A Machine Learning Approach to Gene Expression in Hypertrophic Cardiomyopathy
Abstract
1. Introduction
2. Results
2.1. Gene Expression Analysis
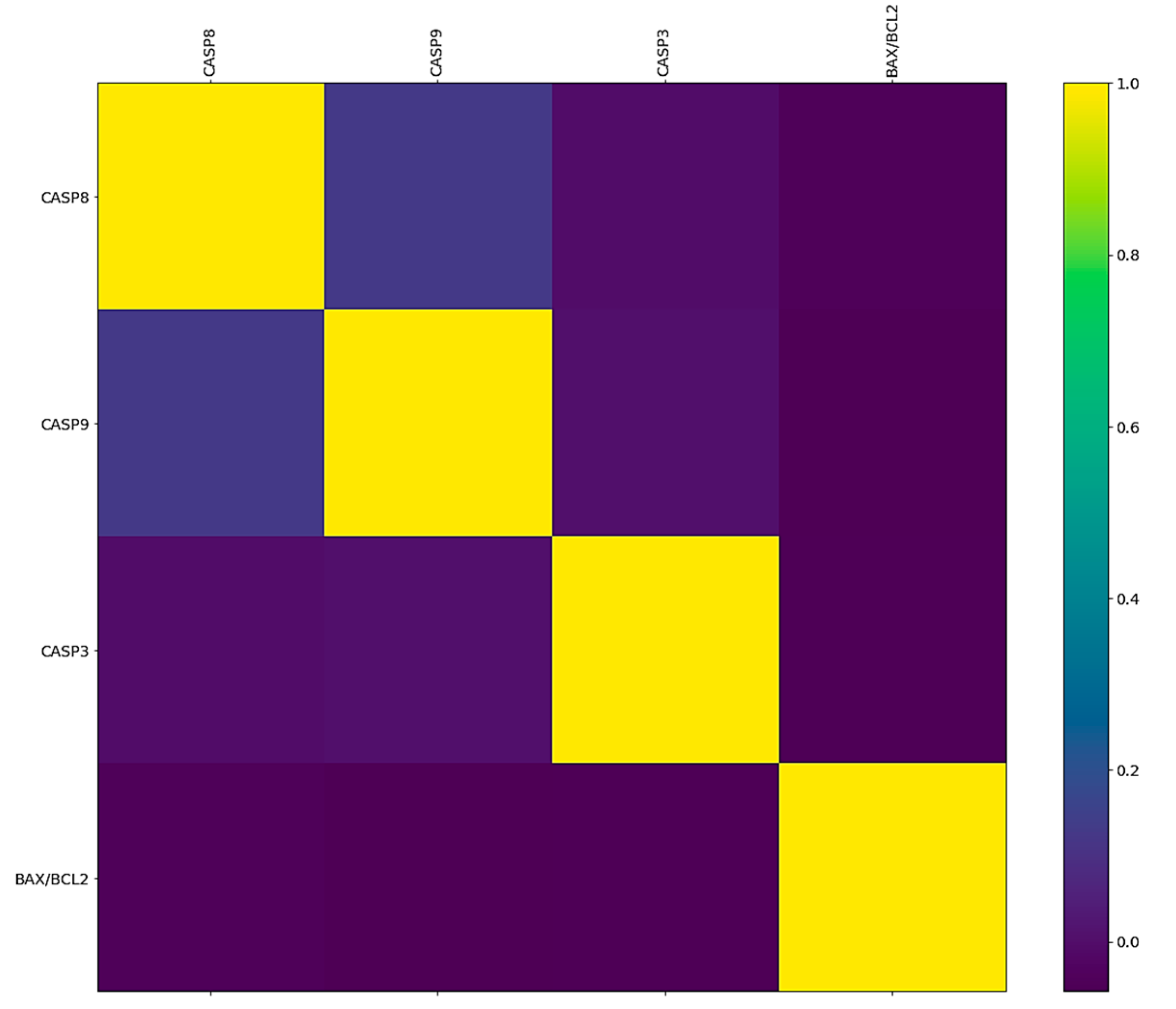
2.2. Correlation between the BAX/BCL2 Ratio and CASP Genes
2.3. The Feature Importance of Analyzed Apoptotic Genes
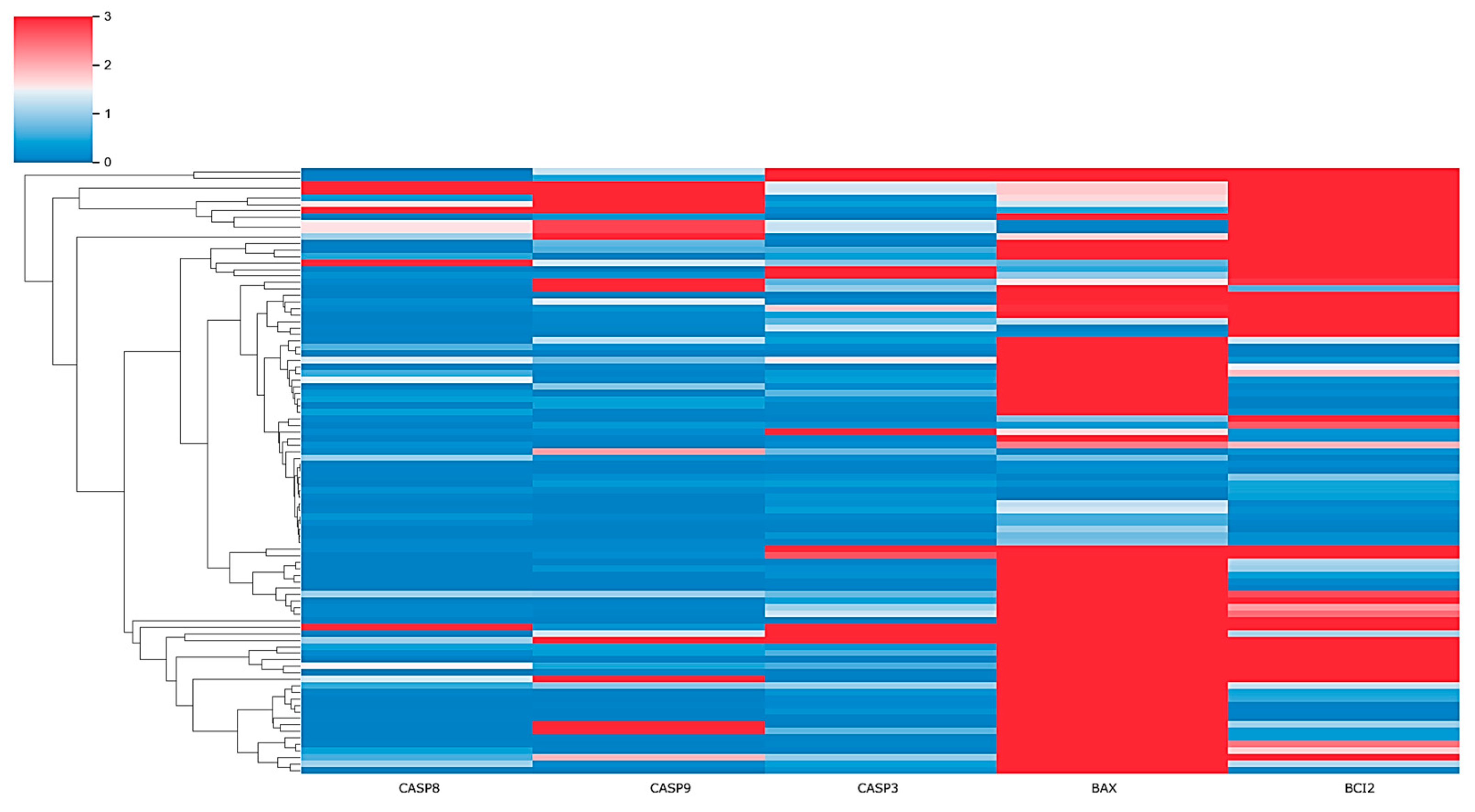
2.4. Risk Profile in HCM Patients through Clustering Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Blood Sampling
4.3. Analysis of Gene Expression by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.3.1. Isolation of RNA
4.3.2. Reverse Transcription (RT-PCR)
4.3.3. Quantification of Relative Gene Expression (qPCR)
4.3.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yusuf, S.; Reddy, S.; Ôunpuu, S.; Anand, S. Global Burden of Cardiovascular Diseases: Part II: Variations in Cardiovascular Disease by Specific Ethnic Groups and Geographic Regions and Prevention Strategies. Circulation 2001, 104, 2855–2864. [Google Scholar] [CrossRef] [PubMed]
- Gaidai, O.; Cao, Y.; Loginov, S. Global Cardiovascular Diseases Death Rate Prediction. Curr. Probl. Cardiol. 2023, 48, 101622. [Google Scholar] [CrossRef] [PubMed]
- Vaduganathan, M.; Mensah, G.A.; Turco, J.V.; Fuster, V.; Roth, G.A. The Global Burden of Cardiovascular Diseases and Risk. J. Am. Coll. Cardiol. 2022, 80, 2361–2371. [Google Scholar] [CrossRef]
- Frąk, W.; Wojtasińska, A.; Lisińska, W.; Młynarska, E.; Franczyk, B.; Rysz, J. Pathophysiology of Cardiovascular Diseases: New Insights into Molecular Mechanisms of Atherosclerosis, Arterial Hypertension, and Coronary Artery Disease. Biomedicines 2022, 10, 1938. [Google Scholar] [CrossRef]
- Lockhart, P.B.; Sun, Y. Diseases of the Cardiovascular System. In Burket’s Oral Medicine; Glick, M., Greenberg, M.S., Lockhart, P.B., Challacombe, S.J., Eds.; Wiley: Oxford, UK, 2021; pp. 505–552. ISBN 978-1-119-59774-2. [Google Scholar]
- Maisch, B.; Bauersachs, J. Cardiomyopathies—Past, Present, Future. Herz 2020, 45, 209–211. [Google Scholar] [CrossRef]
- Charles, J.; Pollack, A.; Miller, G. Cardiomyopathy. Aust. Fam. Physician 2014, 43, 253. [Google Scholar]
- Li, Q.; Gao, X.; Zhou, Z.; Zhang, H.; Li, W.; Gao, Y.; Bo, K.; Wang, H.; Wang, R.; Sun, Z.; et al. Impaired Cardiac Pump Function Assessment with Normalized Cardiac Power Using Cardiac Magnetic Resonance in Patients with Hypertrophic Cardiomyopathy. Quant. Imaging Med. Surg. 2023, 13, 4103–4116. [Google Scholar] [CrossRef]
- Ciarambino, T.; Menna, G.; Sansone, G.; Giordano, M. Cardiomyopathies: An Overview. Int. J. Mol. Sci. 2021, 22, 7722. [Google Scholar] [CrossRef] [PubMed]
- Mattsson, G.; Magnusson, P. Familial Dilated Cardiomyopathy: Risk Stratification for Sudden Cardiac Death. In Sudden Cardiac Death; Magnusson, P., Ann LeQuang, J., Eds.; IntechOpen: London, UK, 2020; ISBN 978-1-83880-069-7. [Google Scholar]
- Makavos, G.; Κairis, C.; Tselegkidi, M.-E.; Karamitsos, T.; Rigopoulos, A.G.; Noutsias, M.; Ikonomidis, I. Hypertrophic Cardiomyopathy: An Updated Review on Diagnosis, Prognosis, and Treatment. Heart Fail. Rev. 2019, 24, 439–459. [Google Scholar] [CrossRef]
- Baenen, O.; Carreño-Martínez, A.C.; Abraham, T.P.; Rugonyi, S. Energetics of Cardiac Blood Flow in Hypertrophic Cardiomyopathy through Individualized Computational Modeling. J. Cardiovasc. Dev. Dis. 2023, 10, 411. [Google Scholar] [CrossRef]
- Keramida, K.; Lazaros, G.; Nihoyannopoulos, P. Right Ventricular Involvement in Hypertrophic Cardiomyopathy: Patterns and Implications. Hell. J. Cardiol. 2020, 61, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.R.; Roma-Rodrigues, C. Genetics of Hypertrophic Cardiomyopathy: Advances and Pitfalls in Molecular Diagnosis and Therapy. TACG 2014, 7, 195–208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McKenna, W.J. Hypertrophic Cardiomyopathy: Management, Risk Stratification, and Prevention of Sudden Death. Heart 2002, 87, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Zaiser, E.; Sehnert, A.J.; Duenas, A.; Saberi, S.; Brookes, E.; Reaney, M. Patient Experiences with Hypertrophic Cardiomyopathy: A Conceptual Model of Symptoms and Impacts on Quality of Life. J. Patient Rep. Outcomes 2020, 4, 102. [Google Scholar] [CrossRef]
- Ashrafian, H.; Watkins, H. Reviews of Translational Medicine and Genomics in Cardiovascular Disease: New Disease Taxonomy and Therapeutic Implications. J. Am. Coll. Cardiol. 2007, 49, 1251–1264. [Google Scholar] [CrossRef]
- Marian, A.J.; Braunwald, E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ. Res. 2017, 121, 749–770. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Weintraub, R. Overview of Cardiomyopathies in Childhood. Front. Pediatr. 2021, 9, 708732. [Google Scholar] [CrossRef]
- Dadson, K.; Hauck, L.; Billia, F. Molecular Mechanisms in Cardiomyopathy. Clin. Sci. 2017, 131, 1375–1392. [Google Scholar] [CrossRef]
- Kimura, A. Molecular Genetics and Pathogenesis of Cardiomyopathy. J. Hum. Genet. 2016, 61, 41–50. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Obeng, E. Apoptosis (Programmed Cell Death) and Its Signals—A Review. Braz. J. Biol. 2021, 81, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting Apoptosis in Cancer Therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, Pyroptosis and Apoptosis: An Intricate Game of Cell Death. Cell. Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef]
- Nosalova, N.; Keselakova, A.; Kello, M.; Martinkova, M.; Fabianova, D.; Pilatova, M.B. Involvement of Both Extrinsic and Intrinsic Apoptotic Pathways in Tridecylpyrrolidine-Diol Derivative-Induced Apoptosis In Vitro. Int. J. Mol. Sci. 2023, 24, 11696. [Google Scholar] [CrossRef]
- Yanumula, A.; Cusick, J.K. Biochemistry, Extrinsic Pathway of Apoptosis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Al-Aamri, H.M.; Irving, H.R.; Bradley, C.; Meehan-Andrews, T. Intrinsic and Extrinsic Apoptosis Responses in Leukaemia Cells Following Daunorubicin Treatment. BMC Cancer 2021, 21, 438. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, G.C.; Schaan, A.P.; Cabral, G.F.; Santana-da-Silva, M.N.; Pinto, P.; Vidal, A.F.; Ribeiro-dos-Santos, Â. A Cell’s Fate: An Overview of the Molecular Biology and Genetics of Apoptosis. Int. J. Mol. Sci. 2019, 20, 4133. [Google Scholar] [CrossRef]
- Judice, W.A.S.; Ferraz, L.S.; Lopes, R.D.M.; Vianna, L.D.S.; Siqueira, F.D.S.; Di Iorio, J.F.; Dalzoto, L.D.A.M.; Trujilho, M.N.R.; Santos, T.D.R.; Machado, M.F.M.; et al. Cysteine Proteases as Potential Targets for Anti-Trypanosomatid Drug Discovery. Bioorg. Med. Chem. 2021, 46, 116365. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, G.; Samal, D.; Khandayataray, P.; Murthy, M.K. A Review on Caspases: Key Regulators of Biological Activities and Apoptosis. Mol. Neurobiol. 2023, 60, 5805–5837. [Google Scholar] [CrossRef]
- Green, D.R. Caspases and Their Substrates. Cold Spring Harb. Perspect. Biol. 2022, 14, a041012. [Google Scholar] [CrossRef]
- Yadav, P.; Yadav, R.; Jain, S.; Vaidya, A. Caspase-3: A Primary Target for Natural and Synthetic Compounds for Cancer Therapy. Chem. Biol. Drug Des. 2021, 98, 144–165. [Google Scholar] [CrossRef]
- Carpenter, R.; Brady, M.F. BAX Gene. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Vucicevic, K.; Jakovljevic, V.; Colovic, N.; Tosic, N.; Kostic, T.; Glumac, I.; Pavlovic, S.; Karan-Djurasevic, T.; Colovic, M. Association of Bax Expression and Bcl2/Bax Ratio with Clinical and Molecular Prognostic Markers in Chronic Lymphocytic Leukemia. J. Med. Biochem. 2016, 35, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, C.J.A.; Cong, S.; Chan, X.; Yap, E.P.; Yu, F.; Hausenloy, D.J. Oxidative Stress in Cardiac Hypertrophy: From Molecular Mechanisms to Novel Therapeutic Targets. Free Radic. Biol. Med. 2021, 166, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Fang, T.; Huang, J.; Guo, Y.; Alam, M.; Qian, H. Hypertrophic Cardiomyopathy: From Phenotype and Pathogenesis to Treatment. Front. Cardiovasc. Med. 2021, 8, 722340. [Google Scholar] [CrossRef] [PubMed]
- Frey, N.; Luedde, M.; Katus, H.A. Mechanisms of Disease: Hypertrophic Cardiomyopathy. Nat. Rev. Cardiol. 2012, 9, 91–100. [Google Scholar] [CrossRef]
- Frontiers Production Office Erratum: Hypertrophic Cardiomyopathy: Mutations to Mechanisms to Therapies. Front. Physiol. 2022, 13, 1111059. [CrossRef]
- Sheng, S.; Li, J.; Hu, X.; Wang, Y. Regulated Cell Death Pathways in Cardiomyopathy. Acta Pharmacol. Sin. 2023, 44, 1521–1535. [Google Scholar] [CrossRef]
- Li, K.; Ma, L.; Lu, Z.; Yan, L.; Chen, W.; Wang, B.; Xu, H.; Asemi, Z. Apoptosis and Heart Failure: The Role of Non-Coding RNAs and Exosomal Non-Coding RNAs. Pathol.—Res. Pract. 2023, 248, 154669. [Google Scholar] [CrossRef]
- Lee, Y.; Gustafsson, Å.B. Role of Apoptosis in Cardiovascular Disease. Apoptosis 2009, 14, 536–548. [Google Scholar] [CrossRef]
- Leri, A.; Rota, M.; Pasqualini, F.S.; Goichberg, P.; Anversa, P. Origin of Cardiomyocytes in the Adult Heart. Circ. Res. 2015, 116, 150–166. [Google Scholar] [CrossRef]
- Kaur, N.; Ruiz-Velasco, A.; Raja, R.; Howell, G.; Miller, J.M.; Abouleisa, R.R.E.; Ou, Q.; Mace, K.; Hille, S.S.; Frey, N.; et al. Paracrine Signal Emanating from Stressed Cardiomyocytes Aggravates Inflammatory Microenvironment in Diabetic Cardiomyopathy. iScience 2022, 25, 103973. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, A.Y.; Blagonravov, M.L.; Neborak, E.V.; Syatkin, S.P.; Sklifasovskaya, A.P.; Semyatov, S.M.; Agostinelli, E. BCL2-regulated Apoptotic Process in Myocardial Ischemia-reperfusion Injury (Review). Int. J. Mol. Med. 2021, 47, 23–36. [Google Scholar] [CrossRef]
- Latif, N.; Khan, M.A.; Birks, E.; O’Farrell, A.; Westbrook, J.; Dunn, M.J.; Yacoub, M.H. Upregulation of the Bcl-2 Family of Proteins in End Stage Heart Failure. J. Am. Coll. Cardiol. 2000, 35, 1769–1777. [Google Scholar] [CrossRef]
- Long, X.; Boluyt, M.O.; Hipolito, M.L.; Lundberg, M.S.; Zheng, J.S.; O’Neill, L.; Cirielli, C.; Lakatta, E.G.; Crow, M.T. P53 and the Hypoxia-Induced Apoptosis of Cultured Neonatal Rat Cardiac Myocytes. J. Clin. Investig. 1997, 99, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Jiang, H.; Zou, Y.; Song, C.; Cao, K.; Chen, S.; Wu, Y.; Zhang, Z.; Geng, D.; Zhang, N.; et al. Programmed Death of Cardiomyocytes in Cardiovascular Disease and New Therapeutic Approaches. Pharmacol. Res. 2024, 206, 107281. [Google Scholar] [CrossRef]
- Yang, Y.; Duan, W.; Jin, Z.; Yi, W.; Yan, J.; Zhang, S.; Wang, N.; Liang, Z.; Li, Y.; Chen, W.; et al. JAK 2/STAT 3 Activation by Melatonin Attenuates the Mitochondrial Oxidative Damage Induced by Myocardial Ischemia/Reperfusion Injury. J. Pineal Res. 2013, 55, 275–286. [Google Scholar] [CrossRef]
- Ma, N.; Bai, J.; Zhang, W.; Luo, H.; Zhang, X.; Liu, D.; Qiao, C. Trimetazidine Protects against Cardiac Ischemia/Reperfusion Injury via Effects on Cardiac miRNA-21 Expression, Akt and the Bcl-2/Bax Pathway. Mol. Med. Rep. 2016, 14, 4216–4222. [Google Scholar] [CrossRef]
- Wolf, C.M. Hypertrophic Cardiomyopathy: Genetics and Clinical Perspectives. Cardiovasc. Diagn. Ther. 2019, 9, S388–S415. [Google Scholar] [CrossRef]
- Li, J.; Feng, X.; Wei, X. Modeling Hypertrophic Cardiomyopathy with Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. Stem Cell Res. Ther. 2022, 13, 232. [Google Scholar] [CrossRef]
- Girolami, F.; Ho, C.Y.; Semsarian, C.; Baldi, M.; Will, M.L.; Baldini, K.; Torricelli, F.; Yeates, L.; Cecchi, F.; Ackerman, M.J.; et al. Clinical Features and Outcome of Hypertrophic Cardiomyopathy Associated with Triple Sarcomere Protein Gene Mutations. J. Am. Coll. Cardiol. 2010, 55, 1444–1453. [Google Scholar] [CrossRef]
- Basit, H.; Alahmadi, M.H.; Rout, P.; Sharma, S. Hypertrophic Cardiomyopathy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Writing Committee Members; Ommen, S.R.; Mital, S.; Burke, M.A.; Day, S.M.; Deswal, A.; Elliott, P.; Evanovich, L.L.; Hung, J.; Joglar, J.A.; et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients with Hypertrophic Cardiomyopathy: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2020, 142, e558–e631. [Google Scholar] [CrossRef] [PubMed]
- Tuohy, C.V.; Kaul, S.; Song, H.K.; Nazer, B.; Heitner, S.B. Hypertrophic Cardiomyopathy: The Future of Treatment. Eur. J. Heart Fail. 2020, 22, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J. Clinical Course and Management of Hypertrophic Cardiomyopathy. N. Engl. J. Med. 2018, 379, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Chomzynski, P.; Sacchi, N. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate–Phenol–Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]


| Location | Patients | Sex (F/M) | Age | NYHA Class (I/II/III) |
|---|---|---|---|---|
| Group 1 | 43 | 13/30 | 60.4 ± 10.3 | 19/20/4 |
| Group 2 | 50 | 22/28 | 53.5 ± 13.1 | 30/18/2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavić, J.; Živanović, M.; Tanasković, I.; Pavić, O.; Stanković, V.; Virijević, K.; Mladenović, T.; Košarić, J.; Milićević, B.; Qamar, S.U.R.; et al. A Machine Learning Approach to Gene Expression in Hypertrophic Cardiomyopathy. Pharmaceuticals 2024, 17, 1364. https://doi.org/10.3390/ph17101364
Pavić J, Živanović M, Tanasković I, Pavić O, Stanković V, Virijević K, Mladenović T, Košarić J, Milićević B, Qamar SUR, et al. A Machine Learning Approach to Gene Expression in Hypertrophic Cardiomyopathy. Pharmaceuticals. 2024; 17(10):1364. https://doi.org/10.3390/ph17101364
Chicago/Turabian StylePavić, Jelena, Marko Živanović, Irena Tanasković, Ognjen Pavić, Vesna Stanković, Katarina Virijević, Tamara Mladenović, Jelena Košarić, Bogdan Milićević, Safi Ur Rehman Qamar, and et al. 2024. "A Machine Learning Approach to Gene Expression in Hypertrophic Cardiomyopathy" Pharmaceuticals 17, no. 10: 1364. https://doi.org/10.3390/ph17101364
APA StylePavić, J., Živanović, M., Tanasković, I., Pavić, O., Stanković, V., Virijević, K., Mladenović, T., Košarić, J., Milićević, B., Qamar, S. U. R., Velicki, L., Novaković, I., Preveden, A., Popović, D., Tesić, M., Seman, S., & Filipović, N. (2024). A Machine Learning Approach to Gene Expression in Hypertrophic Cardiomyopathy. Pharmaceuticals, 17(10), 1364. https://doi.org/10.3390/ph17101364














