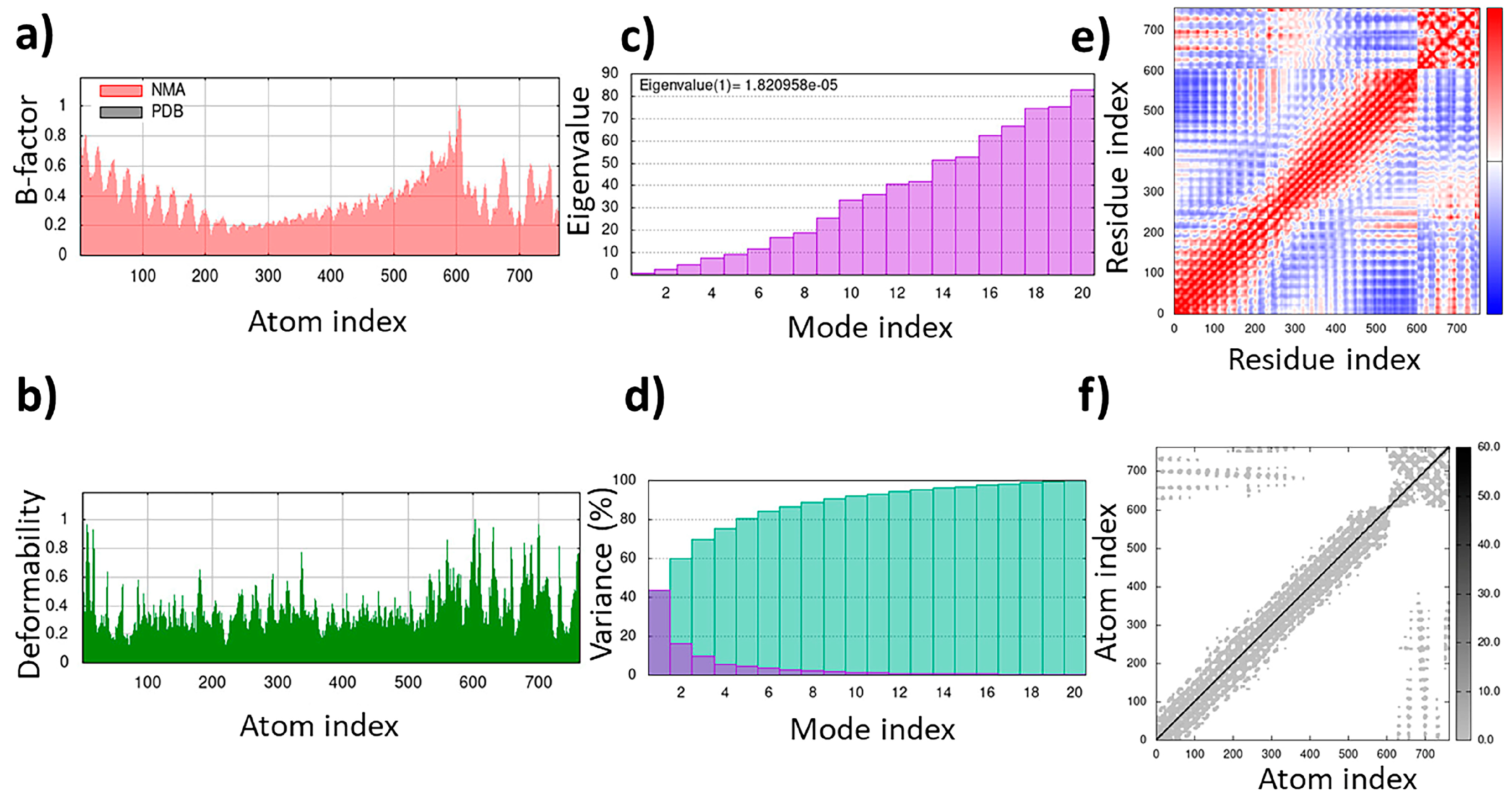
2.8. TLR4/MD2-MRh4-679 Complex as the Universal Immunoadjuvant
2.9. Evaluating the In Vivo Targets of the Identified Immunoadjuvants by Systems Biology
3. Discussion
4. Materials and Methods
4.1. Identification of Immunomodulatory Peptides and Their Biological Functions
4.2. Retrieval of TLR Structures as Receptors
4.3. Receptor and Ligand Preparation
4.4. Molecular Docking and Molecular Dynamics Simulation Studies
4.5. Identification of In Vivo Target Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, C.M.; Ryan, L.K.; Gera, M.; Choudhuri, S.; Lyle, N.; Ali, K.A.; Diamond, G. Antifungals and Drug Resistance. Encyclopedia 2022, 2, 1722–1737. [Google Scholar] [CrossRef]
- Sartelli, M.; Hardcastle, T.C.; Catena, F.; Chichom-Mefire, A.; Coccolini, F.; Dhingra, S.; Haque, M.; Hodonou, A.; Iskandar, K.; Labricciosa, F.M. Antibiotic Use in Low and Middle-Income Countries and the Challenges of Antimicrobial Resistance in Surgery. Antibiotics 2020, 9, 497. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, Adaptive and Acquired Antimicrobial Resistance in Gram-Negative Bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar] [PubMed]
- Lin, J.S.; Bekale, L.A.; Molchanova, N.; Nielsen, J.E.; Wright, M.; Bacacao, B.; Diamond, G.; Jenssen, H.; Santa Maria, P.L.; Barron, A.E. Anti-Persister and Anti-Biofilm Activity of Self-Assembled Antimicrobial Peptoid Ellipsoidal Micelles. ACS Infect. Dis. 2022, 8, 1823–1830. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured Antimicrobial Peptides: The Last Push towards Clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef]
- Ranzani, O.T.; Hitchings, M.D.; Dorion, M.; D’Agostini, T.L.; de Paula, R.C.; de Paula, O.F.P.; de Moura Villela, E.F.; Torres, M.S.S.; de Oliveira, S.B.; Schulz, W. Effectiveness of the CoronaVac Vaccine in Older Adults during a Gamma Variant Associated Epidemic of COVID-19 in Brazil: Test Negative Case-Control Study. BMJ 2021, 374, n2015. [Google Scholar] [CrossRef]
- Reed, S.G.; Orr, M.T.; Fox, C.B. Key Roles of Adjuvants in Modern Vaccines. Nat. Med. 2013, 19, 1597–1608. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, Y.; Li, Y.; Chen, X. Adjuvantation of Influenza Vaccines to Induce Cross-Protective Immunity. Vaccines 2021, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Azmi, F.; Ahmad Fuaad, A.A.H.; Skwarczynski, M.; Toth, I. Recent Progress in Adjuvant Discovery for Peptide-Based Subunit Vaccines. Hum. Vaccines Immunother. 2014, 10, 778–796. [Google Scholar] [CrossRef] [PubMed]
- Singleton, K.L.; Joffe, A.; Leitner, W.W. Current Trends, Challenges, and Success Stories in Adjuvant Research. Front. Immunol. 2023, 14, 1105655. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Vrba, S.M.; Kirk, N.; Liang, Y.; Ly, H. Emerging Concepts and Technologies in Vaccine Development. Front. Immunol. 2020, 11, 583077. [Google Scholar] [CrossRef] [PubMed]
- Marrack, P.; McKee, A.S.; Munks, M.W. Towards an Understanding of the Adjuvant Action of Aluminium. Nat. Rev. Immunol. 2009, 9, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Moni, S.S.; Abdelwahab, S.I.; Jabeen, A.; Elmobark, M.E.; Aqaili, D.; Ghoal, G.; Oraibi, B.; Farasani, A.M.; Jerah, A.A.; Alnajai, M.M.A. Advancements in Vaccine Adjuvants: The Journey from Alum to Nano Formulations. Vaccines 2023, 11, 1704. [Google Scholar] [CrossRef] [PubMed]
- Chatzikleanthous, D.; O’Hagan, D.T.; Adamo, R. Lipid-Based Nanoparticles for Delivery of Vaccine Adjuvants and Antigens: Toward Multicomponent Vaccines. Mol. Pharm. 2021, 18, 2867–2888. [Google Scholar] [CrossRef]
- Hwang, J.S.; Kim, S.G.; Shin, T.H.; Jang, Y.E.; Kwon, D.H.; Lee, G. Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics. Pharmaceutics 2022, 14, 997. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, J. Better Adjuvants for Better Vaccines: Progress in Adjuvant Delivery Systems, Modifications, and Adjuvant-Antigen Codelivery. Vaccines 2020, 8, 128. [Google Scholar] [CrossRef]
- Zhao, T.; Cai, Y.; Jiang, Y.; He, X.; Wei, Y.; Yu, Y.; Tian, X. Vaccine Adjuvants: Mechanisms and Platforms. Signal Transduct. Target. Ther. 2023, 8, 283. [Google Scholar] [CrossRef]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Petrina, M.; Martin, J.; Basta, S. Granulocyte Macrophage Colony-Stimulating Factor Has Come of Age: From a Vaccine Adjuvant to Antiviral Immunotherapy. Cytokine Growth Factor Rev. 2021, 59, 101–110. [Google Scholar] [CrossRef]
- Zupin, L.; Crovella, S. Human Defensins from Antivirals to Vaccine Adjuvants: Rediscovery of the Innate Immunity Arsenal. Protein Pept. Lett. 2022, 29, 121–124. [Google Scholar] [CrossRef]
- Menzel, L.; Chowdhury, H.; Masso-Silva, J.; Ruddick, W.; Falkovsky, K.; Vorona, R.; Malsbary, A.; Cherabuddi, K.; Ryan, L.; DiFranco, K. Potent In Vitro and In Vivo Antifungal Activity of a Small Molecule Host Defense Peptide Mimic through a Membrane-Active Mechanism. Sci. Rep. 2017, 7, 4353. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, B.-H.; Jang, Y.-S. Understanding the Roles of Host Defense Peptides in Immune Modulation: From Antimicrobial Action to Potential as Adjuvants. J. Microbiol. Biotechnol. 2023, 33, 288. [Google Scholar] [CrossRef] [PubMed]
- Temizoz, B.; Kuroda, E.; Ishii, K.J. Vaccine Adjuvants as Potential Cancer Immunotherapeutics. Int. Immunol. 2016, 28, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Q.; Bazin-Lee, H.; Evans, J.T.; Casella, C.R.; Mitchell, T.C. MPL Adjuvant Contains Competitive Antagonists of Human TLR4. Front. Immunol. 2020, 11, 577823. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.C.; Kobinger, G.P. Hypothesis Driven Development of New Adjuvants: Short Peptides as Immunomodulators. Hum. Vaccines Immunother. 2013, 9, 808–811. [Google Scholar] [CrossRef]
- Hemmati, S.; Rasekhi Kazerooni, H. Polypharmacological Cell-Penetrating Peptides from Venomous Marine Animals Based on Immunomodulating, Antimicrobial, and Anticancer Properties. Mar. Drugs 2022, 20, 763. [Google Scholar] [CrossRef]
- Baxter, R.H.; Contet, A.; Krueger, K. Arthropod Innate Immune Systems and Vector-Borne Diseases. Biochemistry 2017, 56, 907–918. [Google Scholar] [CrossRef]
- Ong, G.H.; Lian, B.S.X.; Kawasaki, T.; Kawai, T. Exploration of Pattern Recognition Receptor Agonists as Candidate Adjuvants. Front. Cell. Infect. Microbiol. 2021, 11, 745016. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.A.; Giraldo, P.; Orduz, S. InverPep: A Database of Invertebrate Antimicrobial Peptides. J. Glob. Antimicrob. Resist. 2017, 8, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Kang, T.-B.; Kim, D.-H.; Keum, M.; Lee, S.-H.; Kim, J.-H.; Lee, S.-H.; Kim, J.; Kweon, H.-J.; Park, J.-W. 10-Mer and 9-Mer WALK Peptides with Both Antibacterial and Anti-Inflammatory Activities. Antibiotics 2022, 11, 1588. [Google Scholar] [CrossRef]
- Aiman, S.; Alhamhoom, Y.; Ali, F.; Rahman, N.; Rastrelli, L.; Khan, A.; Ahmed, A.; Khan, A.; Li, C. Multi-Epitope Chimeric Vaccine Design against Emerging Monkeypox Virus via Reverse Vaccinology Techniques-a Bioinformatics and Immunoinformatics Approach. Front. Immunol. 2022, 13, 985450. [Google Scholar] [CrossRef]
- Zaib, S.; Rana, N.; Hussain, N.; Alrbyawi, H.; Dera, A.A.; Khan, I.; Khalid, M.; Khan, A.; Al-Harrasi, A. Designing Multi-Epitope Monkeypox Virus-Specific Vaccine Using Immunoinformatics Approach. J. Infect. Public Health 2023, 16, 107–116. [Google Scholar] [CrossRef]
- Abraham, P.; Jose, L.; Maliekal, T.T.; Kumar, R.A.; Kumar, K.S. B1CTcu5: A Frog-Derived Brevinin-1 Peptide with Anti-Tuberculosis Activity. Peptides 2020, 132, 170373. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, A.; Anisimova, V.; Nikonorova, A.; Babakov, A.; Krause, E.; Bienert, M.; Grishin, E.; Egorov, T. An Antimicrobial Peptide Ar-AMP from Amaranth (Amaranthus retroflexus L.) Seeds. Phytochemistry 2005, 66, 2426–2431. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Li, B.; Chen, X.; Zhang, Q.; Lu, C.; Yang, S.; Long, J.; Ning, L.; Chen, H.; Huang, J. PDL1Binder: Identifying Programmed Cell Death Ligand 1 Binding Peptides by Incorporating next-Generation Phage Display Data and Different Peptide Descriptors. Front. Microbiol. 2022, 13, 928774. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, H.; Huang, J.; He, B. CD47Binder: Identify CD47 Binding Peptides by Combining Next-Generation Phage Display Data and Multiple Peptide Descriptors. Interdiscip. Sci. 2023, 15, 578–589. [Google Scholar] [CrossRef]
- Raeven, R.H.; van Riet, E.; Meiring, H.D.; Metz, B.; Kersten, G.F. Systems Vaccinology and Big Data in the Vaccine Development Chain. Immunology 2019, 156, 33–46. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Vázquez-Maldonado, N.; Kelly, H.R.; Leitner, W.W. Comprehensive Immunoprofiling and Systematic Adjuvant Comparisons for Identifying Suitable Vaccine: Adjuvant Pairings. Hum. Vaccines Immunother. 2023, 19, 2223503. [Google Scholar] [CrossRef]
- Dituri, F.; Gigante, G.; Scialpi, R.; Mancarella, S.; Fabregat, I.; Giannelli, G. Proteoglycans in Cancer: Friends or Enemies? A Special Focus on Hepatocellular Carcinoma. Cancers 2022, 14, 1902. [Google Scholar] [CrossRef]
- Looi, C.-K.; Hii, L.-W.; Ngai, S.C.; Leong, C.-O.; Mai, C.-W. The Role of Ras-Associated Protein 1 (Rap1) in Cancer: Bad Actor or Good Player? Biomedicines 2020, 8, 334. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.; Lam, H.Y.; Yap, K.C.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B. PI3K/AKT/mTOR Signaling Transduction Pathway and Targeted Therapies in Cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Knaut, H. Focal Adhesion-Mediated Cell Anchoring and Migration: From In Vitro to In Vivo. Development 2022, 149, dev200647. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-F.; Yeh, Y.-T.; Su, Y.-C.; Liao, C.-A.; Huang, C.-H.; Cheng, Y.-J.; Jan, J.-S. Cell Adhesion Inhibiting Peptides Exhibit Potent Anticancer Activity and Modulate Intestinal Microbiota. Mater. Des. 2022, 224, 111303. [Google Scholar] [CrossRef]
- Geng, K.; Kumar, S.; Kimani, S.G.; Kholodovych, V.; Kasikara, C.; Mizuno, K.; Sandiford, O.; Rameshwar, P.; Kotenko, S.V.; Birge, R.B. Requirement of Gamma-Carboxyglutamic Acid Modification and Phosphatidylserine Binding for the Activation of Tyro3, Axl, and Mertk Receptors by Growth Arrest-Specific 6. Front. Immunol. 2017, 8, 1521. [Google Scholar] [CrossRef] [PubMed]
- Akalu, Y.T.; Rothlin, C.V.; Ghosh, S. TAM Receptor Tyrosine Kinases as Emerging Targets of Innate Immune Checkpoint Blockade for Cancer Therapy. Immunol. Rev. 2017, 276, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Feuerer, N.; Carvajal Berrio, D.A.; Billing, F.; Segan, S.; Weiss, M.; Rothbauer, U.; Marzi, J.; Schenke-Layland, K. Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in Thp-1-and Pbmc-Derived Macrophages. Biomedicines 2022, 10, 989. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Scott, M.K.; Wimmers, F.; Arunachalam, P.S.; Luo, W.; Fox, C.B.; Tomai, M.; Khatri, P.; Pulendran, B. A Molecular Atlas of Innate Immunity to Adjuvanted and Live Attenuated Vaccines, in Mice. Nat. Commun. 2022, 13, 549. [Google Scholar] [CrossRef]
- Chaudhury, S.; Duncan, E.H.; Atre, T.; Dutta, S.; Spring, M.D.; Leitner, W.W.; Bergmann-Leitner, E.S. Combining Immunoprofiling with Machine Learning to Assess the Effects of Adjuvant Formulation on Human Vaccine-Induced Immunity. Hum. Vaccines Immunother. 2020, 16, 400–411. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C. Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Brubaker, D.K.; Lauffenburger, D.A. Translating Preclinical Models to Humans. Science 2020, 367, 742–743. [Google Scholar] [CrossRef]
- Brubaker, D.K.; Proctor, E.A.; Haigis, K.M.; Lauffenburger, D.A. Computational Translation of Genomic Responses from Experimental Model Systems to Humans. PLoS Comput. Biol. 2019, 15, e1006286. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.C.; Maasch, J.R.; de la Fuente-Nunez, C. Accelerating Antibiotic Discovery through Artificial Intelligence. Commun. Biol. 2021, 4, 1050. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Puga, M.D.C.; Cancelarich, N.L.; Marani, M.M.; de la Fuente-Nunez, C.; Plisson, F. Accelerating the Discovery and Design of Antimicrobial Peptides with Artificial Intelligence. In Computational Drug Discovery and Design; Humana: New York, NY, USA, 2024; Volume 2714. [Google Scholar]
- Szymczak, P.; Szczurek, E. Artificial Intelligence-Driven Antimicrobial Peptide Discovery. Curr. Opin. Struct. Biol. 2023, 83, 102733. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Ovchinnikova, T.V. Immunomodulatory and Allergenic Properties of Antimicrobial Peptides. Int. J. Mol. Sci. 2022, 23, 2499. [Google Scholar] [CrossRef] [PubMed]
- Luchner, M.; Reinke, S.; Milicic, A. TLR Agonists as Vaccine Adjuvants Targeting Cancer and Infectious Diseases. Pharmaceutics 2021, 13, 142. [Google Scholar] [CrossRef]
- Behzadipour, Y.; Gholampour, M.; Pirhadi, S.; Seradj, H.; Khoshneviszadeh, M.; Hemmati, S. Viral 3CLpro as a Target for Antiviral Intervention Using Milk-Derived Bioactive Peptides. Int. J. Pept. Res. Ther. 2021, 27, 2703–2716. [Google Scholar] [CrossRef]
- Kaur, A.; Kaushik, D.; Piplani, S.; Mehta, S.K.; Petrovsky, N.; Salunke, D.B. TLR2 Agonistic Small Molecules: Detailed Structure-Activity Relationship, Applications, and Future Prospects. J. Med. Chem. 2021, 64, 233–278. [Google Scholar] [CrossRef]
- Shepardson, K.M.; Schwarz, B.; Larson, K.; Morton, R.V.; Avera, J.; McCoy, K.; Caffrey, A.; Harmsen, A.; Douglas, T.; Rynda-Apple, A. Induction of Antiviral Immune Response through Recognition of the Repeating Subunit Pattern of Viral Capsids Is Toll-Like Receptor 2 Dependent. mBio 2017, 8, e01356-17. [Google Scholar] [CrossRef]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef]
- Shukla, N.M.; Chan, M.; Hayashi, T.; Carson, D.A.; Cottam, H.B. Recent Advances and Perspectives in Small-Molecule TLR Ligands and Their Modulators. ACS Med. Chem. Lett. 2018, 9, 1156–1159. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, L.; Zhou, H.; Qi, Y. A Meta-Analysis of Th1 and Th2 Cytokine Profiles Differentiating Tuberculous from Malignant Pleural Effusion. Sci. Rep. 2022, 12, 2743. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, X.; Jiang, S.; Lu, L. Immunoengineered Adjuvants for Universal Vaccines against Respiratory Viruses. Fundam. Res. 2021, 1, 189–192. [Google Scholar] [CrossRef]
- Hjertner, B.; Bengtsson, T.; Morein, B.; Paulie, S.; Fossum, C. A Novel Adjuvant G3 Induces Both Th1 and Th2 Related Immune Responses in Mice after Immunization with a Trivalent Inactivated Split-Virion Influenza Vaccine. Vaccine 2018, 36, 3340–3344. [Google Scholar] [CrossRef] [PubMed]
- Behzadipour, Y.; Hemmati, S. Considerations on the Rational Design of Covalently Conjugated Cell-Penetrating Peptides (CPPs) for Intracellular Delivery of Proteins: A Guide to CPP Selection Using Glucarpidase as the Model Cargo Molecule. Molecules 2019, 24, 4318. [Google Scholar] [CrossRef] [PubMed]
- Sadeghian, I.; Khalvati, B.; Ghasemi, Y.; Hemmati, S. TAT-Mediated Intracellular Delivery of Carboxypeptidase G2 Protects against Methotrexate-Induced Cell Death in HepG2 Cells. Toxicol. Appl. Pharmacol. 2018, 346, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Behzadipour, Y.; Sadeghian, I.; Ghaffarian Bahraman, A.; Hemmati, S. Introducing a Delivery System for Melanogenesis Inhibition in Melanoma B16F10 Cells Mediated by the Conjugation of Tyrosine Ammonia-lyase and a TAT-penetrating Peptide. Biotechnol. Prog. 2021, 37, e3071. [Google Scholar] [CrossRef]
- Hemmati, S.; Behzadipour, Y.; Haddad, M. Decoding the Proteome of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) for Cell-Penetrating Peptides Involved in Pathogenesis or Applicable as Drug Delivery Vectors. Infect. Genet. Evol. 2020, 85, 104474. [Google Scholar] [CrossRef]
- Seil, M.; Nagant, C.; Dehaye, J.-P.; Vandenbranden, M.; Lensink, M.F. Spotlight on Human LL-37, an Immunomodulatory Peptide with Promising Cell-Penetrating Properties. Pharmaceuticals 2010, 3, 3435–3460. [Google Scholar] [CrossRef]
- Behzadipour, Y.; Hemmati, S. Viral Prefusion Targeting Using Entry Inhibitor Peptides: The Case of SARS-CoV-2 and Influenza A Virus. Int. J. Pept. Res. Ther. 2022, 28, 42. [Google Scholar] [CrossRef] [PubMed]
- Backlund, C.M.; Holden, R.L.; Moynihan, K.D.; Garafola, D.; Farquhar, C.; Mehta, N.K.; Maiorino, L.; Pham, S.; Iorgulescu, J.B.; Reardon, D.A. Cell-Penetrating Peptides Enhance Peptide Vaccine Accumulation and Persistence in Lymph Nodes to Drive Immunogenicity. Proc. Natl. Acad. Sci. USA 2022, 119, e2204078119. [Google Scholar] [CrossRef] [PubMed]
- von Both, U.; Berk, M.; Agapow, P.-M.; Wright, J.D.; Git, A.; Hamilton, M.S.; Goldgof, G.; Siddiqui, N.; Bellos, E.; Wright, V.J. Mycobacterium tuberculosis Exploits a Molecular off Switch of the Immune System for Intracellular Survival. Sci. Rep. 2018, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Horvati, K.; Fodor, K.; Pályi, B.; Henczko, J.; Balka, G.; Gyulai, G.; Kiss, É.; Biri-Kovacs, B.; Senoner, Z.; Bősze, S. Novel Assay Platform to Evaluate Intracellular Killing of Mycobacterium tuberculosis: In Vitro and In Vivo Validation. Front. Immunol. 2021, 12, 750496. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Sundling, C.; Folkesson, E.; Fröberg, G.; Nobrega, C.; Canto-Gomes, J.; Chambers, B.J.; Lakshmikanth, T.; Brodin, P.; Bruchfeld, J. High Dimensional Immune Profiling Reveals Different Response Patterns in Active and Latent Tuberculosis following Stimulation with Mycobacterial Glycolipids. Front. Immunol. 2021, 12, 727300. [Google Scholar] [CrossRef]
- Hu, W.; Spaink, H.P. The Role of TLR2 in Infectious Diseases Caused by Mycobacteria: From Cell Biology to Therapeutic Target. Biology 2022, 11, 246. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, F.M.; van Vliet, A.H.; Lawton, S.P.; Betson, M. In Silico Design of a Polypeptide as a Vaccine Candidate against Ascariasis. Sci. Rep. 2023, 13, 3504. [Google Scholar] [CrossRef]
- Diamond, G. Antifungal Peptides. J. Fungi 2021, 7, 437. [Google Scholar] [CrossRef]
- Ghanbarzadeh, Z.; Hemmati, S.; Mohagheghzadeh, A. Humanizing Plant-Derived Snakins and Their Encrypted Antimicrobial Peptides. Biochimie 2022, 199, 92–111. [Google Scholar] [CrossRef]
- Traini, G.; Ruiz-de-Angulo, A.; Blanco-Canosa, J.B.; Zamacola Bascaran, K.; Molinaro, A.; Silipo, A.; Escors, D.; Mareque-Rivas, J.C. Cancer Immunotherapy of TLR4 Agonist–Antigen Constructs Enhanced with Pathogen-Mimicking Magnetite Nanoparticles and Checkpoint Blockade of PD-L1. Small 2019, 15, 1803993. [Google Scholar] [CrossRef]
- Yang, D.; Luo, X.; Lian, Q.; Gao, L.; Wang, C.; Qi, X.; Zhang, R.; Liu, Z.; Liao, G. Fully Synthetic Tn-Based Three-Component Cancer Vaccine Using Covalently Linked TLR4 Ligand MPLA and iNKT Cell Agonist KRN-7000 as Built-in Adjuvant Effectively Protects Mice from Tumor Development. Acta Pharm. Sin. B 2022, 12, 4432–4445. [Google Scholar] [CrossRef] [PubMed]
- Pilon, C.; Subang, R.; Lonina, E.; Lesage, S.; Qureshi, S.T.; Charette, E.; Fonseca, G.; Levine, J.S.; Rauch, J. Type I Interferon-Dependent and-Independent TRIF Signaling Are Required for Autoantibody Generation in an Induced Model of Systemic Lupus Erythematosus. J. Immunol. 2023, 210, 78.04. [Google Scholar] [CrossRef]
- Hadianamrei, R.; Tomeh, M.A.; Brown, S.; Wang, J.; Zhao, X. Rationally Designed Short Cationic α-Helical Peptides with Selective Anticancer Activity. J. Colloid Interface Sci. 2022, 607, 488–501. [Google Scholar] [CrossRef]
- Guo, Y.; Yang, Y.; Huang, Y.; Shen, H.-B. Discovering Nuclear Targeting Signal Sequence through Protein Language Learning and Multivariate Analysis. Anal. Biochem. 2020, 591, 113565. [Google Scholar] [CrossRef]
- Mukherjee, N.; Bhunia, D.; Garai, P.K.; Mondal, P.; Barman, S.; Ghosh, S. Designed Novel Nuclear Localizing Anticancer Peptide Targets P53 Negative Regulator MDM2 Protein. J. Pept. Sci. 2023, 30, e3535. [Google Scholar] [CrossRef]
- Li, Z.; Wu, X.; Zhao, Y.; Xiao, Y.; Zhao, Y.; Zhang, T.; Li, H.; Sha, F.; Wang, Y.; Deng, L. Clinical Benefit of Neoadjuvant anti-PD-1/PD-L1 Utilization among Different Tumors. MedComm 2021, 2, 60–68. [Google Scholar] [CrossRef]
- Yang, H.; Xun, Y.; You, H. The Landscape Overview of CD47-Based Immunotherapy for Hematological Malignancies. Biomark. Res. 2023, 11, 15. [Google Scholar] [CrossRef]
- Majidpoor, J.; Mortezaee, K. Interleukin-2 Therapy of Cancer-Clinical Perspectives. Int. Immunopharmacol. 2021, 98, 107836. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zhu, H.; Xia, X.; Liang, Z.; Ma, X.; Sun, B. Vaccine Adjuvants: Understanding the Structure and Mechanism of Adjuvanticity. Vaccine 2019, 37, 3167–3178. [Google Scholar] [CrossRef]
- Knudsen, N.P.H.; Olsen, A.; Buonsanti, C.; Follmann, F.; Zhang, Y.; Coler, R.N.; Fox, C.B.; Meinke, A.; D’Oro, U.; Casini, D. Different Human Vaccine Adjuvants Promote Distinct Antigen-Independent Immunological Signatures Tailored to Different Pathogens. Sci. Rep. 2016, 6, 19570. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, P.; Mirmohammadi, M.; Tehrani, M.H.H. Anticancer Peptides Mechanisms, Simple and Complex. Chem. Biol. Interact. 2022, 368, 110194. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vaisman, I.I.; van Hoek, M.L. Machine Learning Prediction of Antimicrobial Peptides. In Computational Peptide Science: Methods and Protocols; Humana: New York, NY, USA, 2022; Volume 2405. [Google Scholar]
- Sidorczuk, K.; Gagat, P.; Pietluch, F.; Kała, J.; Rafacz, D.; Bąkała, L.; Słowik, J.; Kolenda, R.; Rödiger, S.; Fingerhut, L.C. Benchmarks in Antimicrobial Peptide Prediction Are Biased due to the Selection of Negative Data. Brief. Bioinform. 2022, 23, bbac343. [Google Scholar] [CrossRef] [PubMed]
- Arabnia, H.R.; Daimi, K.; Stahlbock, R.; Soviany, C.; Heilig, L.; Brüssau, K. Principles of Data Science; Springer: Cham, Switzerland, 2020; ISBN 3-030-43980-1. [Google Scholar]
- Ryan, L.K.; Freeman, K.B.; Masso-Silva, J.A.; Falkovsky, K.; Aloyouny, A.; Markowitz, K.; Hise, A.G.; Fatahzadeh, M.; Scott, R.W.; Diamond, G. Activity of Potent and Selective Host Defense Peptide Mimetics in Mouse Models of Oral Candidiasis. Antimicrob. Agents Chemother. 2014, 58, 3820–3827. [Google Scholar] [CrossRef] [PubMed]
- Ryan, L.K.; Wu, J.; Schwartz, K.; Yim, S.; Diamond, G. β-Defensins Coordinate in Vivo to Inhibit Bacterial Infections of the Trachea. Vaccines 2018, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Eibl, M.M.; Wolf, H.M. Vaccination in Patients with Primary Immune Deficiency, Secondary Immune Deficiency and Autoimmunity with Immune Regulatory Abnormalities. Immunotherapy 2015, 7, 1273–1292. [Google Scholar] [CrossRef] [PubMed]
- Scheenstra, M.R.; Van Harten, R.M.; Veldhuizen, E.J.; Haagsman, H.P.; Coorens, M. Cathelicidins Modulate TLR-Activation and Inflammation. Front. Immunol. 2020, 11, 1137. [Google Scholar] [CrossRef] [PubMed]
- Hemshekhar, M.; Choi, K.-Y.G.; Mookherjee, N. Host Defense Peptide LL-37-Mediated Chemoattractant Properties, but Not Anti-Inflammatory Cytokine IL-1RA Production, Is Selectively Controlled by Cdc42 Rho GTPase via G Protein-Coupled Receptors and JNK Mitogen-Activated Protein Kinase. Front. Immunol. 2018, 9, 1871. [Google Scholar] [CrossRef] [PubMed]
- Agier, J.; Różalska, S.; Wiktorska, M.; Żelechowska, P.; Pastwińska, J.; Brzezińska-Błaszczyk, E. The RLR/NLR Expression and pro-Inflammatory Activity of Tissue Mast Cells Are Regulated by Cathelicidin LL-37 and Defensin hBD-2. Sci. Rep. 2018, 8, 11750. [Google Scholar] [CrossRef]
- Yang, B.; Good, D.; Mosaiab, T.; Liu, W.; Ni, G.; Kaur, J.; Liu, X.; Jessop, C.; Yang, L.; Fadhil, R. Significance of LL-37 on Immunomodulation and Disease Outcome. BioMed Res. Int. 2020, 2020, 8349712. [Google Scholar] [CrossRef]
- Cheifet, B. Mining Extinct Proteomes for Antimicrobial Peptides. Nat. Biotechnol. 2023, 41, 1201. [Google Scholar] [CrossRef]
- Maasch, J.R.; Torres, M.D.; Melo, M.C.; de la Fuente-Nunez, C. Molecular De-Extinction of Ancient Antimicrobial Peptides Enabled by Machine Learning. Cell Host Microbe 2023, 31, 1260–1274. [Google Scholar] [CrossRef]
- Ali, A.; Khan, A.; Kaushik, A.C.; Wang, Y.; Ali, S.S.; Junaid, M.; Saleem, S.; Cho, W.C.; Mao, X.; Wei, D.-Q. Immunoinformatic and Systems Biology Approaches to Predict and Validate Peptide Vaccines against Epstein–Barr Virus (EBV). Sci. Rep. 2019, 9, 720. [Google Scholar] [CrossRef]
- Nagpal, G.; Chaudhary, K.; Agrawal, P.; Raghava, G.P. Computer-Aided Prediction of Antigen Presenting Cell Modulators for Designing Peptide-Based Vaccine Adjuvants. J. Transl. Med. 2018, 16, 181. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A Server for in Silico Prediction of Allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Consortium, O.S.D.D.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Subramaniyam, S.; Shin, T.H.; Kim, M.O.; Lee, G. Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy. J. Proteome Res. 2018, 17, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Frøslev, P.; Franzyk, H.; Ozgür, B.; Brodin, B.; Kristensen, M. Highly Cationic Cell-Penetrating Peptides Affect the Barrier Integrity and Facilitates Mannitol Permeation in a Human Stem Cell-Based Blood-Brain Barrier Model. Eur. J. Pharm. Sci. 2022, 168, 106054. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Dhall, A.; Patiyal, S.; Raghava, G.P. IL13Pred: A Method for Predicting Immunoregulatory Cytokine IL-13 Inducing Peptides. Comput. Biol. Med. 2022, 143, 105297. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An Updated Model for Predicting Anticancer Peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Bąkała, M.; Słowik, J.; Gagat, P. Cancergram: An Effective Classifier for Differentiating Anticancer from Antimicrobial Peptides. Pharmaceutics 2020, 12, 1045. [Google Scholar] [CrossRef]
- Asami, J.; Shimizu, T. Structural and Functional Understanding of the Toll-like Receptors. Protein Sci. 2021, 30, 761–772. [Google Scholar] [CrossRef]
- Ishida, H.; Asami, J.; Zhang, Z.; Nishizawa, T.; Shigematsu, H.; Ohto, U.; Shimizu, T. Cryo-EM Structures of Toll-like Receptors in Complex with UNC93B1. Nat. Struct. Mol. Biol. 2021, 28, 173–180. [Google Scholar] [CrossRef]
- Barman, N.; De, A.; Paul, J.; Haldar, S.; Bhattacharya, A.; Pal, K. Strategy to Configure Multi-Epitope Recombinant Immunogens with Weightage on Proinflamatory Response Using SARS-CoV-2 Spike Glycoprotein (S-Protein) and RNA-Dependent RNA Polymerase (RdRp) as Model Targets. J. Pure Appl. Microbiol. 2022, 16, 281–295. [Google Scholar] [CrossRef]
- Su, L.; Wang, Y.; Wang, J.; Mifune, Y.; Morin, M.D.; Jones, B.T.; Moresco, E.M.Y.; Boger, D.L.; Beutler, B.; Zhang, H. Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim. J. Med. Chem. 2019, 62, 2938–2949. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein–Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Krivák, R.; Hoksza, D. P2Rank: Machine Learning Based Tool for Rapid and Accurate Prediction of Ligand Binding Sites from Protein Structure. J. Cheminform. 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Rahmatabadi, S.S.; Sadeghian, I.; Ghasemi, Y.; Sakhteman, A.; Hemmati, S. Identification and Characterization of a Sterically Robust Phenylalanine Ammonia-Lyase among 481 Natural Isoforms through Association of In Silico and In Vitro Studies. Enzym. Microb. Technol. 2019, 122, 36–54. [Google Scholar] [CrossRef]
- Rahmatabadi, S.S.; Sadeghian, I.; Nezafat, N.; Negahdaripour, M.; Hajighahramani, N.; Hemmati, S.; Ghasemi, Y. In Silico Investigation of Pullulanase Enzymes from Various Bacillus Species. Curr. Proteom. 2017, 14, 175–185. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Nowotny, J.; Cao, R.; Cheng, J. 3Drefine: An Interactive Web Server for Efficient Protein Structure Refinement. Nucleic Acids Res. 2016, 44, W406–W409. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Kuhn, M.; Szklarczyk, D.; Pletscher-Frankild, S.; Blicher, T.H.; Von Mering, C.; Jensen, L.J.; Bork, P. STITCH 4: Integration of Protein–Chemical Interactions with User Data. Nucleic Acids Res. 2014, 42, D401–D407. [Google Scholar] [CrossRef]






| Class | Order | Family | Genus | Species |
|---|---|---|---|---|
Arachnida | Ixodida | Ixodidae | Ixodes | Ixodes ricinus |
| Ixodes scapularis | ||||
| Ixodes sinensis | ||||
| Rhipicephalus | Rhipicephalus haemaphysaloides | |||
| Rhipicephalus microplus | ||||
| Dermacentor | Dermacentor silvarum | |||
| Araneae | Theraphosidae | Cyriopagopus | Cyriopagopus hainanus | |
| Acanthoscurria | Acanthoscurria gomesiana | |||
| Oxyopidae | Oxyopes | Oxyopes takobius | ||
| Oxyopes kitabensis | ||||
| Zodariidae | Lachesana | Lachesana tarabaevi | ||
| Lycosidae | Hogna | Hogna carolinensis | ||
| Scorpiones | Buthidae | Parabuthus | Parabuthus schlechteri | |
| Olivierus | Olivierus martensii | |||
| Vaejovis | Vaejovis punctatus | |||
| Androctonus | Androctonus australis | |||
| Mesobuthus | Mesobuthus eupeus | |||
| Scorpionidae | Pandinus | Pandinus imperator | ||
Insecta | Hymenoptera | Vespidae | Vespa | Vespa tropica |
| Mischocyttarus | Mischocyttarus phthisicus | |||
| Eumenes | Eumenes magnifica | |||
| Formicidae | Neoponera | Neoponera goeldii | ||
| Pteromalidae | Pteromalus | Pteromalus puparum | ||
| Melittidae | Macropis | Macropis fulvipes | ||
| Lepidoptera | Saturniidae | Hyalophora | Hyalophora cecropia | |
| Antheraea | Antheraea pernyi | |||
| Noctuidae | Chloridea | Chloridea virescens | ||
| Psychidae | Oiketicus | Oiketicus kirbyi | ||
| Pyralidae | Galleria | Galleria mellonella | ||
| Bombycidae | Bombyx | Bombyx mori | ||
| Sphingidae | Manduca | Manduca sexta | ||
| Erebidae | Hyphantria | Hyphantria cunea | ||
| Diptera | Calliphoridae | Calliphora | Calliphora vicina | |
| Lucilia | Lucilia sericata | |||
| Tephritidae | Ceratitis | Ceratitis capitata | ||
| Bactrocera | Bactrocera dorsalis | |||
| Simuliidae | Simulium | Simulium bannaense | ||
| Drosophilidae | Drosophila | Drosophila melanogaster | ||
| Hemiptera | Cicadidae | Cryptotympana | Cryptotympana dubia | |
| Cicada | Cicada flammata | |||
| Pentatomidae | Podisus | Podisus maculiventris | ||
| Coleoptera | Cerambycidae | Acalolepta | Acalolepta luxuriosa | |
| Chrysomelidae | Chrysomelinae | Chrysomelinae atrocyanea | ||
| Blattodea | Blattidae | Periplaneta | Periplaneta americana | |
| Orthoptera | Acrididae | Locusta | Locusta migratoria | |
Malacostraca | Decapoda | Portunidae | Scylla | Scylla paramamosain |
| Scylla serrata | ||||
| Portunus | Portunus trituberculatus | |||
| Callinectes | Callinectes sapidus | |||
| Penaeidae | Litopenaeus | Litopenaeus vannamei | ||
| Litopenaeus stylirostris | ||||
| Astacidae | Pacifastacus | Pacifastacus leniusculus | ||
| Palaemonidae | Macrobrachium | Macrobrachium rosenbergii | ||
| Oregoniidae | Hyas | Hyas araneus | ||
| Merostomata | Xiphosura | Limulidae | Limulus | Limulus polyphemus |
| Chilopoda | Scolopendromorpha | Scolopendridae | Scolopendra | Scolopendra subspinipes |
| Peptide ID | Sequence | pI | Charge | GRAVY | CPP | BBBp | IL-4 | IL-10 | IL-13 | IL-2 | IL-6 | TNFα | IFN-Ɣ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Universal IM | MRh4-679 | KPAIRRLARR | 12.48 | +5 | −1.16 | ✓ | ✓ | X | X | ✓ 0.78 | ✓ 0.95 | ✓ 0.35 | X | ✓ |
| Antiviral IM | SPalf2-453 | HIRRRPKFRK | 12.49 | +6 | −2.33 | ✓ | ✓ | ✓ 0.30 | X | ✓ 0.30 | ✓ 0.75 | ✓ 0.25 | ✓ 0.56 | ✓ |
| Antifungal IM | LSsty1-174 | PCVQQPCPKC | 8.26 | +1 | −0.40 | X | ✓ | ✓ 0.28 | X | ✓ 0.28 | ✓ 0.85 | ✓ 0.38 | ✓ 0.55 | X |
| Antitubercular IM | PPpp113-266 | RVQERRFKRI | 12.01 | +4 | −1.74 | ✓ | ✓ | ✓ 1.30 | ✓ 0.60 | ✓ 0.06 | ✓ 0.93 | ✓ 0.30 | ✓ 0.58 | X |
| Anticancer IM | SBsib-711 | KLKRGAKKAL | 11.34 | +5 | −0.93 | ✓ | ✓ | X | X | ✓ 0.83 | ✓ 0.87 | ✓ 0.35 | ✓ 0.64 | X |
| Ligand | Receptor Interacting Amino Acids | Type of Interaction | Location of Interaction | Distance (Å) | Binding Energy (kcal/mol) |
|---|---|---|---|---|---|
| N 1 | LEU 269 | H-donor | TLR4 (Chain B) | 3.20 | −2.1 |
| NZ 7 | GLU 266 | H-donor | TLR4 (Chain B) | 2.94 | −5.5 |
| NE 73 | SER 120 | H-donor | MD-2 (Chain D) | 3.38 | −0.6 |
| NH2 76 | PHE 121 | H-donor | MD-2 (Chain D) | 3.01 | −1.9 |
| NH1 99 | ASP 294 | H-donor | TLR4 (Chain B) | 3.08 | −2.4 |
| NH2 100 | ASP 294 | H-donor | TLR4 (Chain B) | 3.29 | −3.0 |
| NE 150 | GLU 92 | H-donor | MD-2 (Chain D) | 2.93 | −4.7 |
| NH1 152 | VAL 93 | H-donor | MD-2 (Chain D) | 3.23 | −1.3 |
| NH2 153 | GLU 92 | H-donor | MD-2 (Chain D) | 2.96 | −5.6 |
| NH2 153 | VAL 93 | H-donor | MD-2 (Chain D) | 3.22 | −1.8 |
| OXT 180 | ARG 264 | H-acceptor | TLR4 (Chain B) | 3.02 | −2.4 |
| OXT 180 | LYS 362 | H-acceptor | TLR4 (Chain B) | 3.25 | −2.4 |
| N 1 | ASP 294 | Ionic | TLR4 (Chain B) | 3.39 | −2.3 |
| NH1 99 | ASP 294 | Ionic | TLR4 (Chain B) | 3.2 | −3.3 |
| NH1 99 | ASP 294 | Ionic | TLR4 (Chain B) | 3.08 | −4.0 |
| NH2 100 | ASP 294 | Ionic | TLR4 (Chain B) | 3.26 | −3.0 |
| NH2 100 | ASP 294 | Ionic | TLR4 (Chain B) | 3.29 | −2.8 |
| NE 150 | GLU 92 | Ionic | MD-2 (Chain D) | 2.93 | −4.9 |
| NH2 153 | GLU 92 | Ionic | MD-2 (Chain D) | 2.96 | −4.8 |
| NH1 176 | ASP 101 | Ionic | MD-2 (Chain D) | 3.42 | −2.2 |
| NH2 177 | ASP 101 | Ionic | MD-2 (Chain D) | 3.50 | −1.9 |
| NH2 177 | ASP 101 | Ionic | MD-2 (Chain D) | 3.71 | −1.2 |
| OXT 180 | ARG 264 | Ionic | MD-2 (Chain D) | 3.49 | −1.9 |
| OXT 180 | ARG 264 | Ionic | MD-2 (Chain D) | 3.02 | −4.3 |
| Ligand | Optimal Complex | Number of Interactions | Binding Energy (kcal/mol) |
|---|---|---|---|
| Positive control | TLR4/MD2-WALK244.04 | 10 | −25.2 |
| Universal IM | TLR4/MD2-MRh4-679 | 24 | −70.3 |
| Antiviral IM | TLR1/2-SPalf2-453 | 20 | −72.1 |
| Antifungal IM | TLR2-LSsty1-174 | 7 | −7.6 |
| Antitubercular IM | TLR2-PPpp113-266 | 16 | −49.6 |
| Anticancer IM | TLR4/MD2-SBsib-711 | 12 | −39.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hemmati, S.; Saeidikia, Z.; Seradj, H.; Mohagheghzadeh, A. Immunomodulatory Peptides as Vaccine Adjuvants and Antimicrobial Agents. Pharmaceuticals 2024, 17, 201. https://doi.org/10.3390/ph17020201
Hemmati S, Saeidikia Z, Seradj H, Mohagheghzadeh A. Immunomodulatory Peptides as Vaccine Adjuvants and Antimicrobial Agents. Pharmaceuticals. 2024; 17(2):201. https://doi.org/10.3390/ph17020201
Chicago/Turabian StyleHemmati, Shiva, Zahra Saeidikia, Hassan Seradj, and Abdolali Mohagheghzadeh. 2024. "Immunomodulatory Peptides as Vaccine Adjuvants and Antimicrobial Agents" Pharmaceuticals 17, no. 2: 201. https://doi.org/10.3390/ph17020201
APA StyleHemmati, S., Saeidikia, Z., Seradj, H., & Mohagheghzadeh, A. (2024). Immunomodulatory Peptides as Vaccine Adjuvants and Antimicrobial Agents. Pharmaceuticals, 17(2), 201. https://doi.org/10.3390/ph17020201





