Abstract
Background/Objectives: Four years after the COVID-19 pandemic, a very limited number of drugs has been marketed; thus, the search for new medications still represents a compelling need. In our previous work on antiviral, antiparasitic, and antiproliferative agents, we described several compounds (1–13 and 16–20) structurally related to clofazimine, chloroquine, and benzimidazole derivatives. Thus, we deemed it worthwhile to test them against the replication of SARS-CoV-2, together with a few other compounds (14, 15 and 21–25), which showed some analogy to miscellaneous anti-coronavirus agents. Methods: Twenty-five structurally assorted compounds were evaluated in vitro for cytotoxicity against Vero E6 and for their ability to inhibit SARS-CoV-2 replication. Results: Several compounds (2, 3, 10, 11, 13–15, 18–20) demonstrated antiviral activity (IC50 range 1.5–28 µM) and six of them exhibited an interesting selectivity index in the range 4.5–20. The chloroquine analogs 10 and 11 were more potent than the reference chloroquine itself and doubled its SI value (20 versus 11). Also, the benzimidazole ring emerged as a valuable scaffold, originating several compounds (13–15 and 18–20) endowed with anti-SARS-CoV-2 activity. Despite the modest activity, the cytisine and the arylamino enone derivatives 23 and 25, respectively, also deserve further consideration as model compounds. Conclusions: The investigated chemotypes may represent valuable hit compounds, deserving further in-depth biological studies to define their mechanisms of action. The derived information will guide the subsequent chemical optimization towards the development of more efficient anti-SARS-CoV-2 agents.
1. Introduction
COVID-19 is an acute respiratory distress syndrome caused by SARS-CoV-2 (wild and mutated strains), which is an enveloped positive-sense, single-stranded RNA β-coronavirus. The disease may be fatal, and indeed, from the onset of the pandemic (November 2019) to the present day (autumn 2024), more than 770 million cases have occurred worldwide, with about 7 million deaths, according to WHO [1].
Many drug treatments have been attempted, but commonly with disappointing or, at the best, contradictory results [2,3]. In fact, even compounds that demonstrated very strong activity against the virus in in vitro assays have shown limited utility in clinical settings, and the reasons for their failure (ADME characteristics, rapid resistance onset, toxicity, etc.) are not fully elucidated [4,5].
At present, only a few known antiviral agents (structurally complex and rather expensive) have been approved for clinical use, even if with many limitations, such as remdesivir, Paxlovid (an association of nirmatrelvir and ritonavir), and molnupiravir (Figure 1A) [3]. Remdesivir was approved by the American FDA in 2020, although data regarding its ability to reduce mortality and length of hospitalization remain controversial [6,7,8]. Similarly, molnupiravir was approved in 2021, but in 2023 was declared to have no beneficial effects by EMA (24 February 2023) [9].
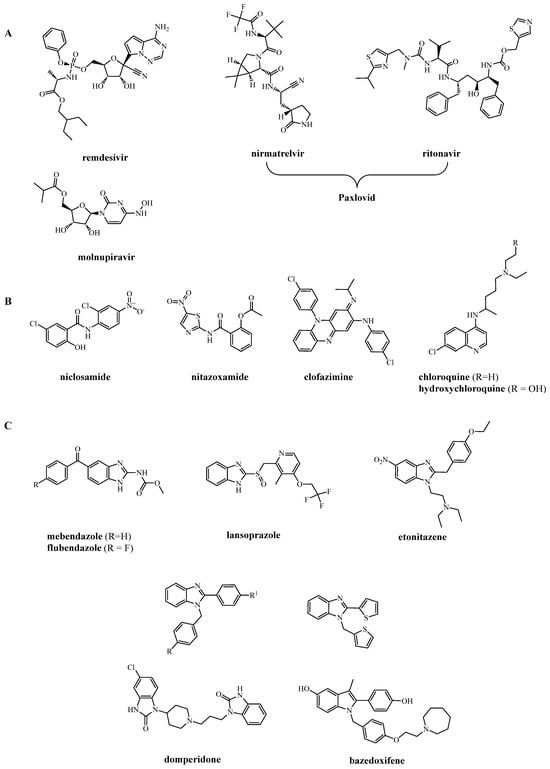
Figure 1.
(A) American FDA- and European EMA-approved drugs for COVID-19 therapy; (B) largely used drugs even if not approved by regulatory agencies; (C) examples of benzimidazole (and similar rings) derivatives active against SARS-CoV-2.
Besides the cited repurposed antiviral drugs (eventually associated with anti-inflammatory agents to alleviate symptoms and slow disease progression), a large number of natural and synthetic compounds have been tested against the virus and/or some of its functional components, leading to the definition of the most promising targets to be hit, such as SARS-CoV-2 main protease (Mpro), pre-fusion spike protein, and papain-like protease (PL) proteins [10,11,12].
From a library of more than 3000 FDA-approved drugs, niclosamide, an old drug for tapeworm infection, emerged as the most potent inhibitor (IC50 = 0.28 µM) [13]. Niclosamide, along with clofazimine (an antibiotic used for the treatment of leprosy and drug-resistant tuberculosis), showed the most effective inhibition of spike-induced TMEM-16 activation (Figure 2), which leads to syncytia formation and thrombotic events [14,15]. Despite its antiviral potency, due to questionable bioavailability issues, niclosamide has currently been replaced by nitazoxanide (Figure 1B), an analogous salicylamide derivative. Recently, niclosamide has been reappraised as an anti-SARS-CoV-2 drug by restricting entry protein CD147 [16] and possessing effective anti-inflammatory activity in the respiratory tract of mice [17] at concentrations comparable to plasma concentrations attainable after oral administration.
During the early management of the pandemic, chloroquine (CQ) and hydroxychloroquine (well-known agents for treating malaria and rheumatoid arthritis) were largely used on the basis of an already known general and potent antiviral activity [18,19,20] (Figure 1B and Figure 2). Against SARS-CoV-2, they exhibited IC50 = 5.47 and 0.72 µM, respectively, but, unfortunately, these molecules provided limited to no therapeutic benefit in COVID-19 treatment because effective concentrations were not reached in vivo [21,22]. However, we feel it is still possible that some analogs of CQ may retain antiviral activity with ADME properties more appropriate for clinical use.
From an extensive screening of more than 12,000 compounds, examined by different physico-chemical and biological points of view, the benzimidazole nucleus resulted in a privileged structure for developing agents against SARS-CoV-2 [23,24,25,26] (Figure 2). Indeed, antimycotics such as mebendazole and flubendazole, proton pump inhibitors such as lansoprazole and analogs, analgesics such as etonitazene, and a miscellanea of 2-aryl-1-arylmethylbenzimidazoles (Figure 1C) exhibit potent activity against SARS-CoV-2 proteases [27,28] and spike proteins [23,29]. It is worth noting the high potency of the D2 antagonist domperidone (used as an antiemetic) and that of the estrogen receptor modulator bazedoxifene (used as an anti-osteoporotic). Although domperidone is a benzimidazolone [30] and bazedoxifene an indole [29] derivative, both possess a molecular geometry very close to that of benzimidazoles.
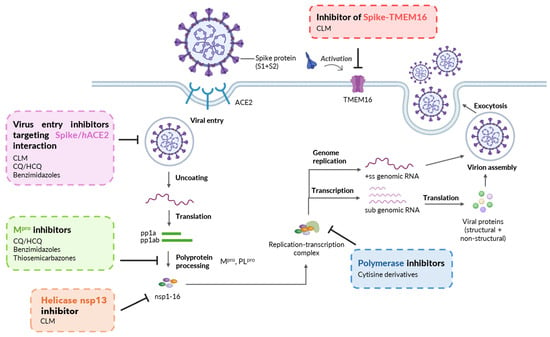
Figure 2.
Mechanisms of SARS-CoV-2 inhibition showed by prototype drugs/molecules, which are structurally related to the chemotypes here investigated. Virus entry inhibitors: CLM [15], CQ/HCQ [19], Benzimidazoles [23,29]; Mpro inhibitors: CQ/HCQ [20], Benzimidazoles [27,28], Thiosemicarbazones [31,32]; Helicase inhibitor: CLM [15]; Polymerase inhibitors: Cytisine derivatives [33]; Inhibitor of Spike-TMEM16 activation: CLM [14]. Created with Biorender.com.
2. Results and Discussion
2.1. Design of the Study
In our previous work on antiviral, antiparasitic, and antiproliferative agents, we described several compounds (1–13 and 16–20) structurally related to the above cited clofazimine, chloroquine, and benzimidazole derivatives. Thus, we deemed it worthwhile to test them against the replication of SARS-CoV-2, together with a few other compounds (14, 15 and 21–25), which showed some analogy to miscellaneous anti-coronavirus agents (Figure 3, Figure 4 and Figure 5).
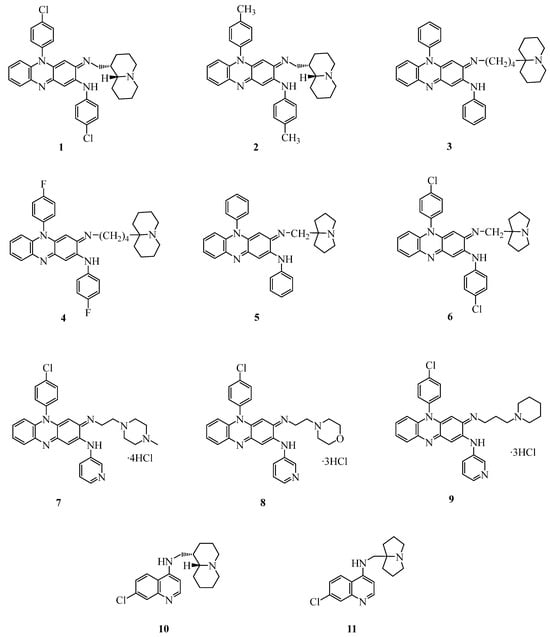
Figure 3.
Structures of clofazimine and chloroquine analogs investigated as anti-SARS-CoV-2 agents.
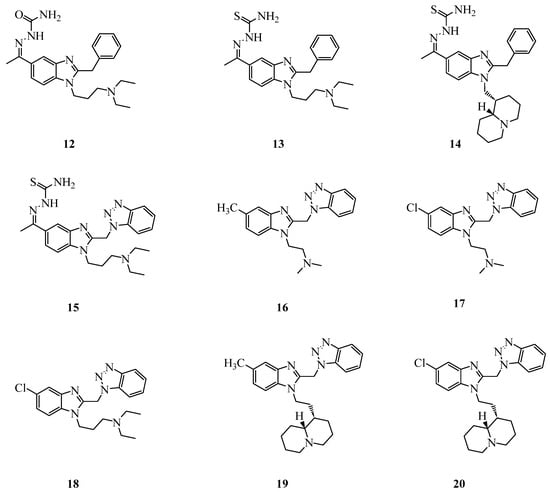
Figure 4.
Structures of benzimidazole derivatives investigated as anti-SARS-CoV-2 agents.
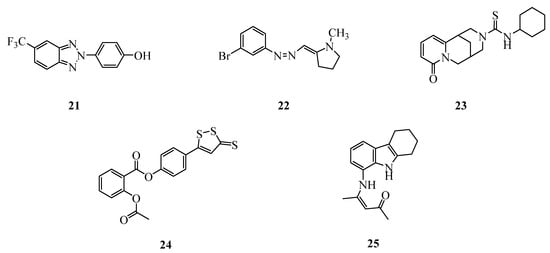
Figure 5.
Structures of miscellaneous compounds investigated as anti-SARS-CoV-2 agents.
In the last ten years, through the introduction of basic moieties in the molecule of clofazimine (CFM), we obtained several CFM analogs with improved activity against Mycobacteria, Plasmodia, Leishmania, and other protozoa [34,35,36]. Among these compounds, a highly water soluble C3-aminopyridinylriminophenazine 7 (known as MU17) was found to also be endowed with improved anti-Wnt and anti-cancer activities (triple-negative breast cancer), and, most notably, to be devoid of drug-related skin coloration [37]. For these valuable characteristics, nine basic clofazimine analogs (1–9) (Figure 3), characterized by different degrees of lipophilicity, were first chosen for testing against SARS-CoV-2.
Through the exchange of the linear basic side chain of CQ with the cumbersome quinolizidinyl- and pyrrolizidinyl-alkyl moieties, we obtained potent anti-plasmodial agents active against CQ-resistant strains of P. falciparum and different species of Leishmania [38,39]. Two of these compounds (10 and 11, Figure 3) displayed favorable preclinical pharmacological profiles [40,41] and, hence, were selected for the present screening.
Among the many benzimidazole derivatives that we investigated as analgesics and antivirals, and somewhat related to the cited etonitazene (Figure 1C), compounds 12–20 (Figure 4) were selected as relevant to the present study. Compounds 12 and 13 exhibited moderate activity against influenza A H1N1 and coronavirus 229E [42], while compounds 16–20 [31,32,43] were active versus several viruses and showed a remarkable potency against RSV, interfering with the F protein-mediated fusion with the host cells [31]. Compounds 14 and 15 are new synthetized compounds that incorporate the thiosemicarbazone Schiff base with quinolizidine (14) or benzotriazole (15) motifs, which have previously shown to provide promising antiviral agents [31,32,43,44]. Thiosemicarbazone-based compounds have recently been demonstrated to inhibit SARS-CoV-2 by targeting Mpro [45,46] (Figure 2).
Finally, to further explore the chemical space important for developing novel anti-SARS-CoV-2 agents, we selected five compounds (21–25, Figure 5) from an in-house library, characterized by very different structures and biological profiles.
Compounds 21 and 22 have previously demonstrated a broad spectrum antiviral activity [44,47], including activity against Dengue virus type 2 (unpublished results, IC50 = 17 and 24 µM, respectively).
Compound 23 is a cytisine derivative that was studied as a nicotinic receptor ligand [48]; it is an analog of the 3-(N-allyl)cytisin thiocarbamide described by Russian authors as a very potent anti-dengue agent with EC50 = 0.1 µM [33]. The same authors have recently shown that other cytisine derivatives are able to inhibit SARS-CoV-2 RNA polymerase (RdRp) (Figure 2) [49].
The acetyl salicylic ester 24 (also known as MZe786) was studied by some of us as a gastric mucosa sparing aspirin analog [50]. The salicylic residue is also present in the cited nitazoxanide and niclosamide (Figure 1), potently active against SARS-CoV-2 [2,13].
Lastly, compound 25 (also known as FSAS-3), firstly prepared in 1968 by one of the authors [51], was subsequently found to possess anti-inflammatory activity. It is worth noting that compound 25 embodies in its structure the tetrahydrocarbazole moiety characteristic of the antiviral agent THC-19 [52], and the 3-phenylamino-2-propen-1-one residue of compound 6877002, a well-known inhibitor of the interaction of CD40/TRAF6 [53] displaying anti-inflammatory and antitumoral activities [54]. Relations between TRAF6 inhibition and anti-SARS-CoV-2 activity are currently under investigation [55,56,57,58].
2.2. Chemistry
Most of the investigated compounds were re-synthesized as described in previous works: 1 and 2 [34], 3–6 [35], 7–9 [36], 10 [38], 11 [39], 12 and 13 [42], 16 and 19 [32], 17 and 18 [43], 20 [31], 21 [44], 22 [47], 23 [48], 24 [50], and 25 [51].
The novel compounds 14 and 15 were prepared (Scheme 1) by reacting thiosemicarbazide with the relevant 5-acetyl benzimidazole previously described [43,59].
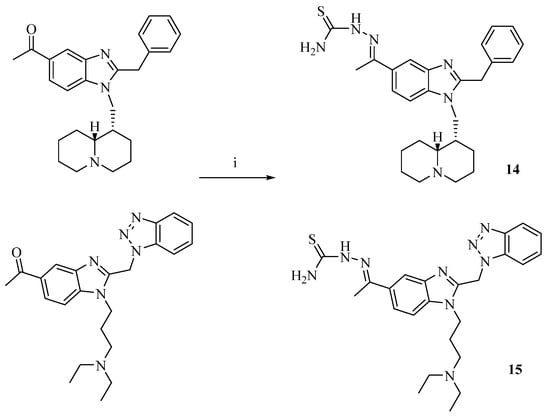
Scheme 1.
Reagents and conditions: (i) thiosemicarbazide in EtOH, H2O and glacial CH3COOH, 3 h at reflux.
The structures of the novel compounds were confirmed using 1H and 13C NMR (see Supplementary Materials), and elemental analysis. The purity of compounds (checked by elemental analysis) was, in all cases, >95%. It is known that thiosemicarbazones can exist in two tautomeric forms due to thione-thiol tautomerism of the thioamide group. In the 1H NMR spectra of the Schiff bases 14 and 15, the signal at 4.00 ppm attributed to the -SH proton is absent, thus suggesting that they adopt the thione form in DMSO [60]. Moreover, they display the secondary -NH- proton signal at 10.16/10.14 ppm, respectively; thus, it is reasonable that they are E isomers (range 9–12 ppm [60]). Finally, the C(S)NH2 protons appear as two signals, both because of the possibility of tautomerism which prevents the free rotation around the N-C bond, and because one of the protons can establish an intramolecular hydrogen bond with the sp2 nitrogen atom (forming a pseudo five-member ring), which would shift the corresponding signal to lower fields [61].
2.3. Biological Studies and SARS-CoV-2
Compounds 1–25 (Figure 3, Figure 4 and Figure 5) were evaluated in vitro for their antiviral activity against SARS-CoV-2 through real-time PCR, which measures the presence of the virus genome in the supernatants of control and treated infected cells. To determine non-cytotoxic doses suitable for use on SARS-CoV-2-infected cells, cytotoxicity against Vero E6 cells was assessed using the MTT assay. Non-toxic concentrations were then selected for antiviral screening against SARS-CoV-2. Results (IC50) are summarized in Table 1, which also includes cytotoxicity against Vero E6 cells (CC50) and the selectivity index (SI = CC50/IC50). Chloroquine was used as the reference drug.

Table 1.
Cytotoxicity (CC50), antiviral activity (IC50), and SI of compounds 1–25 and CQ.
In line with previous observations [34,35,36,37], the riminophenazine derivatives 1–9 exhibited relatively high cytotoxicity (CC50 = 1.74–10.5 µM), higher than that observed for all the other subsets of tested compounds (CC50 = 19–190 µM). On the other hand, different from the reference compound clofazimine (IC50 = 0.31 µM, [15]), these compounds exhibited only a low inhibition of virus replication with the best IC50 values of 1.5 and 4.0 µM for compounds 2 and 3 (SI = 1.9 and 2.6, respectively) (Figure 6). From the data presently available (particularly comparing compounds 3 and 7), the molecular hydrophilicity, which is important to improve the anti-cancer and anti-Wnt activities of riminophenazines [37], seems to play an unfavorable role in anti-SARS-CoV-2 activity. It is worth noting that, in a parallel study [62], contrary to previous data, the inhibition of Wnt signaling by clofazimine and other inhibitors (like 7, MU-17) did not result in a significant reduction in viral RNA load in the lung epithelium cell line used; clofazimine itself only achieved a 25% reduction in the tests. These issues surely deserve more in-depth studies.
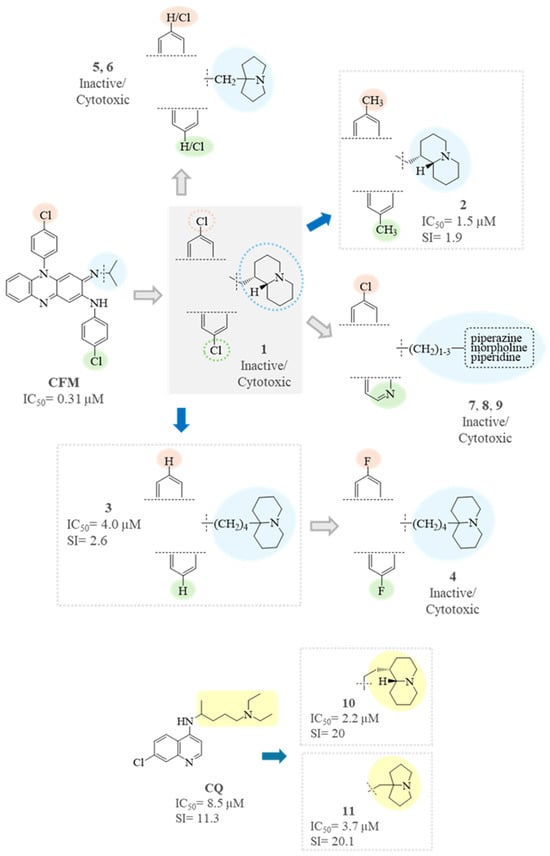
Figure 6.
SAR analysis of CFM analogs (1–9) and CQ analogs (10–11).
Very interesting were the results for the chloroquine analogs 10 and 11, which demonstrated dose-dependent activity and exhibited an SI (SI = 20) that was nearly double that of the reference drug CQ (SI = 11) (Figure 6). Both compounds have been previously studied for their in vitro and in vivo antimalarial activity and ADME-Tox profile [40,41], showing an excellent oral bioavailability with a high volume of distribution, good metabolic stability in microsomes and hepatocytes of different species, and low toxicity in mice (with a maximum tolerated dose, MTD, greater than 100 mg/Kg, i.p.), rats (oral MTD for 11 equal to 120 mg/Kg/day), and non-human primates (oral MTD for 11 equal to 50 mg/Kg/day). These results are in line with or even better than those obtained with chloroquine and are indicative of a good developability profile of the two compounds. Hence, the in-depth investigation of the activity of compounds 10 and 11 against SARS-CoV-2 is warranted, as is, on the other hand, the antiviral screening of other quinolizidine- and pyrrolizidine-derived chloroquine analogs synthetized in the past [38,39,63,64], which sometimes are endowed with even higher bulkiness and lipophilicity of the basic side chains.
In the subset of benzimidazole derivatives (12–20), six compounds (13–15, 18–20) were moderately active and showed an SI in the range 2.8–5.7 (Figure 7). The net different activity observed for compounds 12 and 13, despite the strict similarity of their structures, should mainly rely on the presence of the thiosemicarbazone group (Figure 7). Indeed, the thiosemicarbazone moiety and the embodied thiourea group are well-known antiviral pharmacophores, with studies addressing the activity versus different viruses [33,42,49,65,66,67], including SARS-CoV-2 [45,46,68], in relation to the molecular scaffold to which they are linked. Therefore, other thiocarbamido derivatives (analogs of 13–15 and 23) are worthy of investigation.
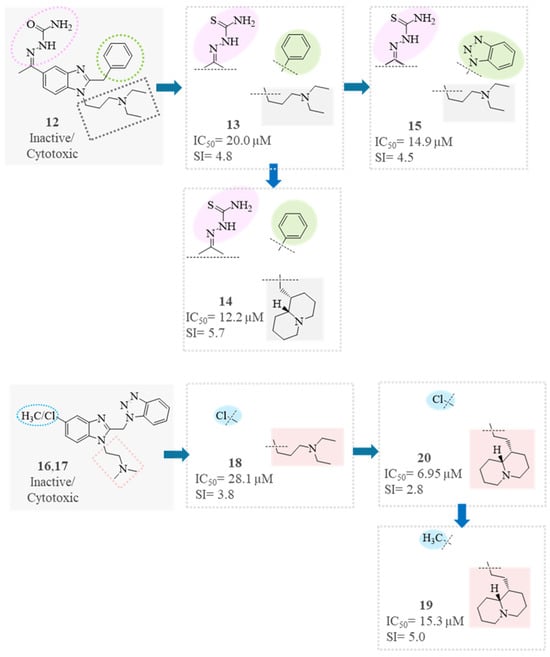
Figure 7.
SAR analysis of benzimidazole derivatives (12–20).
In compounds 17, 18, and 20, which differ in the increasing length of the basic side chain, both antiviral activity and toxicity increased accordingly (Figure 7). The exchange of the chlorine atom in position 5 (20) with a methyl group (19) reduced the activity but, even more, the toxicity; thus, compound 19 was found to be a better antiviral agent, with SI = 5 (Figure 7). On the contrary, the same exchange of groups for compounds 17 and 16 led to a strong increase in toxicity (CC50 = 191.5 µM → 37.7 µM).
Regarding the subset of miscellaneous compounds, it was observed that compounds 21, 22, and 24 exhibited no activity against SARS-CoV-2 when tested at concentrations close to their corresponding CC50 values. Despite the poor results obtained for compound 21, the investigation of further benzotriazole derivatives is still worthwhile. Indeed, besides our previous work [44], many others illustrated their antiviral activities [69,70,71,72], and benzotriazole itself has been shown to interact with SARS-CoV-2 proteins [73].
Also, the even modest activity of 23 and 25 still suggests the screening of some other cytisine and arylamino enone derivatives, respectively. Indeed, the activity of 23 may be related to the affinity to the α4β2 subtype of the nicotinic receptor (Ki = 2.3 µM [48]), and the interaction of the SARS-CoV-2 spike protein with the nicotinic receptor is increasingly recognized [74,75]. Compound 25 has been shown to act favorably against prostate and triple-negative breast cancer [76,77], probably via TRAF6 inhibition.
Finally, to confirm the results obtained through real-time PCR, the two most active compounds with the highest SI, along with two compounds from the benzimidazole derivative group, were tested using the plaque assay, a method that assesses the actual number of infectious viral particles present in the samples. The compounds 10, 11, and chloroquine were tested at concentrations of 50–25-16.7–5.6 and 1.9 µM, while the benzimidazole derivatives were tested at concentrations of 40–20-6.7–2.2–0.7 µM. The results (Table 2), expressed as the mean number of plaques (PFU/mL) and as the percentage of viral replication (in brackets), confirm that all tested compounds can inhibit viral replication and infectivity in a dose-dependent manner.

Table 2.
Virucidal activity a of tested compounds and CQ.
3. Materials and Methods
3.1. Chemistry
3.1.1. General Information
Chemicals and solvents were purchased from Sigma-Aldrich (Milan, Italy). Mps: Büchi apparatus, uncorrected. 1H NMR spectra and 13C NMR spectra were recorded using a Jeol instrument at 400 and 101 MHz, respectively; chemical shifts are reported as δ (ppm) and are referenced to the solvent signal: DMSO-d6, quintet at 2.5 ppm (1H), septet at 39.5 ppm (13C); J in Hz. Elemental analyses were performed on a Flash 2000 CHNS (Thermo Scientific, Waltham, MA, USA) instrument in the Microanalysis Laboratory of the Department of Pharmacy, University of Genova. Results of elemental analyses indicated that the purity of all compounds was ≥95%. Benz: benzimidazole ring; Q: quinolizidine ring.
3.1.2. General Procedure for the Preparation of Thiosemicarbazones
To a solution of the proper 5-acetyl benzimidazole (0.80 mmol) [43,59] in ethanol (2 mL), a solution of thiosemicarbazide (0.85 mmol) in water (2.8 mL) and glacial acetic acid (0.22 mL) was added. The mixture was refluxed for 3 h under stirring. The reaction mixture was then evaporated to dryness, affording an oily residue that was purified by CC (SiO2, CH2Cl2+2% DEA), affording the final product as a white solid.
2-(1-{2-Benzyl-1-[((1S,9aR)-octahydro-2H-quinolizin-1-yl)methyl]-1H-benzo[d]imidazol-5-yl}-ethylidene)hydrazine-1-carbothioamide (14) Yield: 78%; m.p. 205–207 °C. 1H NMR (400 MHz, DMSO-D6) δ 10.16 (s, 1H, NH), 8.21 (s, 1H, NH2), 8.11 (d, J = 1.7 Hz, 1H, Benz.), 7.93 (s, 1H, NH2), 7.90 (dd, J = 8.6, 1.8 Hz, 1H, Benz.), 7.40 (d, J = 8.7 Hz, 1H, Benz.), 7.37–7.20 (m, 5H, Ar), 4.35–4.23 (m, 2H, CH2-Q and 4.28, s, 2H, CH2-Ar), 2.82–2.75 (m, 2H, Hα N of Q), 2.35 (s, 3H, CH3), 2.12–2.07 (m, 1H, Q), 1.99 (s, 1H, Q), 1.93–1.80 (m, 3H, Q), 1.76–1.68 (m, 1H, Q), 1.60–1.50 (m, 3H, Q), 1.45–1.30 (m, 2H, Q), 1.27–1.11 (m, 3H, Q). 13C NMR (101 MHz, DMSO-D6) δ 178.63, 154.52, 148.85, 142.25, 136.76, 136.55, 131.37, 128.77 (2C), 128.50 (2C), 126.64, 120.88, 117.57, 109.82, 64.11, 56.72, 56.54, 41.60, 38.17, 33.30, 28.8–5, 25.60, 25.09, 24.60, 20.61, 14.35. Anal. calcd. for C27H34N6S: % C 68.32, H 7.22, N 17.71, S 6.75; found: % C 67.98, H 7.18, N 17.96, S 6.87.
2-(1-{2-[(1H-Benzo[d][1,2,3]triazol-1-yl)methyl]-1-[3-(N,N-diethylamino)propyl]-1H-benzo-[d]imidazol-5-yl}ethylidene)hydrazine-1-carbothioamide (15) Yield: 64%; m.p. 195–197 °C. 1H NMR (400 MHz, DMSO-D6) δ 10.14 (s, 1H, NH), 8.21 (s, 1H, NH2), 8.12 (d, J = 1.7 Hz, 1 arom. H), 8.08 (d, J = 8.3 Hz, 1 arom. H), 7.98 (dd, J = 8.7, 1.8 Hz, 2H, 1 arom. H + 1 NH2), 7.84 (d, J = 8.3 Hz, 1 arom. H), 7.60–7.50 (m, 2 arom. H.), 7.46–7.38 (m, 1 arom. H), 6.44 (s, 2H, CH2-Ar), 4.38 (t, J = 7.3 Hz, 2H,CH2CH2CH2N(Et)2), 2.39 (q, J = 7.1 Hz, 4H, N(CH2CH3)2), 2.32 (s, 3H, CH3), 2.28 (t, J = 6.8 Hz, 2H, CH2CH2CH2N(Et)2), 1.72 (p, J = 7.0 Hz, 2H, CH2CH2CH2N(Et)2), 0.89 (t, J = 7.1 Hz, 6H, N(CH2CH3)2). 13C NMR (101 MHz, DMSO-D6) δ 178.65, 149.23, 148.61, 145.34, 141.92, 135.98, 133.24, 131.97, 127.63, 124.20, 121.77, 119.28, 118.09, 111.01, 110.22, 48.94, 45.84 (2C), 44.56, 41.70, 26.53, 14.29, 11.38 (2C). Anal. calcd. for C24H31N9S: % C 60.35, H 6.54, N 26.39, S 6.71; found % C 60.48, H 6.34, N 26.31, S 6.58.
3.2. Cytotoxicity Assay
Vero E6 cells (Monkey Kidney Epithelial Cells, ATCC C1008) were maintained in DMEM medium (EuroClone, Milan, Italy) supplemented with 10% heat-inactivated fetal calf serum (EuroClone, Milan, Italy), 2 mM glutamine (EuroClone, Milan, Italy), 100 units/mL of penicillin, and 100 μg/mL of streptomycin (EuroClone, Milan, Italy). For the cytotoxicity assay, Vero E6 cells were seeded into 96-well plates at concentration of 1 × 104 cells/well. After 24 h of incubation, the cells were treated with serial 2-fold dilutions of compounds, in a final volume of 200 μL, in duplicate. After incubation for 72 h at 37 °C in 5% CO2, cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) (Merk, Darmstadt, Germany) assay, as previously described [78]. Percentage of viable cells was calculated using untreated cells as control (100% viability) using the formula [(sample absorbance—cell free sample blank)/mean media control absorbance] × 100. The 50% cytotoxic concentration (CC50) causing 50% reduction in Vero E6 cells’ viability with respect to untreated control cells was determined using Gene5 software. Morphological changes of Vero E6 cells were also observed by light microscopy.
3.3. Vero E6 Cells Infection, Treatment and Evaluation of the Antiviral Activity
SARS-CoV-2 belonging to the B1.1 lineage was isolated from the nasal-pharyngeal swab of an Italian patient, as described [79]. Vero E6 cells were seeded into 96-well plates at a density of 1.3 × 104 cells/well and were incubated for 24 h at 37 °C, 5% CO2. Cells were infected with an MOI of 0.05 (1000 PFU/well/30 µL) and incubated for 2 h at 37 °C, 5% CO2. After removal of virus inoculum, cells were treated with the compounds (dose range 2.2–60 µM; 200 µL/well final volume) and incubated for 72 h at 37 °C, 5% CO2. Chloroquine was used as the control drug.
SARS-CoV-2 replication was evaluated by RNA isolation from cells’ supernatants followed by specific qRT-PCR, targeting the N1 gene [80,81]. SARS-CoV-2 viral load data (copies/µL) were normalized versus untreated infected controls according to the following formula: % SARS-CoV-2 replication = 100 × (viral load treated sample/viral load untreated control). Data were plotted as a function of drug concentration and curve fitting was obtained by non-linear regression analysis using a four-parameter logistic method (software GraphPad Prism v. 6.0, GraphPad, La Jolla, CA, USA). The IC50 value was extrapolated as the concentration that induced a 50% inhibition of viral replication.
Antiviral activity of the most active compounds and of the reference drug chloroquine was also evaluated by plaque assay. Vero E6 cells were seeded in 6-well plates (400,000 cells/well) for 24 h at 37 °C. Virus inoculum (50 PFU/well) was added to the wells for 2 h at 37 °C. Subsequently, virus inoculum was removed, and cells were covered with 0.3% agarose dissolved in cell medium in the presence or not of different concentrations of compounds (range 0.7 µM–50 µM) at 37 °C for 72 h. Cells were fixed with 4% formaldehyde solution (Merk, Darmstadt, Germany) and, after agarose removal, stained with methylene blue (Merk, Darmstadt, Germany). Results were expressed as Plaque Forming Unit (PFU)/mL, and as percentage of virus replication, compared to untreated infected cells.
4. Conclusions
Many chemical classes of compounds are able to inhibit the in vitro replication of SARS-CoV-2 (the virus responsible for COVID-19 disease), with valuable potencies and various mechanisms of action. However, the current COVID-19 therapy is far from being satisfactory, and the search for novel, more effective compounds continues to be a priority.
Thus, we deemed it worthwhile to test, against SARS-CoV-2 replication, several compounds previously identified by our group as endowed with potent antiviral and/or anti-parasitic activity, and which are structurally related to some privileged scaffolds discussed in the Introduction (riminophenazines, 4-aminoquinolines, benzimidazole derivatives, etc.), and in most cases characterized by the presence of the bulky quinolizidine and pyrrolizidine rings.
Indeed, many of the twenty-five tested compounds (2, 3, 10, 11, 13–15, and 18–20) displayed valuable antiviral activity (IC50 in the range 1.5–28 µM), and, in relation to their cytotoxicity versus Vero E6 cells, only six of them (10, 11, 13–15, and 19) exhibited a promising selectivity index (SI). Evidence in the literature indicates that drugs and compounds tested against SARS-CoV-2 and related coronaviruses display a variable SI, which can be higher or lower, depending on the sensitivity of the animal or human cell line used as host cells for viral replication [82,83]. Therefore, the present compounds require a more in-depth analysis to better define their antiviral efficacy.
In particular, the bulkier and lipophilic compounds 10 and 11 were found to be more potent than the reference drug chloroquine, even having a doubled SI value (20 versus 11). These compounds have been shown to exhibit favorable preclinical pharmacological profiles [40,41], and therefore other chloroquine analogs bearing an even bulkier basic side chain will be investigated.
On the other hand, the benzimidazole derivatives 13–15 and 18–20 warrant further investigation of some analogs to, potentially, furnish improved antiviral agents. Similarly, despite the modest activity, compounds 23 and 25 also deserve further consideration as model compounds, given the recently shown antiviral activity of some cytisine (nicotinic ligand) analogs [49,74] and the promising anti-TRAF6 and anticancer activities of 25, which are currently under investigation by some of the authors.
The definition of the mechanism of action of the diverse chemotypes identified will be investigated as a further extension of this exploratory work. Moreover, due to the mutational landscape of SARS-CoV-2 that impacts its transmissibility and antigenicity, the most promising compounds will be then tested against different SARS-CoV-2 variants with a view to probing their potential broad-spectrum antiviral activity and mutation-induced drug resistance leading to targeting failure.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph17121668/s1, 1H and 13C NMR spectra of the newly synthesized compounds 14 and 15.
Author Contributions
Conceptualization, F.S.; methodology and validation, M.T., A.S. and N.B.; participated to methodology, K.K.M., S.D. (Serena Delbue), S.D. (Sarah D’Alessandro) and S.P.; investigation, I.B., V.F., M.T., A.S. and N.B.; resources, M.T., A.S., S.D. (Serena Delbue) and N.B.; writing—original draft preparation, F.S. and M.T.; writing—review and editing, F.S., M.T., A.S. and N.B.; project administration, F.S., M.T., A.S. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the University of Genoa (FRA2023) and by Ministero dell’Università e della Ricerca (PRIN) Projects PRIN2022Y8FZCP.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- WHO. Director-General’s Opening Remarks at the Media Briefing. 5 May 2023. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-2023 (accessed on 20 October 2024).
- Aboul-Fotouh, S.; Mahmoud, A.N.; Elnahas, E.M.; Habib, M.Z.; Abdelraouf, S.M. What Are the Current Anti-COVID-19 Drugs? From Traditional to Smart Molecular Mechanisms. Virol. J. 2023, 20, 241. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.C.; Liew, D.F.L.; Tanner, H.L.; Grainger, J.R.; Dwek, R.A.; Reisler, R.B.; Steinman, L.; Feldmann, M.; Ho, L.-P.; Hussell, T.; et al. COVID-19 Therapeutics: Challenges and Directions for the Future. Proc. Natl. Acad. Sci. USA 2022, 119, e2119893119. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents. Chemother. 2020, 64, e00819-20. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic Strategies for COVID-19: Progress and Lessons Learned. Nat. Rev. Drug. Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.; Chan-Tack, K.; Farley, J.; Sherwat, A. FDA Approval of Remdesivir—A Step in the Right Direction. N. Engl. J. Med. 2020, 383, 2598–2600. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Alonso, R.; Camon, A.M.; Cardozo, C.; Albiach, L.; Agüero, D.; Marcos, M.A.; Ambrosioni, J.; Bodro, M.; Chumbita, M.; et al. Impact of Remdesivir According to the Pre-Admission Symptom Duration in Patients with COVID-19. J. Antimicrob. Chemother. 2021, 76, 3296–3302. [Google Scholar] [CrossRef]
- Padilla, S.; Polotskaya, K.; Fernández, M.; Gonzalo-Jiménez, N.; de la Rica, A.; García, J.A.; García-Abellán, J.; Mascarell, P.; Gutiérrez, F.; Masiá, M. Survival Benefit of Remdesivir in Hospitalized COVID-19 Patients with High SARS-CoV-2 Viral Loads and Low-Grade Systemic Inflammation. J. Antimicrob. Chemother. 2022, 77, 2257–2264. [Google Scholar] [CrossRef] [PubMed]
- Lagevrio|European Medicines Agency (EMA). Questions and Answers on the Withdrawal of Application for the Marketing Authorisation of Lagevrio (Molnupiravir) Reference Number: EMA/290450/2023. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/lagevrio (accessed on 20 October 2024).
- Farahani, M.; Niknam, Z.; Mohammadi Amirabad, L.; Amiri-Dashatan, N.; Koushki, M.; Nemati, M.; Danesh Pouya, F.; Rezaei-Tavirani, M.; Rasmi, Y.; Tayebi, L. Molecular Pathways Involved in COVID-19 and Potential Pathway-Based Therapeutic Targets. Biomed. Pharmacother. 2022, 145, 112420. [Google Scholar] [CrossRef]
- Saber, M.M.; Salama, M.M.; Badary, O.A. The Potential of Natural Products in the Management of COVID-19. Adv. Exp. Med. Biol. 2024, 1457, 215–235. [Google Scholar] [CrossRef]
- Zhong, L.; Zhao, Z.; Peng, X.; Zou, J.; Yang, S. Recent Advances in Small-Molecular Therapeutics for COVID-19. Precis. Clin. Med. 2022, 5, pbac024. [Google Scholar] [CrossRef]
- Braga, L.; Ali, H.; Secco, I.; Chiavacci, E.; Neves, G.; Goldhill, D.; Penn, R.; Jimenez-Guardeño, J.M.; Ortega-Prieto, A.M.; Bussani, R.; et al. Drugs That Inhibit TMEM16 Proteins Block SARS-CoV-2 Spike-Induced Syncytia. Nature 2021, 594, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Cappelletto, A.; Allan, H.E.; Crescente, M.; Schneider, E.; Bussani, R.; Ali, H.; Secco, I.; Vodret, S.; Simeone, R.; Mascaretti, L.; et al. SARS-CoV-2 Spike Protein Activates TMEM16F-Mediated Platelet Procoagulant Activity. Front. Cardiovasc. Med. 2022, 9, 1013262. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yin, X.; Meng, X.; Chan, J.F.-W.; Ye, Z.-W.; Riva, L.; Pache, L.; Chan, C.C.-Y.; Lai, P.-M.; Chan, C.C.-S.; et al. Clofazimine Broadly Inhibits Coronaviruses Including SARS-CoV-2. Nature 2021, 593, 418–423. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Q.; Wu, X.; Hao, S.; Hao, X.; Jones, E.; Zhang, Y.; Qiu, J.; Xu, L. Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147. Biomedicines 2023, 11, 2019. [Google Scholar] [CrossRef] [PubMed]
- Ousingsawat, J.; Centeio, R.; Schreiber, R.; Kunzelmann, K. Niclosamide, but Not Ivermectin, Inhibits Anoctamin 1 and 6 and Attenuates Inflammation of the Respiratory Tract. Pflugers. Arch. 2024, 476, 211–227. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of Chloroquine on Viral Infections: An Old Drug against Today’s Diseases? Lancet. Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Sansone, N.M.S.; Boschiero, M.N.; Marson, F.A.L. Efficacy of Ivermectin, Chloroquine/Hydroxychloroquine, and Azithromycin in Managing COVID-19: A Systematic Review of Phase III Clinical Trials. Biomedicines 2024, 12, 2206. [Google Scholar] [CrossRef]
- Nazir, M.S.; Ahmad, M.; Aslam, S.; Rafiq, A.; Al-Hussain, S.A.; Zaki, M.E.A. A Comprehensive Update of Anti-COVID-19 Activity of Heterocyclic Compounds. Drug. Des. Devel. Ther. 2024, 18, 1547–1571. [Google Scholar] [CrossRef]
- Aljadeed, R. The Rise and Fall of Hydroxychloroquine and Chloroquine in COVID-19. J. Pharm. Pract. 2022, 35, 971–978. [Google Scholar] [CrossRef]
- Altulea, D.; Maassen, S.; Baranov, M.V.; van den Bogaart, G. What Makes (Hydroxy)Chloroquine Ineffective against COVID-19: Insights from Cell Biology. J. Mol. Cell Biol. 2021, 13, 175–184. [Google Scholar] [CrossRef]
- Yele, V.; Sanapalli, B.K.R.; Mohammed, A.A. Imidazoles and Benzimidazoles as Putative Inhibitors of SARS-CoV-2 B.1.1.7 (Alpha) and P.1 (Gamma) Variant Spike Glycoproteins: A Computational Approach. Chem. Zvesti 2022, 76, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Mudi, P.K.; Mahato, R.K.; Verma, H.; Panda, S.J.; Purohit, C.S.; Silakari, O.; Biswas, B. In Silico Anti-SARS-CoV-2 Activities of Five-Membered Heterocycle-Substituted Benzimidazoles. J. Mol. Struct. 2022, 1261, 132869. [Google Scholar] [CrossRef] [PubMed]
- Anjaneyulu, B. DFT and Molecular Docking Studies of 1, 2 Disubstituted Benzimidazole Derivatives with COVID-19 Receptors: An Approach for Medications to Treat COVID-19. Letters Org. Chem. 2023, 20, 818–828. [Google Scholar] [CrossRef]
- Ananta, M.F.; Saha, P.; Rahman, F.I.; Spriha, S.E.; Chowdhury, A.K.A.; Rahman, S.M.A. Design, Synthesis and Computational Study of Benzimidazole Derivatives as Potential Anti-SARS-CoV-2 Agents. J. Mol. Struct. 2024, 1306, 137940. [Google Scholar] [CrossRef]
- Chaibi, F.-Z.; Brier, L.; Carré, P.; Landry, V.; Desmarets, L.; Tarricone, A.; Cantrelle, F.-X.; Moschidi, D.; Herledan, A.; Biela, A.; et al. N-Acylbenzimidazoles as Selective Acylators of the Catalytic Cystein of the Coronavirus 3CL Protease. Eur. J. Med. Chem. 2024, 276, 116707. [Google Scholar] [CrossRef]
- Mudi, P.K.; Mahanty, A.K.; Kotakonda, M.; Prasad, S.; Bhattacharyya, S.; Biswas, B. A Benzimidazole Scaffold as a Promising Inhibitor against SARS-CoV-2. J. Biomol. Struct. Dyn. 2023, 41, 1798–1810. [Google Scholar] [CrossRef]
- Omotuyi, O.; Olatunji, O.M.; Nash, O.; Oyinloye, B.; Soremekun, O.; Ijagbuji, A.; Fatumo, S. Benzimidazole Compound Abrogates SARS-CoV-2 Receptor-Binding Domain (RBD)/ACE2 Interaction In Vitro. Microb. Pathog. 2023, 176, 105994. [Google Scholar] [CrossRef]
- Balaramnavar, V.M.; Ahmad, K.; Saeed, M.; Ahmad, I.; Kamal, M.; Jawed, T. Pharmacophore-Based Approaches in the Rational Repurposing Technique for FDA Approved Drugs Targeting SARS-CoV-2 Mpro. RSC Adv. 2020, 10, 40264–40275. [Google Scholar] [CrossRef]
- Tonelli, M.; Paglietti, G.; Boido, V.; Sparatore, F.; Marongiu, F.; Marongiu, E.; La Colla, P.; Loddo, R. Antiviral Activity of Benzimidazole Derivatives. I. Antiviral Activity of 1-Substituted-2-[(Benzotriazol-1/2-Yl)Methyl]Benzimidazoles. Chem. Biodivers. 2008, 5, 2386–2401. [Google Scholar] [CrossRef]
- Cichero, E.; Tonelli, M.; Novelli, F.; Tasso, B.; Delogu, I.; Loddo, R.; Bruno, O.; Fossa, P. Benzimidazole-Based Derivatives as Privileged Scaffold Developed for the Treatment of the RSV Infection: A Computational Study Exploring the Potency and Cytotoxicity Profiles. J. Enzyme Inhib. Med. Chem. 2017, 32, 375–402. [Google Scholar] [CrossRef][Green Version]
- Tsypysheva, I.P.; Lai, H.-C.; Kiu, Y.-T.; Koval’skaya, A.V.; Tsypyshev, D.O.; Huang, S.-H.; Lin, C.-W. Synthesis and Antiviral Evaluation of Cytisine Derivatives against Dengue Virus Types 1 and 2. Bioorg. Med. Chem. Lett. 2021, 54, 128437. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, M.; Novelli, F.; Tasso, B.; Sparatore, A.; Boido, V.; Sparatore, F.; Cannas, S.; Molicotti, P.; Zanetti, S.; Parapini, S.; et al. Antitubercular Activity of Quinolizidinyl/Pyrrolizidinylalkyliminophenazines. Bioorg. Med. Chem. 2014, 22, 6837–6845. [Google Scholar] [CrossRef] [PubMed]
- Barteselli, A.; Casagrande, M.; Basilico, N.; Parapini, S.; Rusconi, C.M.; Tonelli, M.; Boido, V.; Taramelli, D.; Sparatore, F.; Sparatore, A. Clofazimine Analogs with Antileishmanial and Antiplasmodial Activity. Bioorg. Med. Chem. 2015, 23, 55–65. [Google Scholar] [CrossRef]
- Bassanini, I.; Parapini, S.; Basilico, N.; Sparatore, A. Novel Hydrophilic Riminophenazines as Potent Antiprotozoal Agents. ChemMedChem 2019, 14, 1940–1949. [Google Scholar] [CrossRef]
- Koval, A.; Bassanini, I.; Xu, J.; Tonelli, M.; Boido, V.; Sparatore, F.; Amant, F.; Annibali, D.; Leucci, E.; Sparatore, A.; et al. Optimization of the Clofazimine Structure Leads to a Highly Water-Soluble C3-Aminopyridinyl Riminophenazine Endowed with Improved Anti-Wnt and Anti-Cancer Activity in Vitro and in Vivo. Eur. J. Med. Chem. 2021, 222, 113562. [Google Scholar] [CrossRef]
- Sparatore, A.; Basilico, N.; Parapini, S.; Romeo, S.; Novelli, F.; Sparatore, F.; Taramelli, D. 4-Aminoquinoline Quinolizidinyl- and Quinolizidinylalkyl-Derivatives with Antimalarial Activity. Bioorg. Med. Chem. 2005, 13, 5338–5345. [Google Scholar] [CrossRef]
- Sparatore, A.; Basilico, N.; Casagrande, M.; Parapini, S.; Taramelli, D.; Brun, R.; Wittlin, S.; Sparatore, F. Antimalarial Activity of Novel Pyrrolizidinyl Derivatives of 4-Aminoquinoline. Bioorg. Med. Chem. Lett. 2008, 18, 3737–3740. [Google Scholar] [CrossRef] [PubMed]
- Basilico, N.; Parapini, S.; Sparatore, A.; Romeo, S.; Misiano, P.; Vivas, L.; Yardley, V.; Croft, S.L.; Habluetzel, A.; Lucantoni, L.; et al. In Vivo and In Vitro Activities and ADME-Tox Profile of a Quinolizidine-Modified 4-Aminoquinoline: A Potent Anti-P. Falciparum and Anti-P. Vivax Blood-Stage Antimalarial. Molecules 2017, 22, 2102. [Google Scholar] [CrossRef]
- Basilico, N.; Parapini, S.; D’Alessandro, S.; Misiano, P.; Romeo, S.; Dondio, G.; Yardley, V.; Vivas, L.; Nasser, S.; Rénia, L.; et al. Favorable Preclinical Pharmacological Profile of a Novel Antimalarial Pyrrolizidinylmethyl Derivative of 4-Amino-7-Chloroquinoline with Potent In Vitro and In Vivo Activities. Biomolecules 2023, 13, 836. [Google Scholar] [CrossRef]
- Francesconi, V.; Cichero, E.; Schenone, S.; Naesens, L.; Tonelli, M. Synthesis and Biological Evaluation of Novel (Thio)Semicarbazone-Based Benzimidazoles as Antiviral Agents against Human Respiratory Viruses. Molecules 2020, 25, 1487. [Google Scholar] [CrossRef]
- Paglietti, G.; Boido, V.; Sparatore, F. Dialkylaminoalkylbenzimidazoles of pharmacological importance. IV. Il Farmaco Ed. Sci. 1975, 30, 505–511. [Google Scholar]
- Loddo, R.; Novelli, F.; Sparatore, A.; Tasso, B.; Tonelli, M.; Boido, V.; Sparatore, F.; Collu, G.; Delogu, I.; Giliberti, G.; et al. Antiviral Activity of Benzotriazole Derivatives. 5-[4-(Benzotriazol-2-Yl)Phenoxy]-2,2-Dimethylpentanoic Acids Potently and Selectively Inhibit Coxsackie Virus B5. Bioorg. Med. Chem. 2015, 23, 7024–7034. [Google Scholar] [CrossRef] [PubMed]
- Maltarollo, V.G.; da Silva, E.B.; Kronenberger, T.; Sena Andrade, M.M.; de Lima Marques, G.V.; Cândido Oliveira, N.J.; Santos, L.H.; de Oliveira Rezende Júnior, C.; Cassiano Martinho, A.C.; Skinner, D.; et al. Structure-Based Discovery of Thiosemicarbazones as SARS-CoV-2 Main Protease Inhibitors. Future Med. Chem. 2023, 15, 959–985. [Google Scholar] [CrossRef]
- Xu, Y.-S.; Chigan, J.-Z.; Li, J.-Q.; Ding, H.-H.; Sun, L.-Y.; Liu, L.; Hu, Z.; Yang, K.-W. Hydroxamate and Thiosemicarbazone: Two Highly Promising Scaffolds for the Development of SARS-CoV-2 Antivirals. Bioorg. Chem. 2022, 124, 105799. [Google Scholar] [CrossRef]
- Tonelli, M.; Boido, V.; Canu, C.; Sparatore, A.; Sparatore, F.; Paneni, M.S.; Fermeglia, M.; Pricl, S.; La Colla, P.; Casula, L.; et al. Antimicrobial and Cytotoxic Arylazoenamines. Part III: Antiviral Activity of Selected Classes of Arylazoenamines. Bioorg. Med. Chem. 2008, 16, 8447–8465. [Google Scholar] [CrossRef]
- Nicolotti, O.; Canu Boido, C.; Sparatore, F.; Carotti, A. Cytisine Derivatives as High Affinity nAChR Ligands: Synthesis and Comparative Molecular Field Analysis. Farmaco 2002, 57, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hour, M.-J.; Lin, C.-S.; Chang, Y.-S.; Chen, Z.-Y.; Koval’skaya, A.V.; Su, W.-C.; Tsypysheva, I.P.; Lin, C.-W. Assessing the Inhibitory Effects of Some Secondary Amines, Thioureas and 1,3-Dimethyluracil Conjugates of (-)-Cytisine and Thermopsine on the RNA-Dependent RNA Polymerase of SARS-CoV-1 and SARS-CoV-2. Bioorg. Med. Chem. Lett. 2024, 113, 129950. [Google Scholar] [CrossRef]
- Sparatore, A.; Perrino, E.; Tazzari, V.; Giustarini, D.; Rossi, R.; Rossoni, G.; Erdmann, K.; Schröder, H.; Del Soldato, P. Pharmacological Profile of a Novel H(2)S-Releasing Aspirin. Free. Radic. Biol. Med. 2009, 46, 586–592. [Google Scholar] [CrossRef]
- Sparatore, F.; Cerri, R. Condensation of β-Diketones with 7-Amino-2,3-Polymethyleneindoles. Formation of Pyrroloquinolines Instead of Diazepinoindoles. Ann. Chim. 1968, 58, 1477–1490. [Google Scholar]
- Yamada, K.; Koyama, H.; Hagiwara, K.; Ueda, A.; Sasaki, Y.; Kanesashi, S.-N.; Ueno, R.; Nakamura, H.K.; Kuwata, K.; Shimizu, K.; et al. Identification of a Novel Compound with Antiviral Activity against Influenza A Virus Depending on PA Subunit of Viral RNA Polymerase. Microbes Infect. 2012, 14, 740–747. [Google Scholar] [CrossRef]
- Zarzycka, B.; Seijkens, T.; Nabuurs, S.B.; Ritschel, T.; Grommes, J.; Soehnlein, O.; Schrijver, R.; van Tiel, C.M.; Hackeng, T.M.; Weber, C.; et al. Discovery of Small Molecule CD40-TRAF6 Inhibitors. J. Chem. Inf. Model. 2015, 55, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lei, Z.; Wei, L.; Yang, K.; Shen, J.; Hu, L. Tumor Necrosis Factor Receptor-Associated Factor 6 and Human Cancer: A Systematic Review of Mechanistic Insights, Functional Roles, and Therapeutic Potential. J. Cancer 2024, 15, 560–576. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Elaish, M.; Wong, C.P.; Hassan, B.B.; Lopez-Orozco, J.; Felix-Lopez, A.; Ogando, N.S.; Nagata, L.; Mahal, L.K.; Kumar, A.; et al. The Wnt/β-Catenin Pathway Is Important for Replication of SARS-CoV-2 and Other Pathogenic RNA Viruses. npj Viruses 2024, 2, 6. [Google Scholar] [CrossRef]
- Chatterjee, S.; Keshry, S.S.; Ghosh, S.; Ray, A.; Chattopadhyay, S. Versatile β-Catenin Is Crucial for SARS-CoV-2 Infection. Microbiol. Spectr. 2022, 10, e0167022. [Google Scholar] [CrossRef]
- Yamamoto, M.; Gohda, J.; Akiyama, T.; Inoue, J.-I. TNF Receptor-Associated Factor 6 (TRAF6) Plays Crucial Roles in Multiple Biological Systems through Polyubiquitination-Mediated NF-κB Activation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021, 97, 145–160. [Google Scholar] [CrossRef]
- Li, S.-W.; Wang, C.-Y.; Jou, Y.-J.; Huang, S.-H.; Hsiao, L.-H.; Wan, L.; Lin, Y.-J.; Kung, S.-H.; Lin, C.-W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016, 17, 678. [Google Scholar] [CrossRef]
- Paglietti, G.; Sparatore, F. Dialkylaminoalkylbenzimidazoles of pharmacological interest. 3. Farm. Sci 1972, 27, 333–342. [Google Scholar]
- Ali, A.A.; Nimir, H.; Aktas, C.; Huch, V.; Rauch, U.; Schäfer, K.-H.; Veith, M. Organoplatinum(II) Complexes with 2-Acetylthiophene Thiosemicarbazone: Synthesis, Characterization, Crystal Structures, and in Vitro Antitumor Activity. Organometallics 2012, 31, 2256–2262. [Google Scholar] [CrossRef]
- Gastaca, B.; Sánchez, H.R.; Menestrina, F.; Caputo, M.; de las Mercedes Schiavoni, M.; Furlong, J.J.P. Thiosemicarbazones Synthesized from Acetophenones: Tautomerism, Spectrometric Data, Reactivity and Theoretical Calculations. Int. J. Anal. Mass Spectrom. Chromatogr. 2019, 7, 19–34. [Google Scholar] [CrossRef]
- Koval, A.; Xu, J.; Williams, N.; Schmolke, M.; Krause, K.-H.; Katanaev, V.L. Wnt-Independent SARS-CoV-2 Infection in Pulmonary Epithelial Cells. Microbiol. Spectr. 2023, 11, e0482722. [Google Scholar] [CrossRef]
- Tasso, B.; Novelli, F.; Tonelli, M.; Barteselli, A.; Basilico, N.; Parapini, S.; Taramelli, D.; Sparatore, A.; Sparatore, F. Synthesis and Antiplasmodial Activity of Novel Chloroquine Analogues with Bulky Basic Side Chains. ChemMedChem 2015, 10, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Ayoka, T.O.; Uchegbu, U.J.; Alabuike, C.C.; Nnadi, C.O. Efficient Classification and Regression Models for the QSAR of Chloroquine Analogues against Chloroquine- Sensitive and Chloroquine-Resistant Plasmodium Falciparum. Lett. Appl. NanoBioSci. 2024, 13, 90. [Google Scholar] [CrossRef]
- Shim, J.; Jyothi, N.R.; Farook, N.A.M. Biological Applications of Thiosemicarbazones and Their Metal Complexes. Asian J. Chem. 2013, 25, 5838–5840. [Google Scholar] [CrossRef]
- Rogolino, D.; Bacchi, A.; De Luca, L.; Rispoli, G.; Sechi, M.; Stevaert, A.; Naesens, L.; Carcelli, M. Investigation of the Salicylaldehyde Thiosemicarbazone Scaffold for Inhibition of Influenza Virus PA Endonuclease. J. Biol. Inorg. Chem. 2015, 20, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Soraires Santacruz, M.C.; Fabiani, M.; Castro, E.F.; Cavallaro, L.V.; Finkielsztein, L.M. Synthesis, Antiviral Evaluation and Molecular Docking Studies of N4-Aryl Substituted/Unsubstituted Thiosemicarbazones Derived from 1-Indanones as Potent Anti-Bovine Viral Diarrhea Virus Agents. Bioorg. Med. Chem. 2017, 25, 4055–4063. [Google Scholar] [CrossRef]
- Souza, G.B.; Sens, L.; Hammerschmidt, S.J.; de Sousa, N.F.; de Carvalho, M.A.G.; Dos Santos, C.V.D.; Tizziani, T.; Moreira, M.A.; Pollo, L.A.E.; Martin, E.F.; et al. Inhibitory Effects of 190 Compounds against SARS-CoV-2 Mpr o Protein: Molecular Docking Interactions. Arch. Pharm. 2023, 356, e2300207. [Google Scholar] [CrossRef] [PubMed]
- Bollikolla, H.B.; Boddapati, S.N.M.; Thangamani, S.; Mutchu, B.R.; Alam, M.M.; Hussien, M.; Jonnalagadda, S.B. Advances in Synthesis and Biological Activities of Benzotriazole Analogues: A Micro Review. J. Heterocycl. Chem. 2023, 60, 705–742. [Google Scholar] [CrossRef]
- Piras, S.; Sanna, G.; Carta, A.; Corona, P.; Ibba, R.; Loddo, R.; Madeddu, S.; Caria, P.; Aulic, S.; Laurini, E.; et al. Dichloro-Phenyl-Benzotriazoles: A New Selective Class of Human Respiratory Syncytial Virus Entry Inhibitors. Front. Chem. 2019, 7, 247. [Google Scholar] [CrossRef]
- Bajaj, K.; Sakhuja, R. Benzotriazole: Much More Than Just Synthetic Heterocyclic Chemistry. In The Chemistry of Benzotriazole Derivatives: A Tribute to Alan Roy Katritzky; Monbaliu, J.-C.M., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 235–283. ISBN 978-3-319-31554-6. [Google Scholar]
- Briguglio, I.; Piras, S.; Corona, P.; Gavini, E.; Nieddu, M.; Boatto, G.; Carta, A. Benzotriazole: An Overview on Its Versatile Biological Behavior. Eur. J. Med. Chem. 2015, 97, 612–648. [Google Scholar] [CrossRef]
- Manikandan, S.; Sundareswaran, T.; Jayamoorthy, K.; Sasikumar, G. Investigation on Hirshfeld Surface Analysis, Molecular Geometry, DFT, MEP, and Molecular Docking Analysis on Benzotriazole Oxalate against SARS-CoV-2 Virus. J. Mol. Struct. 2024, 1316, 138961. [Google Scholar] [CrossRef]
- Carlson, E.C.; Macsai, M.; Bertrand, S.; Bertrand, D.; Nau, J. The SARS-CoV-2 Virus and the Cholinergic System: Spike Protein Interaction with Human Nicotinic Acetylcholine Receptors and the Nicotinic Agonist Varenicline. Int. J. Mol. Sci. 2023, 24, 5597. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.-P.; Amoura, Z.; Rey, F.A.; Miyara, M. A Nicotinic Hypothesis for COVID-19 with Preventive and Therapeutic Implications. C. R. Biol. 2020, 343, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, G.; Bishop, R.; Maurizi, A.; Capulli, M.; Sparatore, A.; Marino, S.; Idris, A.I. Disruption of Prostate Cancer Cell-Macrophage-Osteoclast Crosstalk by a Novel TRAF6 Inhibitor. Calcif. Tissue Int. 2019, 104, S84. [Google Scholar]
- Zeng, F.; Mario, S.; Bassanini, I.; Conrad, S.; Carrasco, G.; Li, B.; Mollat, P.; Sophocleous, A.; Meli, M.; Ferrandi, E.; et al. Validation of a Novel Inhibitor of TRAF6/NFkB Axis in Models of Breast Cancer Metastasis. Abstracts. Mol. Oncol. 2023, 17, 448. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Gelati, M.; Basilico, N.; Parati, E.A.; Haynes, R.K.; Taramelli, D. Differential Effects on Angiogenesis of Two Antimalarial Compounds, Dihydroartemisinin and Artemisone: Implications for Embryotoxicity. Toxicology 2007, 241, 66–74. [Google Scholar] [CrossRef]
- Delbue, S.; D’Alessandro, S.; Signorini, L.; Dolci, M.; Pariani, E.; Bianchi, M.; Fattori, S.; Modenese, A.; Galli, C.; Eberini, I.; et al. Isolation of SARS-CoV-2 Strains Carrying a Nucleotide Mutation, Leading to a Stop Codon in the ORF 6 Protein. Emerg. Microbes Infect. 2021, 10, 252–255. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef]
- Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases. Available online: https://www.who.int/publications-detail-redirect/10665-331501 (accessed on 19 March 2020).
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug Repurposing Screens Reveal Cell-Type-Specific Entry Pathways and FDA-Approved Drugs Active against SARS-CoV-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef]
- Xiao, X.; Wang, C.; Chang, D.; Wang, Y.; Dong, X.; Jiao, T.; Zhao, Z.; Ren, L.; Dela Cruz, C.S.; Sharma, L.; et al. Identification of Potent and Safe Antiviral Therapeutic Candidates Against SARS-CoV-2. Front. Immunol. 2020, 11, 586572. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).