New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies
Abstract
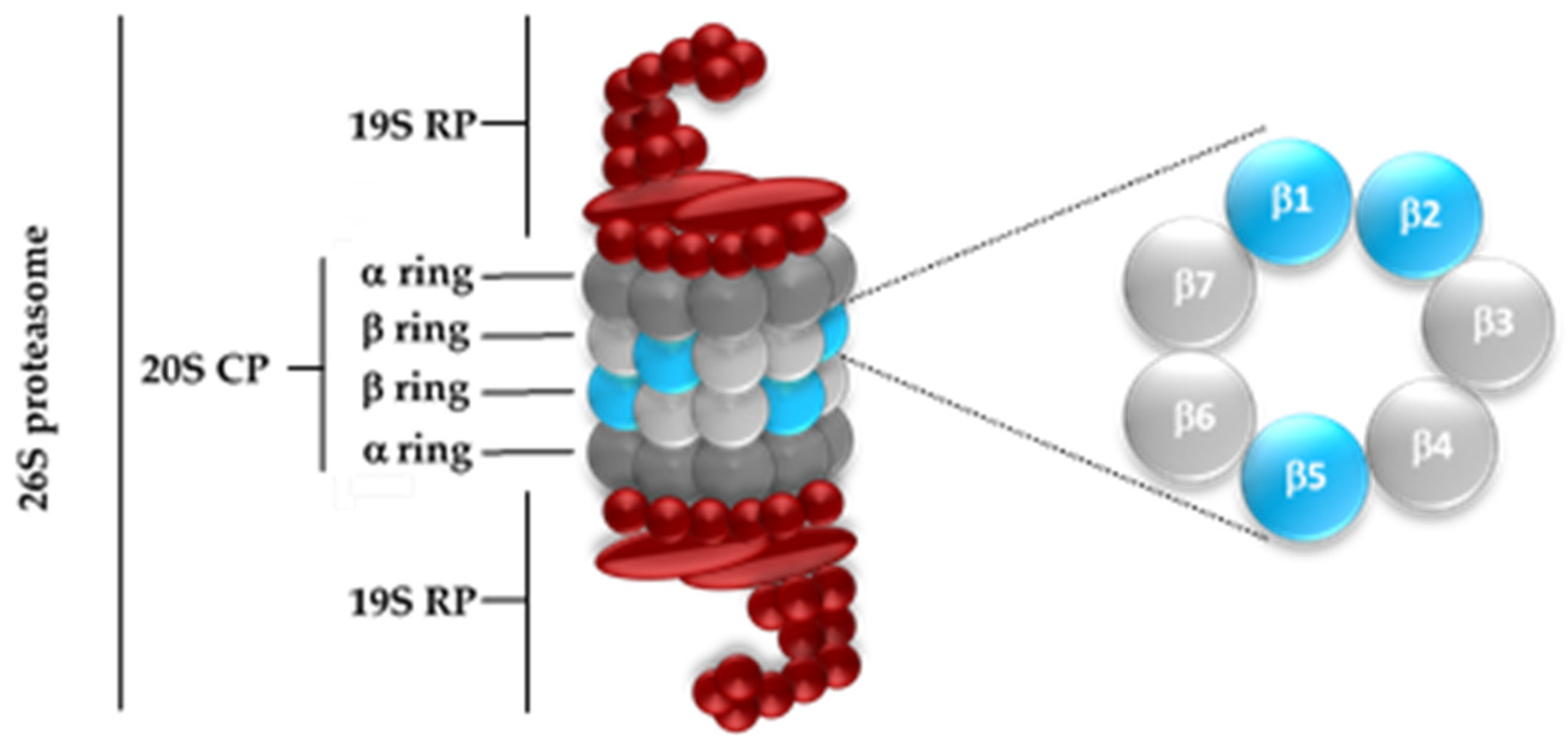
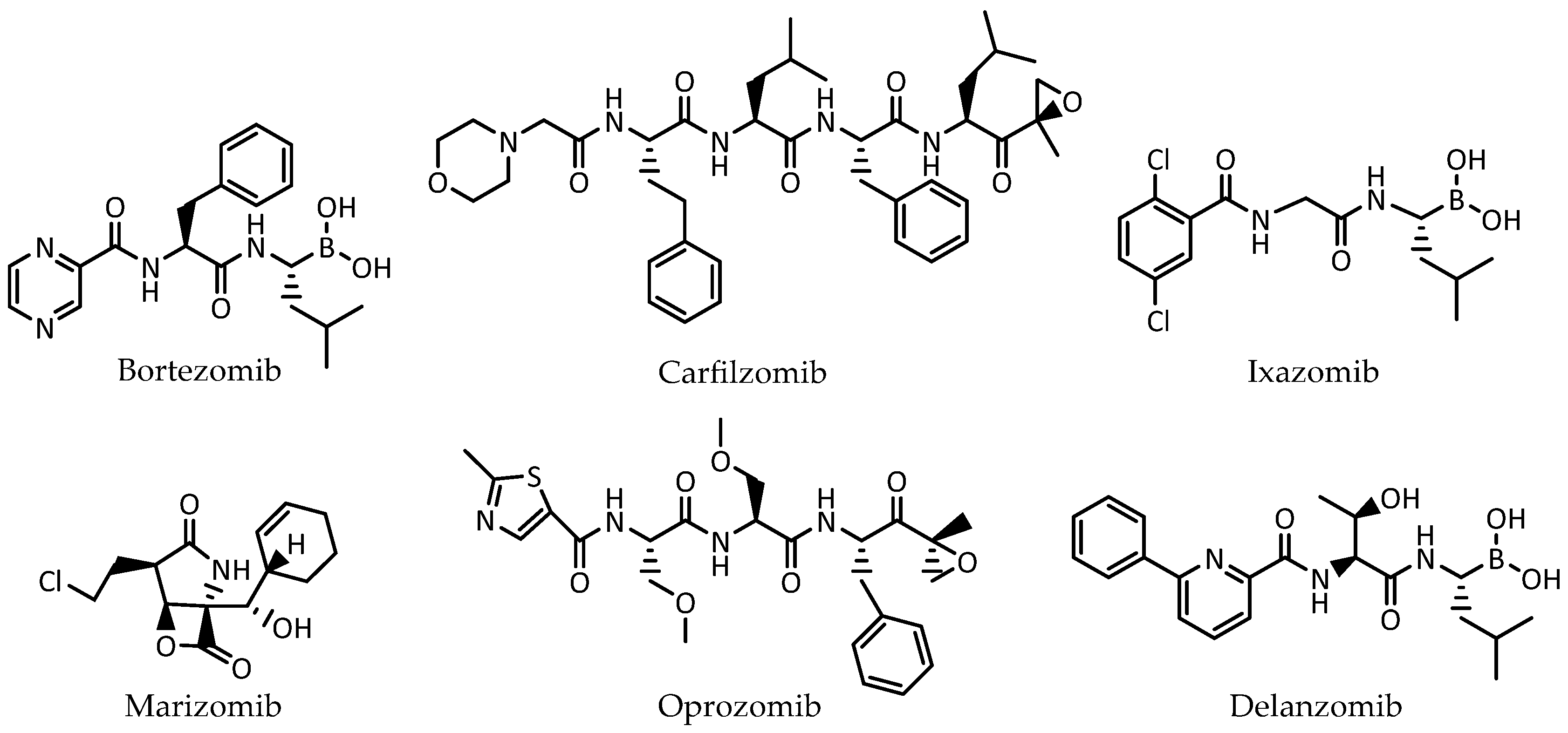
1. Introduction
2. Results and Discussion
2.1. Structure-Based Virtual Screening
2.1.1. Databases
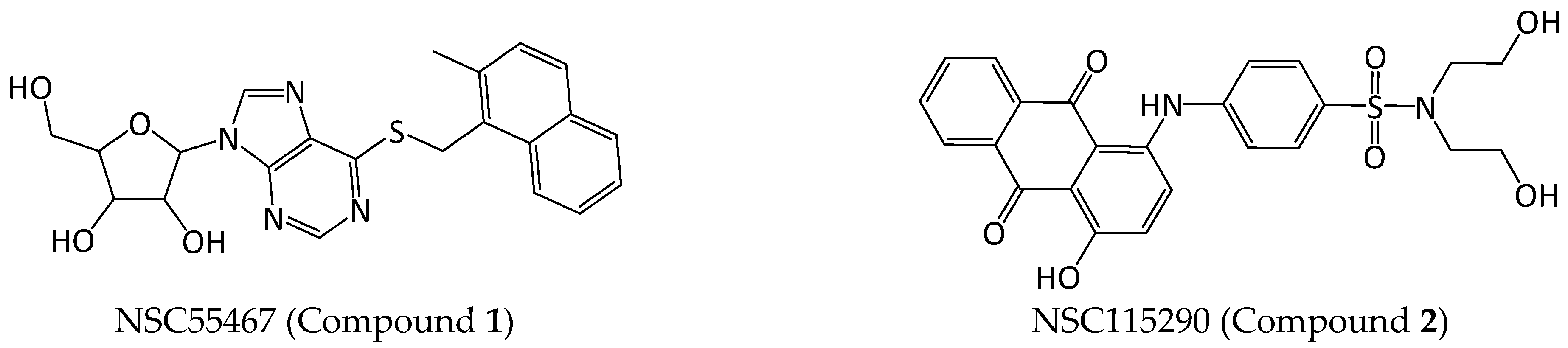
2.1.2. Compound Selection
2.2. In Vitro Studies
2.2.1. Proteasome Inhibition Assays in Isolated Human 20S Proteasomes
2.2.2. Proteasome Inhibition Assays in Cell Lysates
2.2.3. Cell Viability Assays
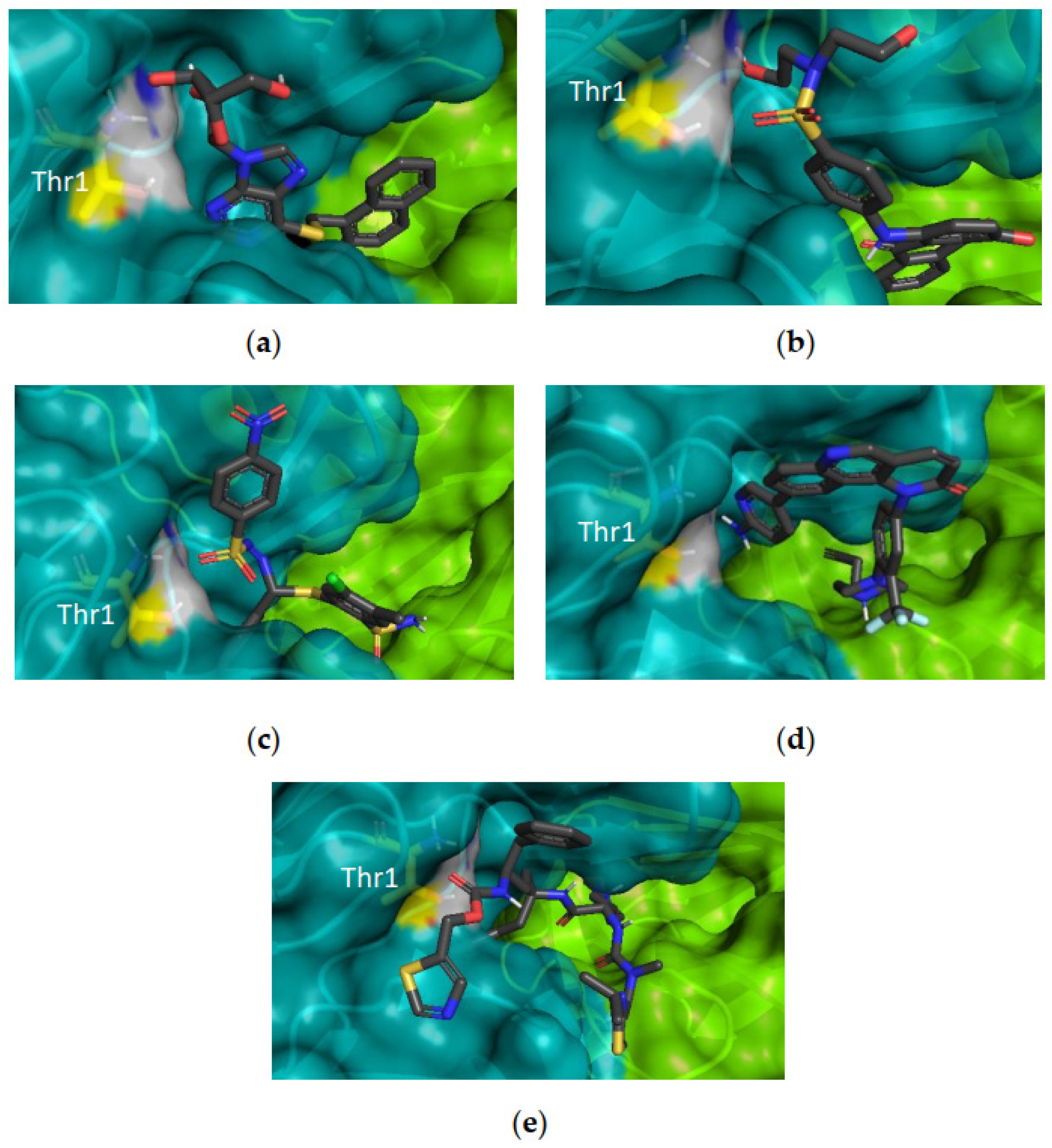
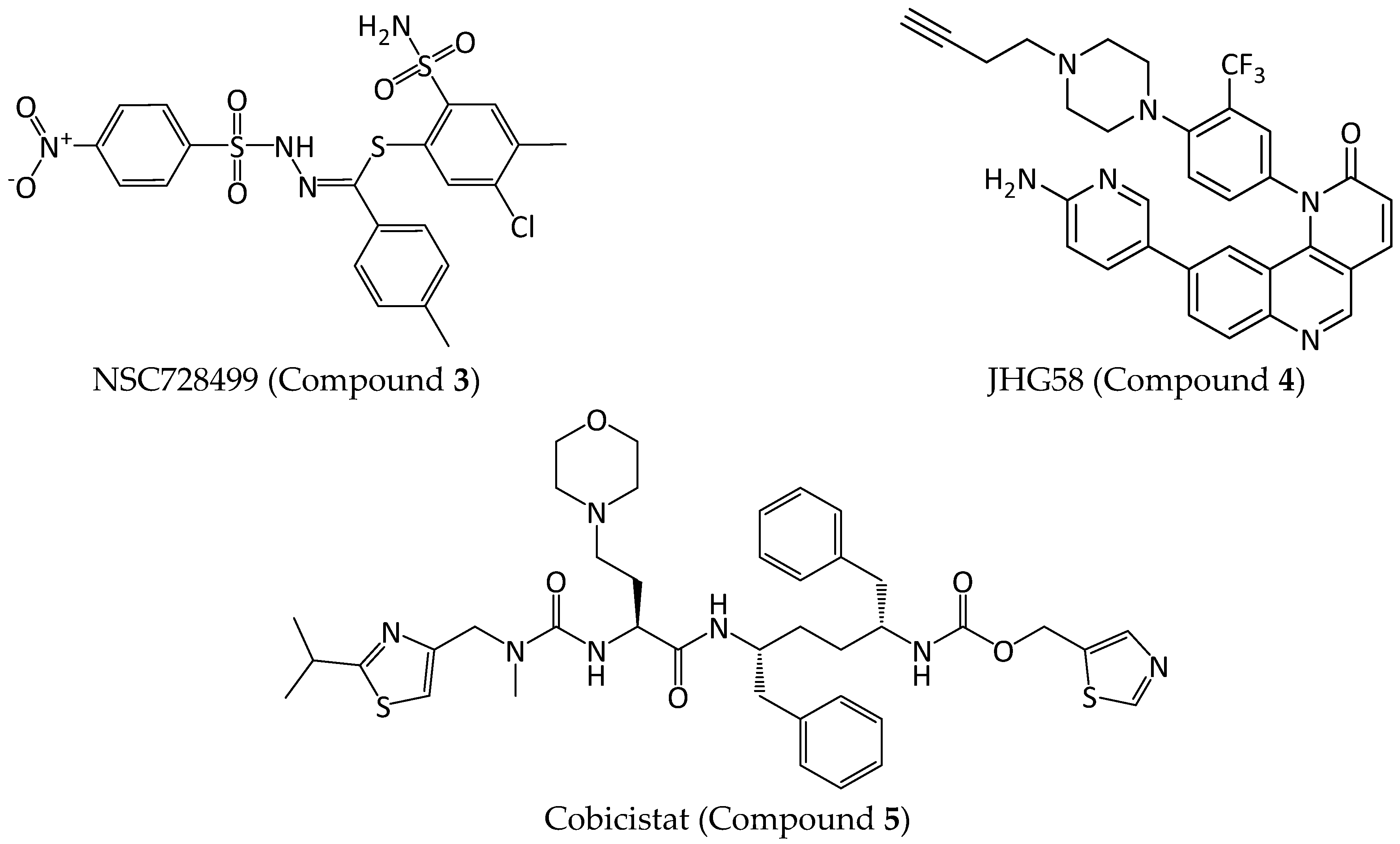
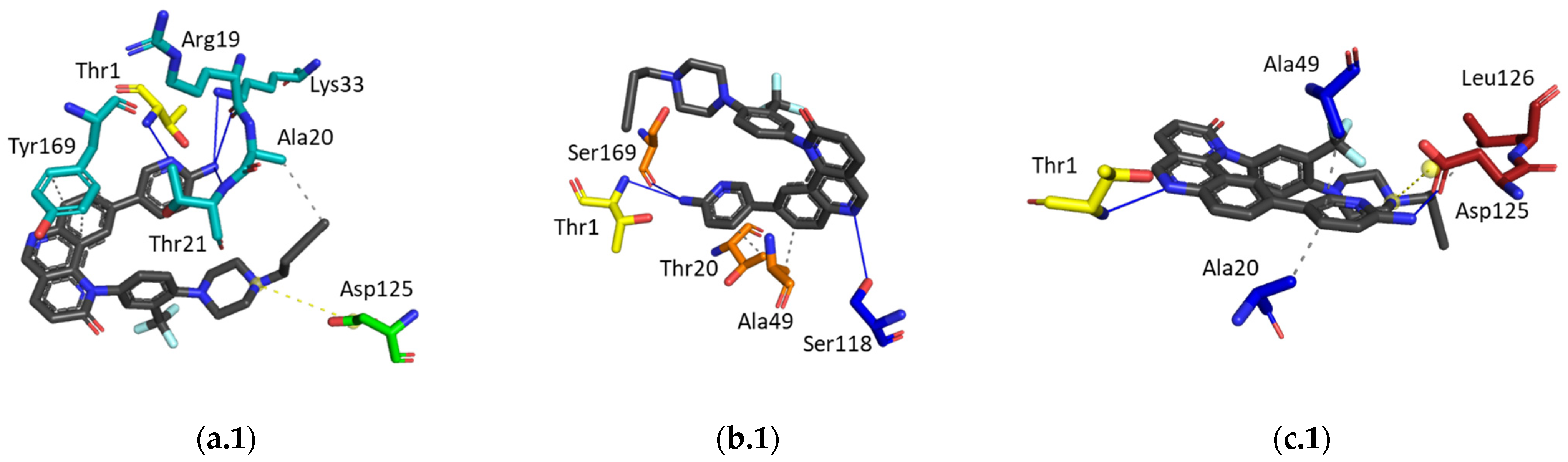
2.2.4. Hit Compound Selection and Docking Studies
3. Materials and Methods
3.1. In Silico Studies
3.1.1. Protein Structure Preparation
3.1.2. Databases Preparation
3.1.3. Molecular Docking for Structure-based Virtual Screening and for the Compounds Evaluated in Biological Assays
3.1.4. Ligand Interaction Patterns
Noncovalent Interaction Patterns for Individual Ligands
3.1.5. Assessment of the Presence of Potential Aggregators and PAINS
3.2. Preparation of 9-(6-Aminopyridin-3-yl)-1-(4-(4-(but-3-yn-1-yl)piperazin-1-yl)-3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (JHG58, Compound 4)
3.3. In Vitro Assays
3.3.1. Proteasome Inhibition Assays in Isolated 20S Proteasomes
3.3.2. Cell Culture
3.3.3. Proteasome Inhibition Assay in Cell Lysates
3.3.4. Assessment of Cell Viability using the MTT Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization|Cancer. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cancer (accessed on 12 December 2022).
- Brancolini, C. Inhibitors of the Ubiquitin-Proteasome System and the cell death machinery: How many pathways are activated? Curr. Mol. Pharmacol. 2008, 1, 24–37. [Google Scholar] [CrossRef]
- Jung, T.; Grune, T. Structure of the Proteasome, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2012; Volume 109. [Google Scholar]
- Kim, H.M.; Yu, Y.; Cheng, Y. Structure characterization of the 26S proteasome. Biochim. Biophys. Acta 2011, 1809, 67–79. [Google Scholar] [CrossRef]
- Ciechanover, A. Intracellular protein degradation from a vague idea through the lysosome and the ubiquitin-proteasome system and on to Human Diseases and Drug Targeting. Ann. N. Y. Acad. Sci. 2007, 1116, 1–28. [Google Scholar] [CrossRef]
- Ciechanover, A. The ubiquitin–proteasome pathway: On protein death and cell life. EMBO J. 1998, 17, 7151–7160. [Google Scholar] [CrossRef]
- da Fonseca, P.C.A.; He, J.; Morris, E.P. Molecular model of the human 26S proteasome. Mol. Cell 2012, 46, 54–66. [Google Scholar] [CrossRef]
- Borissenko, L.; Groll, M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem. Rev. 2007, 107, 687–717. [Google Scholar] [CrossRef]
- Kao, A.; Randall, A.; Yang, Y.; Patel, V.R.; Kandur, W.; Guan, S.; Rychnovsky, S.D.; Baldi, P.; Huang, L. Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. MCP Pap. Press 2012, 11, 1566–1577. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Endo, A.; Saeki, Y. Multi-step ubiquitin decoding mechanism for proteasomal degradation. Pharmaceuticals 2020, 13, 128. [Google Scholar] [CrossRef]
- Hochstrasser, M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 1995, 7, 215–223. [Google Scholar] [CrossRef]
- Adams, J. The proteasome: Structure, function, and role in the cell. Cancer Treat. Rev. 2003, 29, 3–9. [Google Scholar] [CrossRef]
- Jung, T.; Catalgol, B.; Grune, T. The proteasomal system. Mol. Aspects Med. 2009, 30, 191–296. [Google Scholar] [CrossRef]
- Orabi, K.Y.; Abaza, M.S.; El Sayed, K.A.; Elnagar, A.Y.; Al-Attiyah, R.; Guleri, R.P. Selective growth inhibition of human malignant melanoma cells by syringic acid-derived proteasome inhibitors. Cancer Cell Int. 2013, 13, 82. [Google Scholar] [CrossRef]
- Teicher, B.A.; Tomaszewski, J.E. Proteasome inhibitors. Biochem. Pharmacol. 2015, 96, 1–9. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Groettrup, M. Subunit specific inhibitors of proteasomes and their potential for immunomodulation. Curr. Opin. Chem. Biol. 2014, 23, 16–22. [Google Scholar] [CrossRef]
- Packham, G. The role of NF-κB in lymphoid malignancies. Br. J. Haematol. 2008, 143, 3–15. [Google Scholar] [CrossRef]
- da Fonseca, P.C.A.; Morris, E.P. Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 2008, 283, 23305–23314. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Garcia-Calvo, M.; Overkleeft, H.S.; Peterson, E.; Pennington, M.W.; Ploegh, H.L.; Thornberry, N.A.; Goldberg, A.L. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 2003, 278, 35869–35877. [Google Scholar] [CrossRef]
- Blackburn, C.; Gigstad, K.M.; Hales, P.; Garcia, K.; Jones, M.; Bruzzese, F.J.; Barrett, C.; Liu, J.X.; Soucy, T.A.; Sappal, D.S.; et al. Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit. Biochem. J. 2010, 430, 461–476. [Google Scholar] [CrossRef]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer Drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Huber, E.M.; Groll, M. Inhibitors for the immuno- and constitutive proteasome: Current and future trends in drug development. Angew. Chem. Int. Ed. Engl. 2012, 51, 8708–8720. [Google Scholar] [CrossRef]
- Groll, M.; Koguchi, Y.; Huber, R.; Kohno, J. Crystal structure of the 20 S proteasome:TMC-95A complex: A non-covalent proteasome inhibitor. J. Mol. Biol. 2001, 311, 543–548. [Google Scholar] [CrossRef]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Garrigues, L.; Amalric, F.; Vigneron, N.; Menneteau, T.; Stella, A.; Monsarrat, B.; Eynde, B. Van Den Deciphering preferential interactions within supramolecular protein complexes: The proteasome case. Mol. Syst. Biol. 2015, 11, 771. [Google Scholar] [CrossRef]
- Dahlmann, B. Mammalian proteasome subtypes: Their diversity in structure and function. Arch. Biochem. Biophys. 2016, 591, 132–140. [Google Scholar] [CrossRef]
- Loizidou, E.Z.; Zeinalipour-Yazdi, C.D. Computational inhibition studies of the human proteasome by argyrin-based analogues with subunit specificity. Chem. Biol. Drug Des. 2014, 84, 99–107. [Google Scholar] [CrossRef]
- Oinonen, C.; Rouvinen, J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000, 9, 2329–2337. [Google Scholar] [CrossRef]
- Verbrugge, S.E.; Scheper, R.J.; Lems, W.F.; de Gruijl, T.D.; Jansen, G. Proteasome inhibitors as experimental therapeutics of autoimmune diseases. Arthritis Res. Ther. 2015, 17, 17. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Diez-Rivero, C.M.; Lafuente, E.M.; Reche, P.A. Computational analysis and modeling of cleavage by the immunoproteasome and the constitutive proteasome. BMC Bioinform. 2010, 11, 479. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, X.; Zhu, X.; Wu, G.; Li, Y.; Ma, Y.; Yuan, Y.; Yang, J.; Hu, Y.; Ai, L.; et al. Design, synthesis, biological evaluation, and structure-activity relationship (SAR) discussion of dipeptidyl boronate proteasome inhibitors, part I: Comprehensive understanding of the SAR of alpha-amino acid boronates. J. Med. Chem. 2009, 52, 4192–4199. [Google Scholar] [CrossRef]
- Beck, P.; Dubiella, C.; Groll, M. Covalent and non-covalent reversible proteasome inhibition. Biol. Chem. 2012, 393, 1101–1120. [Google Scholar] [CrossRef]
- Demo, S.D.; Kirk, C.J.; Aujay, M.A.; Buchholz, T.J.; Dajee, M.; Ho, M.N.; Jiang, J.; Laidig, G.J.; Lewis, E.R.; Parlati, F.; et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007, 67, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Kupperman, E.; Lee, E.C.; Cao, Y.; Bannerman, B.; Fitzgerald, M.; Berger, A.; Yu, J.; Yang, Y.; Hales, P.; Bruzzese, F.; et al. Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res. 2010, 70, 1970–1980. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Catley, L.; Li, G.; Podar, K.; Hideshima, T.; Velankar, M.; Mitsiades, C.; Mitsiades, N.; Yasui, H.; Letai, A.; et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell 2005, 8, 407–419. [Google Scholar] [CrossRef]
- European Medicines Agency. Available online: https://www.ema.europa.eu/en (accessed on 27 July 2019).
- U.S. Food and Drug Administration. Available online: https://www.fda.gov/ (accessed on 27 July 2019).
- Jayaweera, S.P.E.; Wanigasinghe Kanakanamge, S.P.; Rajalingam, D.; Silva, G.N. Carfilzomib: A promising proteasome inhibitor for the treatment of relapsed and refractory multiple myeloma. Front. Oncol. 2021, 11, 740796. [Google Scholar] [CrossRef]
- Elbezanti, W.O.; Challagundla, K.B.; Jonnalagadda, S.C.; Budak-alpdogan, T.; Pandey, M.K. Past, present, and a glance into the future of multiple myeloma treatment. Pharmaceuticals 2023, 16, 415. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein−ligand docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- NCI Database Download Page. Available online: https://cactus.nci.nih.gov/download/nci/ (accessed on 17 September 2019).
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef]
- Piva, R.; Ruggeri, B.; Williams, M.; Costa, G.; Tamagno, M.; Ferrero, D.; Giai, V.; Coscia, M.; Peola, S.; Massaia, M.; et al. CEP-18770: A novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood 2008, 111, 2765–2775. [Google Scholar] [CrossRef]
- Desvergne, A.; Genin, E.; Maréchal, X.; Gallastegui, N.; Dufau, L.; Richy, N.; Groll, M.; Vidal, J.; Reboud-Ravaux, M. Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S Proteasome. J. Med. Chem. 2013, 56, 3367–3378. [Google Scholar] [CrossRef]
- Hines, J.; Groll, M.; Fahnestock, M.; Crews, C.M. Proteasome inhibition by Fellutamide B induces Nerve Growth Factor synthesis. Chem. Biol. 2008, 15, 501–512. [Google Scholar] [CrossRef]
- Zhou, H.J.; Aujay, M.A.; Bennett, M.K.; Dajee, M.; Demo, S.D.; Fang, Y.; Ho, M.N.; Jiang, J.; Kirk, C.J.; Laidig, G.J.; et al. Design and synthesis of an orally bioavailable and selective peptide epoxyketone proteasome inhibitor (PR-047). J. Med. Chem. 2009, 52, 3028–3038. [Google Scholar] [CrossRef]
- Marastoni, M.; Baldisserotto, A.; Trapella, C.; Gavioli, R.; Tomatis, R. P3 and P4 position analysis of vinyl ester pseudopeptide proteasome inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 3125–3130. [Google Scholar] [CrossRef]
- Chen, D.; Daniel, K.G.; Chen, M.S.; Kuhn, D.J.; Landis-Piwowar, K.R.; Dou, Q.P. Dietary flavonoids as proteasome inhibitors and apoptosis inducers in human leukemia cells. Biochem. Pharmacol. 2005, 69, 1421–1432. [Google Scholar] [CrossRef]
- Dreiseitel, A.; Schreier, P.; Oehme, A.; Locher, S.; Rogler, G.; Piberger, H.; Hajak, G.; Sand, P.G. Inhibition of proteasome activity by anthocyanins and anthocyanidins. Biochem. Biophys. Res. Commun. 2008, 372, 57–61. [Google Scholar] [CrossRef]
- DeGoey, D.A.; Chen, H.-J.; Cox, P.B.; Wendt, M.D. Beyond the Rule of 5: Lessons learned from AbbVie’s drugs and compound collection. J. Med. Chem. 2018, 61, 2636–2651. [Google Scholar] [CrossRef]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef]
- Waszkowycz, B. Towards improving compound selection in structure-based virtual screening. Drug Discov. Today 2008, 13, 219–226. [Google Scholar] [CrossRef]
- Vigneron, N.; Habib, J.A.; Van den Eynde, B.J. The capture proteasome assay (CAPA): A method to measure proteasome activity in vitro. Anal. Biochem. 2015, 482, 7–15. [Google Scholar] [CrossRef]
- Promega Proteasome-Glo™ Assays. Available online: https://worldwide.promega.com/products/protein-quantitation-and-detection/protease-assays/proteasome_glo-assays/?catNum=G8621 (accessed on 26 September 2019).
- Adams, J. Proteasome inhibitors as new anticancer drugs. Curr. Opin. Oncol. 2002, 14, 628–634. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Smith, D.M. A practical review of proteasome pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef]
- Unno, M.; Mizushima, T.; Morimoto, Y.; Tomisugi, Y.; Tanaka, K.; Yasuoka, N.; Tsukihara, T. The structure of the mammalian 20S Proteasome at 2.75 A Resolution. Structure 2002, 10, 609–618. [Google Scholar] [CrossRef]
- Harshbarger, W.; Miller, C.; Diedrich, C.; Sacchettini, J. Crystal structure of the human 20S Proteasome in complex with carfilzomib. Structure 2015, 23, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Hartshorn, M.J.; Verdonk, M.L.; Chessari, G.; Brewerton, S.C.; Mooij, W.T.M.; Mortenson, P.N.; Murray, C.W. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007, 50, 726–741. [Google Scholar] [CrossRef]
- Genetic Optimisation for Ligand Docking (GOLD). The Cambridge Crystallographic Data Centre (CCDC). Available online: https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/ (accessed on 26 August 2019).
- Cambridge Crystallographic Data Centre-CCDC Software Ltd. GOLD User Guide; Cambridge Crystallographic Data Centre-CCDC: Cambridge, UK, 2019. [Google Scholar]
- Irwin, J.J.; Duan, D.; Torosyan, H.; Doak, A.K.; Ziebart, K.T.; Sterling, T.; Tumanian, G.; Shoichet, B.K. An aggregation advisor for ligand discovery. J. Med. Chem. 2015, 58, 7076–7087. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef]
- Gavioli, R.; Vertuani, S.; Masucci, M.G. Proteasome inhibitors reconstitute the presentation of cytotoxic T-cell epitopes in Epstein-Barr virus-associated tumors. Int. J. Cancer 2002, 101, 532–538. [Google Scholar] [CrossRef]

| Inhibition of the Isolated Human 20S Proteasome | |||
|---|---|---|---|
| Compound ID | CT-L | C-L | T-L |
| 1 | 38% | 55% | 52% |
| 2 | 98% 5.60 ± 2.56 µM | 98% 2.42 ± 4.74 µM | 100% 25.84 ± 4.11 µM |
| 3 | 91% 49.26 ± 4.55 µM | 88% 51.14 ± 2.94 µM | 95% 27.52 ± 3.78 µM |
| 4 | 70% 51.13 ± 2.66 µM | 78% 41.18 ± 1.76 µM | 74% 48.36 ± 2.63 µM |
| 5 | 36% | 34% | 5% |
| Bortezomib | 100% 12.90 ± 1.53 nM | 93% 168.80 ± 1.46 nM | 34% 1.89 ± 1.74 µM |
| Inhibition of the 20S Proteasome in Cell Lysates of Human Cancer Cells | |||||
|---|---|---|---|---|---|
| Cell Line | |||||
| Compound ID | Active Site | Jurkat | K562 | HT-29 | HCT116 |
| 1 | CT-L | 54% | 63% | 62% 90.77 ± 4.76 µM | 90% 76.36 ± 3.50 µM |
| C-L | 19% | 28% | 76% 79.36 ± 2.38 µM | 71% 75.65 ± 2.87 µM | |
| T-L | 16% | NI | NI | NI | |
| 2 | CT-L | 72% 73.15 ± 3.18 µM | 40% | 62% 73.83 ± 1.06 µM | 74% 50.92 ± 1.50 µM |
| C-L | 74% 42.37 ± 2.67µM | 63% | 69% 62.06 ± 3.90 µM | 74% 43.47 ± 2.89 µM | |
| T-L | 56% 93.48 ± 1.16 µM | 42% | 52% 89.26 ± 1.23 µM | 44% | |
| 3 | CT-L | 33% | 59% | 31% | 33% |
| C-L | 25% | 39% | 19% | NI | |
| T-L | 33% | 12% | 39% | NI | |
| 4 | CT-L | 98% 24.25 ± 5.14 µM | ND | 98% 31.30 ± 2.31 µM | 98% 35.72 ± 2.60 µM |
| C-L | 99% 27.05 ± 3.02 µM | ND | 82% 35.23 ± 1.56 µM | 90% 28.91 ± 1.44 µM | |
| T-L | NI | ND | NI | NI | |
| 5 | CT-L | 64% | ND | 81% 84.26 ± 3.78 | 67% 95.90 ± 12.48 µM |
| C-L | 36% | ND | 29% | 28% | |
| T-L | NI | ND | 10% | NI | |
| Bortezomib | CT-L | 99% 19.05 ± 1.77 nM | 100% | 99% 87.36 ± 1.98 nM | 99% 80.17 ± 1.78 nM |
| C-L | 94% 489.4 ± 1.91 nM | 97% | 99% 640.7 ± 1.82 nM | 99% 752.9 ± 1.83 nM | |
| T-L | 41% 3.35 ± 1.90 µM | 46% | 66% 21.04 ± 1.95 µM | 45% 131.35 ± 1.99 µM | |
| Cell Viability Assays, MTT | ||||
|---|---|---|---|---|
| Cell Line | ||||
| Compound ID | Jurkat | K562 | HT-29 | HCT116 |
| 1 | 21% 36.84 ± 5.65 µM | 18% | 23% 61.71 ± 4.04 µM | 24% 65.69 ± 8.16 µM |
| 2 | 77% | 60% | 65% | 89% |
| 3 | 86% | 98% | 100% | 95% |
| 4 | 0% 1.37 ± 0.97 µM | ND | 34% 42.96 ± 6.81 µM | 0% 1.02 ± 0.55 µM |
| 5 | 0% 43.83 ± 2.27 µM | ND | 25% 70.47 ± 1.42 µM | 24% 58.41 ± 3.89 µM |
| Bortezomib | 0% 6.48 ± 3.22 nM | 0% ND | 0% 9.73 ± 2.65 nM | 0% 10.70 ± 1.14 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guedes, R.A.; Grilo, J.H.; Carvalho, A.N.; Fernandes, P.M.P.; Ressurreição, A.S.; Brito, V.; Santos, A.O.; Silvestre, S.; Gallerani, E.; Gama, M.J.; et al. New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies. Pharmaceuticals 2023, 16, 1096. https://doi.org/10.3390/ph16081096
Guedes RA, Grilo JH, Carvalho AN, Fernandes PMP, Ressurreição AS, Brito V, Santos AO, Silvestre S, Gallerani E, Gama MJ, et al. New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies. Pharmaceuticals. 2023; 16(8):1096. https://doi.org/10.3390/ph16081096
Chicago/Turabian StyleGuedes, Romina A., Jorge H. Grilo, Andreia N. Carvalho, Pedro M. P. Fernandes, Ana S. Ressurreição, Vanessa Brito, Adriana O. Santos, Samuel Silvestre, Eleonora Gallerani, Maria João Gama, and et al. 2023. "New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies" Pharmaceuticals 16, no. 8: 1096. https://doi.org/10.3390/ph16081096
APA StyleGuedes, R. A., Grilo, J. H., Carvalho, A. N., Fernandes, P. M. P., Ressurreição, A. S., Brito, V., Santos, A. O., Silvestre, S., Gallerani, E., Gama, M. J., Gavioli, R., Salvador, J. A. R., & Guedes, R. C. (2023). New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies. Pharmaceuticals, 16(8), 1096. https://doi.org/10.3390/ph16081096










