Cytocidal Effect of Irradiation on Gastric Cancer Cells Infected with a Recombinant Mammalian Orthoreovirus Expressing a Membrane-Targeted KillerRed
Abstract
1. Introduction
2. Results
2.1. Generation of a Recombinant MRV-Expressing KRmem
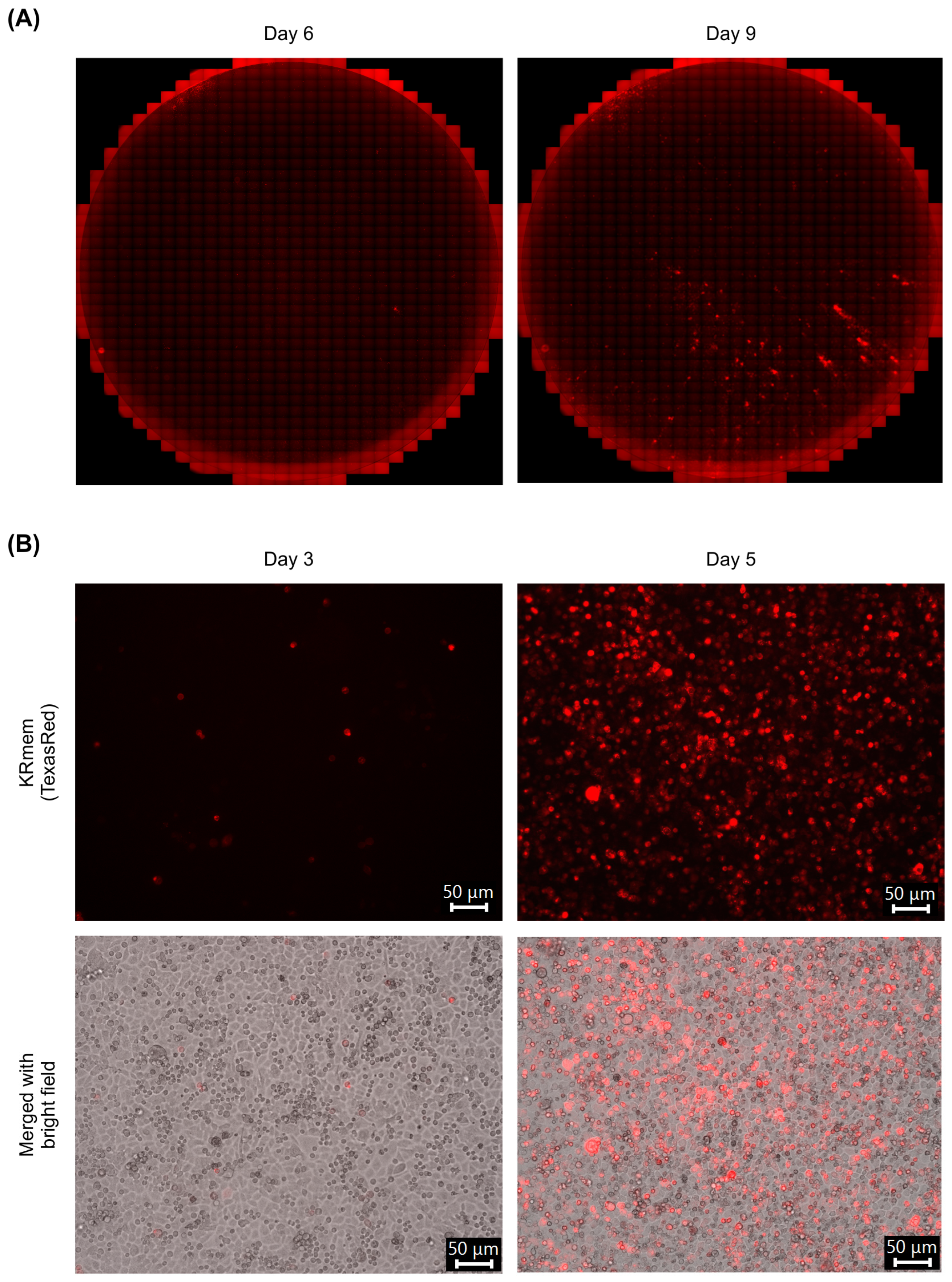
2.2. Characterization of the Recombinant MRV-Expressing KRmem
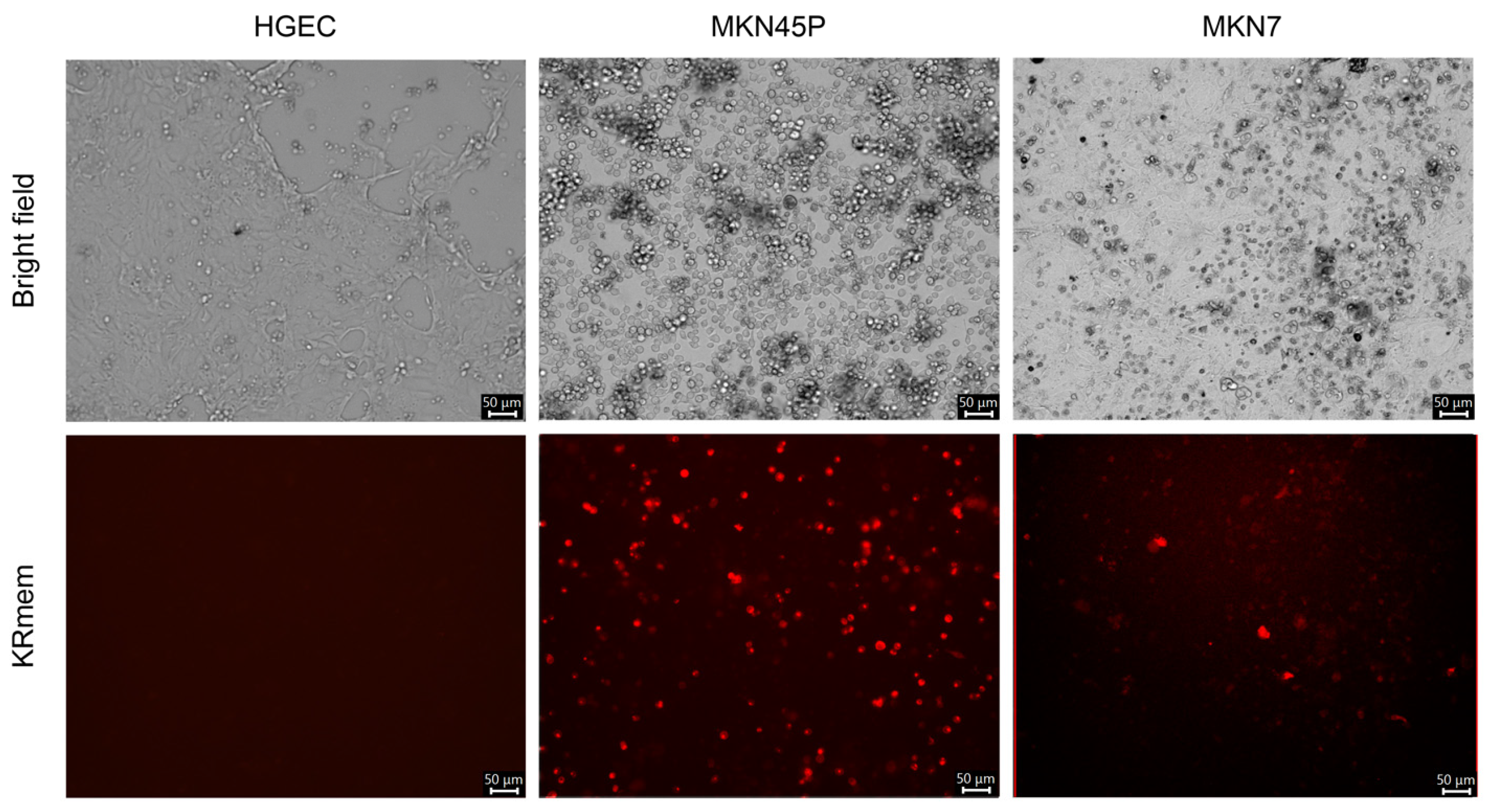
2.3. Infection of Cell Lines with the Recombinant MRV-Expressing KRmem
2.4. Cytocidal Effect of Irradiation on MKN45P Cells Infected with the Recombinant MRV-Expressing KRmem
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. Plasmids
4.3. Recovery of Recombinant MRV
4.4. Virus Titration
4.5. RNA Electrophoresis
4.6. Immunofluorescent Assay
4.7. Western Blot
4.8. Virus Growth in Cells
4.9. Infection of HGECs with Recombinant MRV
4.10. Infection of the GC Cell Lines with Recombinant MRV
4.11. Photodynamic Effect on the Recombinant MRV-Infected GC Cell Line
4.12. Statistical Analysis
4.13. Image Processing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Ohtsu, A.; Shimada, Y.; Shirao, K.; Boku, N.; Hyodo, I.; Saito, H.; Yamamichi, N.; Miyata, Y.; Ikeda, N.; Yamamoto, S.; et al. Randomized phase III trial of fluorouracil alone versus fluorouracil plus cisplatin versus uracil and tegafur plus mitomycin in patients with unresectable, advanced gastric cancer: The Japan Clinical Oncology Group Study (JCOG9205). J. Clin. Oncol. 2003, 21, 54–59. [Google Scholar] [CrossRef]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastrooesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Valentini, V.; Cellini, F.; Minsky, B.D.; Mattiucci, G.C.; Balducci, M.; D’Agostino, G.; D’Angelo, E.; Dinapoli, N.; Nicolotti, N.; Valentini, C.; et al. Survival after radiotherapy in gastric cancer: Systematic review and meta-analysis. Radiother. Oncol. 2009, 92, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, T.J.; Gomer, C.J.; Henderson, B.W.; Jori, G.; Kessel, D.; Korbelik, M.; Moan, J.; Peng, Q. Photodynamic therapy. J. Natl. Cancer Inst. 1998, 90, 889–905. [Google Scholar] [CrossRef]
- Chen, J.; Keltner, L.; Christophersen, J.; Zheng, F.; Krouse, M.; Singhal, A.; Wang, S.S. New technology for deep light distribution in tissue for phototherapy. Cancer J. 2002, 8, 154–163. [Google Scholar] [CrossRef]
- Sibille, A.; Lambert, R.; Souquet, J.C.; Sabben, G.; Descos, F. Long-term survival after photodynamic therapy for esophageal cancer. Gastroenterology 1995, 108, 337–344. [Google Scholar] [CrossRef]
- Kato, H. Photodynamic therapy for lung cancer—A review of 19 years’ experience. J. Photochem. Photobiol. B 1998, 42, 96–99. [Google Scholar] [CrossRef]
- Muroya, T.; Sugishita, T.; Tenjin, Y.; Kunugi, T.; Umayahara, K.; Akiya, T.; Iwabuchi, H.; Sakunaga, H.; Sakamoto, M.; Sugishita, T.; et al. Application and prospect of PDT for cervical cancer. Porphyrins 1997, 7, 187–192. [Google Scholar]
- Chen, Q.; Xu, L.; Liang, C.; Wang, C.; Peng, R.; Liu, Z. Photothermal therapy with immune-adjuvant nanoparticles together with checkpoint blockade for effective cancer immunotherapy. Nat. Commun. 2016, 7, 13193. [Google Scholar] [CrossRef]
- Takenaka, F.; Kobayashi, K.; Kimura, S.; Ozeki, E.; Ohtsuki, T.; Kobuchi, H.; Matsuura, E. PET imaging utilizing 89Zr-labeled human antibody variant and theranostic technologies provided by a novel DDS carrier. Drug Deliv. Syst. 2018, 33, 214–222. [Google Scholar] [CrossRef]
- Fountzilas, C.; Patel, S.; Mahalingam, D. Review: Oncolytic virotherapy, updates and future directions. Oncotarget 2017, 8, 102617–102639. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Maciorowski, D. Advancing oncolytic virus therapy by understanding the biology. Nat. Rev. Clin. Oncol. 2021, 18, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Macedo, N.; Miller, D.M.; Haq, R.; Kaufman, H.L. Clinical landscape of oncolytic virus research in 2020. J. Immunother. Cancer 2020, 8, e001486. [Google Scholar] [CrossRef]
- Heise, C.; Sampson-Johannes, A.; Williams, A.; McCormick, F.; Von Hoff, D.D.; Kirn, D.H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat. Med. 1997, 3, 639–645. [Google Scholar] [CrossRef]
- Coffin, R. Interview with Robert Coffin, inventor of T-VEC: The first oncolytic immunotherapy approved for the treatment of cancer. Immunotherapy 2016, 8, 103–106. [Google Scholar] [CrossRef]
- Etoh, T.; Himeno, Y.; Matsumoto, T.; Aramaki, M.; Kawano, K.; Nishizono, A.; Kitano, S. Oncolytic viral therapy for human pancreatic cancer cells by reovirus. Clin. Cancer Res. 2003, 9, 1218–1223. [Google Scholar]
- Coffey, M.C.; Strong, J.E.; Forsyth, P.A.; Lee, P.W. Reovirus therapy of tumors with activated Ras pathway. Science 1998, 282, 1332–1334. [Google Scholar] [CrossRef]
- Twigger, K.; Roulstone, V.; Kyula, J.; Karapanagiotou, E.M.; Syrigos, K.N.; Morgan, R.; White, C.; Bhide, S.; Nuovo, G.; Coffey, M.; et al. Reovirus exerts potent oncolytic effects in head and neck cancer cell lines that are independent of signalling in the EGFR pathway. BMC Cancer 2012, 12, 368. [Google Scholar] [CrossRef]
- Errington, F.; White, C.L.; Twigger, K.R.; Rose, A.; Scott, K.; Steele, L.; Ilett, L.J.; Prestwich, R.; Pandha, H.S.; Coffey, M.; et al. Inflammatory tumour cell killing by oncolytic reovirus for the treatment of melanoma. Gene Ther. 2008, 15, 1257–1270. [Google Scholar] [CrossRef]
- Adair, R.A.; Scott, K.J.; Fraser, S.; Errington-Mais, F.; Pandha, H.; Coffey, M.; Selby, P.; Cook, G.P.; Vile, R.; Harrington, K.J.; et al. Cytotoxic and immune-mediated killing of human colorectal cancer by reovirus-loaded blood and liver mononuclear cells. Int. J. Cancer 2013, 132, 2327–2338. [Google Scholar] [CrossRef]
- Hirasawa, K.; Nishikawa, S.G.; Norman, K.L.; Alain, T.; Kossakowska, A.; Lee, P.W. Oncolytic reovirus against ovarian and colon cancer. Cancer Res. 2002, 62, 1696–1701. [Google Scholar]
- Norman, K.L.; Coffey, M.C.; Hirasawa, K.; Demetrick, D.J.; Nishikawa, S.G.; Difrancesco, L.M.; Strong, J.E.; Lee, P.W. Reovirus oncolysis of human breast cancer. Hum. Gene Ther. 2002, 13, 641–652. [Google Scholar] [CrossRef]
- Sei, S.; Mussio, J.K.; Yang, Q.E.; Nagashima, K.; Parchment, R.E.; Coffey, M.C.; Shoemaker, R.H.; Tomaszewski, J.E. Synergistic antitumor activity of oncolytic reovirus and chemotherapeutic agents in non-small cell lung cancer cells. Mol. Cancer 2009, 8, 47. [Google Scholar] [CrossRef]
- Thirukkumaran, C.M.; Nodwell, M.J.; Hirasawa, K.; Shi, Z.Q.; Diaz, R.; Luider, J.; Johnston, R.N.; Forsyth, P.A.; Magliocco, A.M.; Lee, P.; et al. Oncolytic viral therapy for prostate cancer: Efficacy of reovirus as a biological therapeutic. Cancer Res. 2010, 70, 2435–2444. [Google Scholar] [CrossRef]
- Hata, Y.; Etoh, T.; Inomata, M.; Shiraishi, N.; Nishizono, A.; Kitano, S. Efficacy of oncolytic reovirus against human breast cancer cells. Oncol. Rep. 2008, 19, 1395–1398. [Google Scholar] [PubMed][Green Version]
- Himeno, Y.; Etoh, T.; Matsumoto, T.; Ohta, M.; Nishizono, A.; Kitano, S. Efficacy of oncolytic reovirus against liver metastasis from pancreatic cancer in immunocompetent models. Int. J. Oncol. 2005, 27, 901–906. [Google Scholar] [CrossRef]
- Hirano, S.; Etoh, T.; Okunaga, R.; Shibata, K.; Ohta, M.; Nishizono, A.; Kitano, S. Reovirus inhibits the peritoneal dissemination of pancreatic cancer cells in an immunocompetent animal model. Oncol. Rep. 2009, 21, 1381–1384. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Etoh, T.; Suzuki, K.; Mitui, M.T.; Nishizono, A.; Shiraishi, N.; Kitano, S. Efficacy of oncolytic reovirus against human gastric cancer with peritoneal metastasis in experimental animal model. Int. J. Oncol. 2010, 37, 1433–1438. [Google Scholar] [CrossRef]
- Kobayashi, T.; Antar, A.A.; Boehme, K.W.; Danthi, P.; Eby, E.A.; Guglielmi, K.M.; Holm, G.H.; Johnson, E.M.; Maginnis, M.S.; Naik, S.; et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 2007, 1, 147–157. [Google Scholar] [CrossRef]
- Kanai, Y.; Kawagishi, T.; Matsuura, Y.; Kobayashi, T. In vivo live imaging of oncolytic mammalian Orthoreovirus expressing NanoLuc luciferase in tumor xenograft mice. J. Virol. 2019, 93, e00401–e00419. [Google Scholar] [CrossRef]
- Kemp, V.; van den Wollenberg, D.J.M.; Camps, M.G.M.; van Hall, T.; Kinderman, P.; Pronk-van Montfoort, N.; Hoeben, R.C. Arming oncolytic reovirus with GM-CSF gene to enhance immunity. Cancer Gene Ther. 2019, 26, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Yamada, K.; Etoh, T.; Kitagawa, M.; Shirasaka, Y.; Noguchi, K.; Kobayashi, T.; Nishizono, A.; Inomata, M. Development of an oncolytic mammalian Orthoreovirus expressing the near-infrared fluorescent protein iRFP720. J. Virol. Methods 2022, 308, 114574. [Google Scholar] [CrossRef]
- Bulina, M.E.; Chudakov, D.M.; Britanova, O.V.; Yanushevich, Y.G.; Staroverov, D.B.; Chepurnykh, T.V.; Merzlyak, E.M.; Shkrob, M.A.; Lukyanov, S.; Lukyanov, K.A. A genetically encoded photosensitizer. Nat. Biotechnol. 2006, 24, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Takehara, K.; Tazawa, H.; Okada, N.; Hashimoto, Y.; Kikuchi, S.; Kuroda, S.; Kishimoto, H.; Shirakawa, Y.; Narii, N.; Mizuguchi, H.; et al. Targeted photodynamic virotherapy armed with a genetically encoded photosensitizer. Mol. Cancer Ther. 2016, 15, 199–208. [Google Scholar] [CrossRef]
- Takehara, K.; Yano, S.; Tazawa, H.; Kishimoto, H.; Narii, N.; Mizuguchi, H.; Urata, Y.; Kagawa, S.; Fujiwara, T.; Hoffman, R.M. Eradication of melanoma in vitro and in vivo via targeting with a Killer-Red-containing telomerase-dependent adenovirus. Cell Cycle 2017, 16, 1502–1508. [Google Scholar] [CrossRef]
- Tran, V.; Moser, L.A.; Poole, D.S.; Mehle, A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 2013, 87, 13321–13329. [Google Scholar] [CrossRef]
- Fénéant, L.; Leske, A.; Günther, K.; Groseth, A. Generation of reporter-expressing New World arenaviruses: A systematic comparison. Viruses 2022, 14, 1563. [Google Scholar] [CrossRef]
- Demidenko, A.A.; Blattman, J.N.; Blattman, N.N.; Greenberg, P.D.; Nibert, M.L. Engineering recombinant reoviruses with tandem repeats and a tetravirus 2A-like element for exogenous polypeptide expression. Proc. Natl. Acad. Sci. USA 2013, 110, E1867–E1876. [Google Scholar] [CrossRef]
- Roner, M.R.; Steele, B.G. Features of the mammalian Orthoreovirus 3 Dearing l1 single-stranded RNA that direct packaging and serotype restriction. J. Gen. Virol. 2007, 88, 3401–3412. [Google Scholar] [CrossRef]
- Roner, M.R.; Steele, B.G. Localizing the reovirus packaging signals using an engineered m1 and S2 ssRNA. Virology 2007, 358, 89–97. [Google Scholar] [CrossRef]
- Martin, S.J.; Reutelingsperger, C.P.; McGahon, A.J.; Rader, J.A.; van Schie, R.C.; Laface, D.M.; Green, D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: Inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995, 182, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.; Richardson-Burns, S.M.; Debiasi, R.L.; Tyler, K.L. Mechanisms of apoptosis during reovirus infection. Curr. Top. Microbiol. Immunol. 2005, 289, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Qin, Y.; Feng, X. Advances in the genetically engineered KillerRed for photodynamic therapy applications. Int. J. Mol. Sci. 2021, 22, 10130. [Google Scholar] [CrossRef] [PubMed]
- Bulina, M.E.; Lukyanov, K.A.; Britanova, O.V.; Onichtchouk, D.; Lukyanov, S.; Chudakov, D.M. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat. Protoc. 2006, 1, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.A.; van Gemert, M.J.C. Lasers in medicine. In Electro-Optics Handbook, 2nd ed.; Waynant, R.W., Ediger, M.N., Eds.; McGraw-Hill Inc.: New York, NY, USA, 1993; pp. 1–32. [Google Scholar]
- Barton, E.S.; Forrest, J.C.; Connolly, J.L.; Chappell, J.D.; Liu, Y.; Schnell, F.J.; Nusrat, A.; Parkos, C.A.; Dermody, T.S. Junction adhesion molecule is a receptor for reovirus. Cell 2001, 104, 441–451. [Google Scholar] [CrossRef]
- Kirchner, E.; Guglielmi, K.M.; Strauss, H.M.; Dermody, T.S.; Stehle, T. Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A. PLoS Pathog. 2008, 4, e1000235. [Google Scholar] [CrossRef]
- Zhao, C.; Lu, F.; Chen, H.; Zhao, X.; Sun, J.; Chen, H. Dysregulation of JAM-A plays an important role in human tumor progression. Int. J. Clin. Exp. Pathol. 2014, 7, 7242–7248. [Google Scholar]
- Kawagishi, T.; Kanai, Y.; Nouda, R.; Fukui, I.; Nurdin, J.A.; Matsuura, Y.; Kobayashi, T. Generation of genetically RGD σ1-modified oncolytic reovirus that enhances JAM-A-independent infection of tumor cells. J. Virol. 2020, 94, e01703-20. [Google Scholar] [CrossRef]
- van den Wollenberg, D.J.; Dautzenberg, I.J.; van den Hengel, S.K.; Cramer, S.J.; de Groot, R.J.; Hoeben, R.C. Isolation of reovirus T3D mutants capable of infecting human tumor cells independent of junction adhesion molecule-A. PLoS ONE 2012, 7, e48064. [Google Scholar] [CrossRef]
- Stiff, A.; Caserta, E.; Sborov, D.W.; Nuovo, G.J.; Mo, X.; Schlotter, S.Y.; Canella, A.; Smith, E.; Badway, J.; Old, M.; et al. Histone Deacetylase Inhibitors Enhance the Therapeutic Potential of Reovirus in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 830–841. [Google Scholar] [CrossRef]
- Yang, L.; Gu, X.; Yu, J.; Ge, S.; Fan, X. Oncolytic virotherapy: From bench to bedside. Front. Cell Dev. Biol. 2021, 9, 790150. [Google Scholar] [CrossRef]
- Ito, N.; Takayama-Ito, M.; Yamada, K.; Hosokawa, J.; Sugiyama, M.; Minamoto, N. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 2003, 47, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transp. 2013, 48, 452–458. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirasaka, Y.; Yamada, K.; Etoh, T.; Noguchi, K.; Hasegawa, T.; Ogawa, K.; Kobayashi, T.; Nishizono, A.; Inomata, M. Cytocidal Effect of Irradiation on Gastric Cancer Cells Infected with a Recombinant Mammalian Orthoreovirus Expressing a Membrane-Targeted KillerRed. Pharmaceuticals 2024, 17, 79. https://doi.org/10.3390/ph17010079
Shirasaka Y, Yamada K, Etoh T, Noguchi K, Hasegawa T, Ogawa K, Kobayashi T, Nishizono A, Inomata M. Cytocidal Effect of Irradiation on Gastric Cancer Cells Infected with a Recombinant Mammalian Orthoreovirus Expressing a Membrane-Targeted KillerRed. Pharmaceuticals. 2024; 17(1):79. https://doi.org/10.3390/ph17010079
Chicago/Turabian StyleShirasaka, Yoshinori, Kentaro Yamada, Tsuyoshi Etoh, Kazuko Noguchi, Takumi Hasegawa, Katsuhiro Ogawa, Takeshi Kobayashi, Akira Nishizono, and Masafumi Inomata. 2024. "Cytocidal Effect of Irradiation on Gastric Cancer Cells Infected with a Recombinant Mammalian Orthoreovirus Expressing a Membrane-Targeted KillerRed" Pharmaceuticals 17, no. 1: 79. https://doi.org/10.3390/ph17010079
APA StyleShirasaka, Y., Yamada, K., Etoh, T., Noguchi, K., Hasegawa, T., Ogawa, K., Kobayashi, T., Nishizono, A., & Inomata, M. (2024). Cytocidal Effect of Irradiation on Gastric Cancer Cells Infected with a Recombinant Mammalian Orthoreovirus Expressing a Membrane-Targeted KillerRed. Pharmaceuticals, 17(1), 79. https://doi.org/10.3390/ph17010079





