Ancistrocladinium A Induces Apoptosis in Proteasome Inhibitor-Resistant Multiple Myeloma Cells: A Promising Therapeutic Agent Candidate
Abstract
:1. Introduction
2. Results
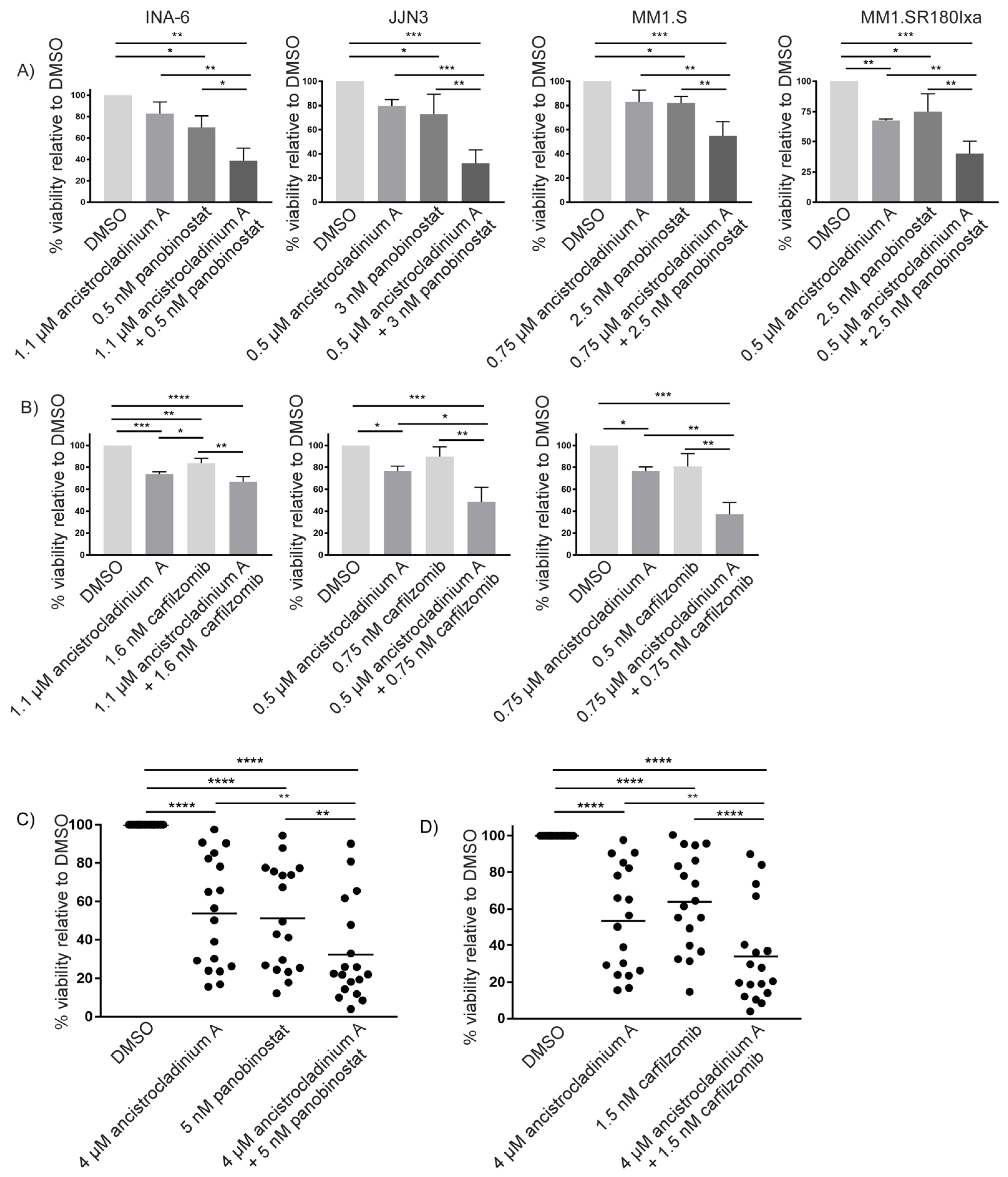
2.1. Treatment with Ancistrocladinium A Shows Activity against MM Cells, including PI-Resistant MM Cells, but Not in Non-Malignant Peripheral Blood Mononuclear Cells (PBMCs)
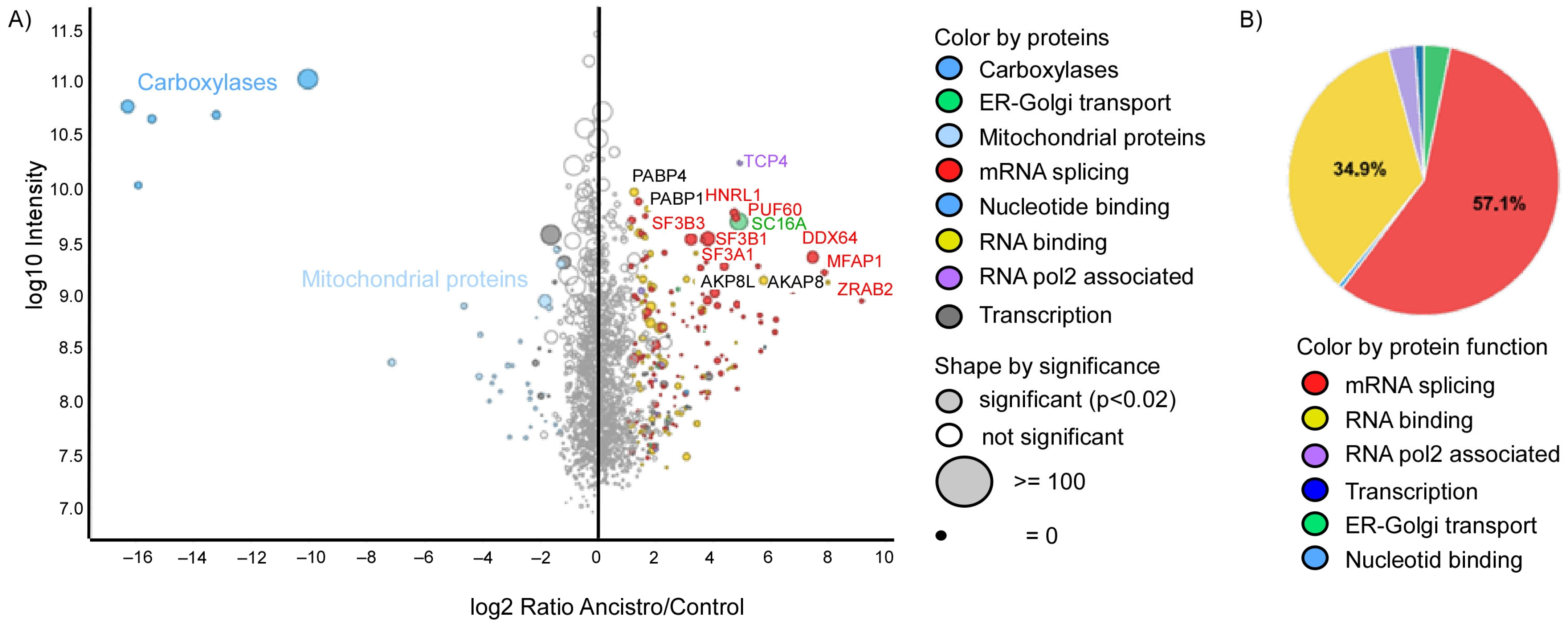
2.2. Ancistrocladinium A Targets the RNA Splicing Machinery
2.3. Ancistrocladinium A Differentially Regulates the RNA Binding and RNA Splicing Associated Genes
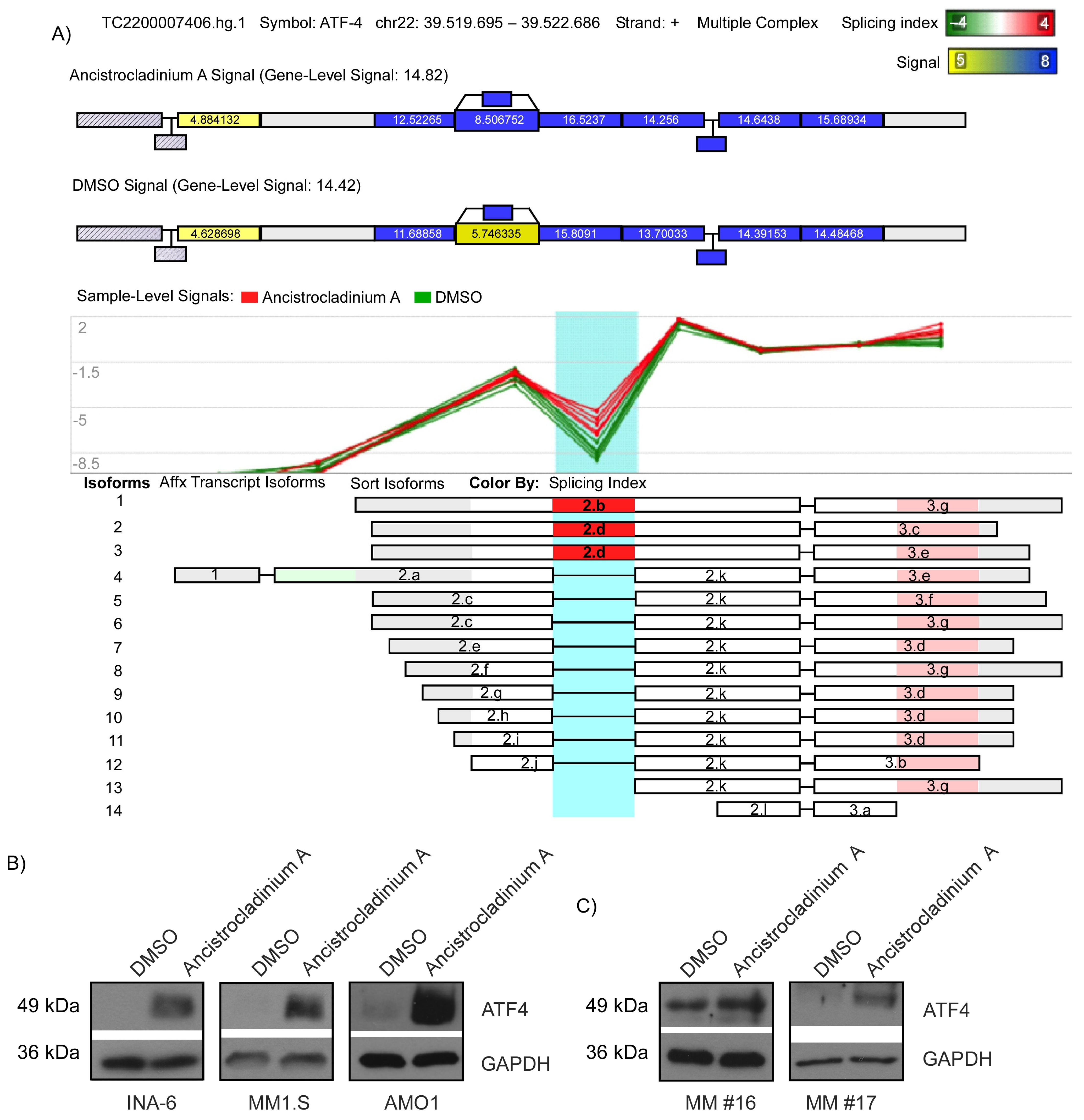
2.4. Ancistrocladinium A Induces RNA Splicing
2.5. Ancistrocladinium A Induces ATF4 Protein Expression
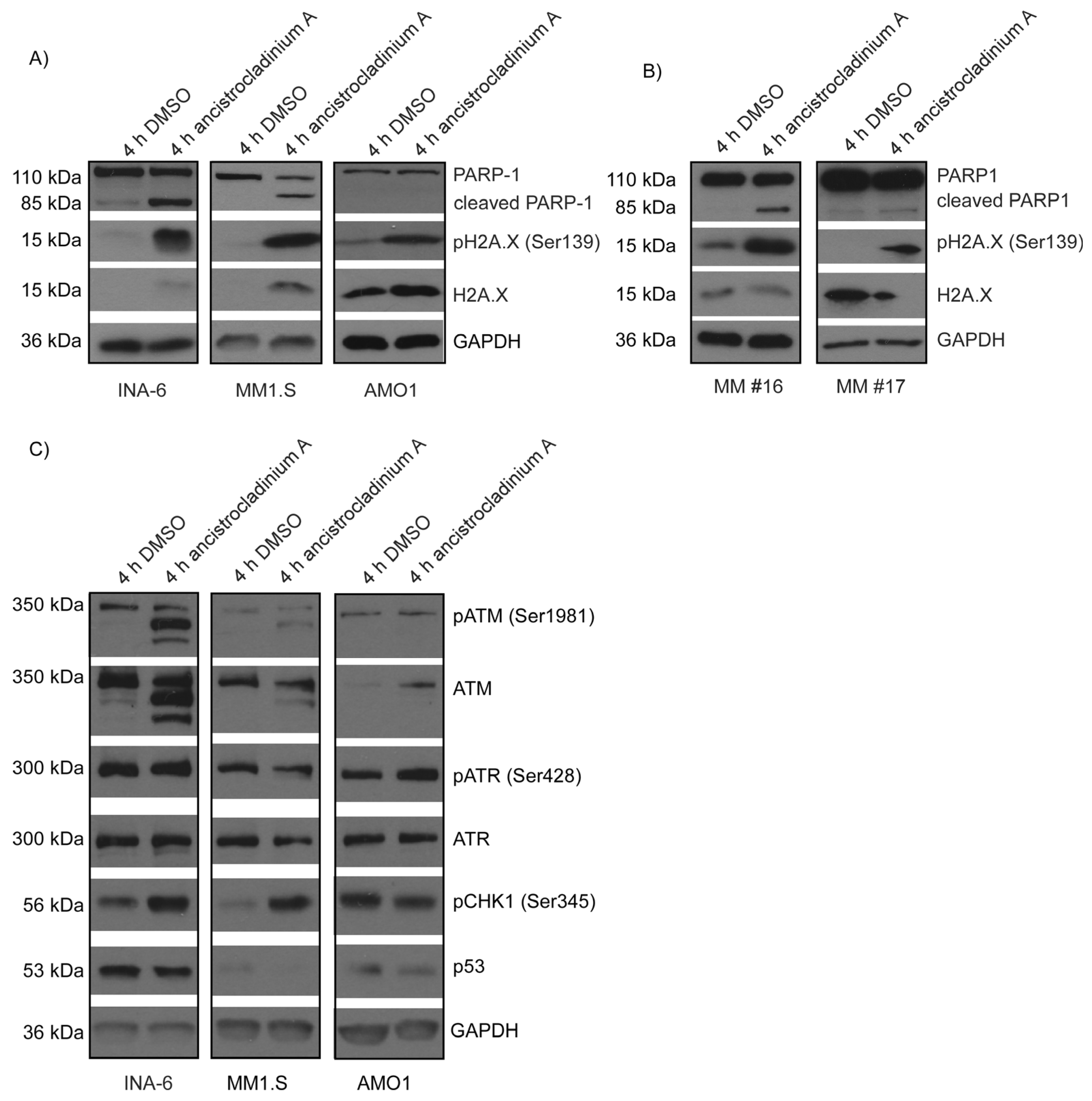
2.6. Ancistrocladinum A Induces H2A.X Phosphorylation
2.7. Concomitant Treatment with Ancistrocladinium A Enhances the Apoptotic Effects of the HDAC Inhibitor Panobinostat
2.8. Carfilzomib-Induced Apoptosis Is Strongly Increased by Concomitant Treatment with Ancistrocladinium A
3. Discussion
4. Materials and Methods
4.1. Pharmacological Inhibitors
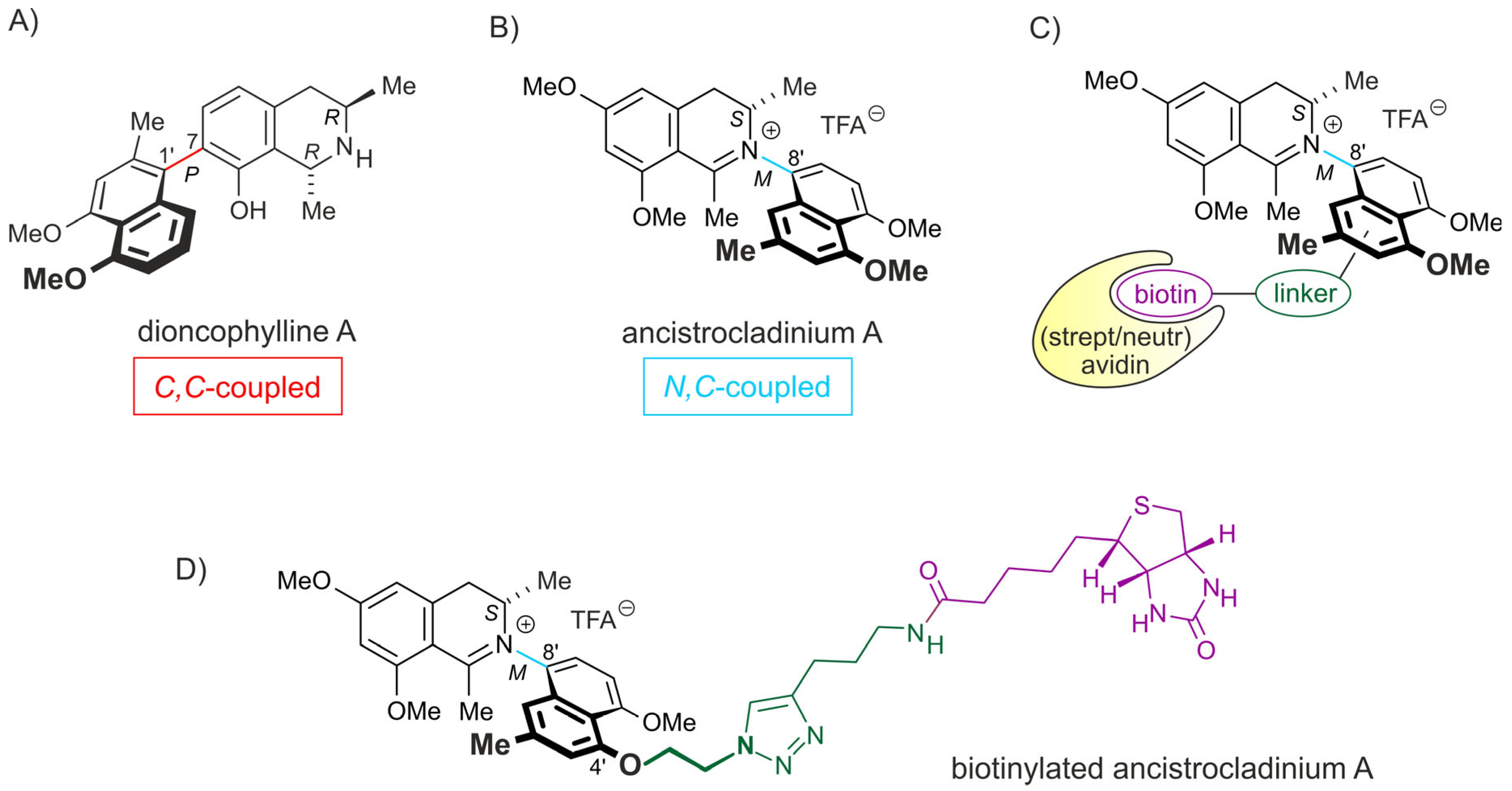
4.2. Synthesis of Biotinylated Ancistrocladinium A
4.3. MS-Based Spectroscopy and Data Analysis
4.4. Cell Culture
4.5. Isolation of CD138+ Primary MM Cells
4.6. Viability Analysis
4.7. RNA Isolation
4.8. GeneChip Microarray Assay
4.9. Microarray Data Analysis
4.10. Gene Set Enrichment Analysis
4.11. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and management of multiple myeloma: A review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Guang, M.H.Z.; Kavanagh, E.L.; Dunne, L.P.; Dowling, P.; Zhang, L.; Lindsay, S.; Bazou, D.; Goh, C.Y.; Hanley, C.; Bianchi, G.; et al. Targeting proteotoxic stress in cancer: A review of the role that protein quality control pathways play in oncogenesis. Cancers 2019, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Aronson, L.I.; Davies, F.E. DangER: Protein ovERload. Targeting protein degradation to treat myeloma. Haematologica 2012, 97, 1119–1130. [Google Scholar] [CrossRef]
- Bross, P.F.; Kane, R.; Farrell, A.T.; Abraham, S.; Benson, K.; Brower, M.E.; Bradley, S.; Gobburu, J.V.; Goheer, A.; Lee, S.L.; et al. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin. Cancer Res. 2004, 10, 3954–3964. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.; Masszi, T.; Grzasko, N.; Bahlis, N.J.; Hansson, M.; Pour, L.; Sandhu, I.; Ganly, P.; Baker, B.W.; Jackson, S.R.; et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med. 2016, 374, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.S.; Martin, T.; Wang, M.; Vij, R.; Jakubowiak, A.J.; Lonial, S.; Trudel, S.; Kukreti, V.; Bahlis, N.; Alsina, M.; et al. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood 2012, 120, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Richardson, P.G.; Schlossman, R.L.; Alsina, M.; Weber, D.M.; Coutre, S.E.; Gasparetto, C.; Mukhopadhyay, S.; Ondovik, M.S.; Khan, M.; Paley, C.S.; et al. PANORAMA 2: Panobinostat in combination with bortezomib and dexamethasone in patients with relapsed and bortezomib-refractory myeloma. Blood 2013, 122, 2331–2337. [Google Scholar] [CrossRef]
- Besse, A.; Stolze, S.C.; Rasche, L.; Weinhold, N.; Morgan, G.J.; Kraus, M.; Bader, J.; Overkleeft, H.S.; Besse, L.; Driessen, C. Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma. Leukemia 2018, 32, 391–401. [Google Scholar] [CrossRef]
- Barrio, S.; Stühmer, T.; Da-Via, M.; Barrio-Garcia, C.; Lehners, N.; Besse, A.; Cuenca, I.; Garitano-Trojaola, A.; Fink, S.; Leich, E.; et al. Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia 2019, 33, 447–456. [Google Scholar] [CrossRef]
- Brünnert, D.; Kraus, M.; Stühmer, T.; Kirner, S.; Heiden, R.; Goyal, P.; Driessen, C.; Bargou, R.C.; Chatterjee, M. Novel cell line models to study mechanisms and overcoming strategies of proteasome inhibitor resistance in multiple myeloma. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1666–1676. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; Schor, I.E.; Alló, M.; Dujardin, G.; Petrillo, E.; Muñoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef]
- Kim, E.; Goren, A.; Ast, G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008, 24, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Dong, H.; Shi, Y.; Bian, L. Mutually exclusive alternative splicing of pre-mRNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1468. [Google Scholar] [CrossRef] [PubMed]
- Kaida, D.; Motoyoshi, H.; Tashiro, E.; Nojima, T.; Hagiwara, M.; Ishigami, K.; Watanabe, H.; Kitahara, T.; Yoshida, T.; Nakajima, H.; et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007, 3, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, M.K.; Kumar, D.; Villa, R.; La Clair, J.J.; Benner, C.; Sasik, R.; Jones, H.; Ghia, E.M.; Rassenti, L.Z.; Kipps, T.J.; et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica 2015, 100, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.A.; Ashby, C.; Wardell, C.; Boyle, E.M.; Ortiz, M.; Flynt, E.; Thakurta, A.; Morgan, G.; Walker, B.A. Differential RNA splicing as a potentially important driver mechanism in multiple myeloma. Haematologica 2021, 106, 736–745. [Google Scholar] [CrossRef]
- Aktas Samur, A.; Fulciniti, M.; Avet-Loiseau, H.; Lopez, M.A.; Derebail, S.; Corre, J.; Minvielle, S.; Magrangeas, F.; Moreau, P.; Anderson, K.C.; et al. In-depth analysis of alternative splicing landscape in multiple myeloma and potential role of dysregulated splicing factors. Blood Cancer J. 2022, 12, 171. [Google Scholar] [CrossRef]
- Song, S.; Zhang, W.; Li, Q.; Wang, Z.; Su, Q.; Zhang, X.; Li, B.; Zhuang, W. Dysregulation of alternative splicing contributes to multiple myeloma pathogenesis. Br. J. Cancer 2023, 128, 1086–1094. [Google Scholar] [CrossRef]
- Huang, H.H.; Ferguson, I.D.; Thornton, A.M.; Bastola, P.; Lam, C.; Lin, Y.T.; Choudhry, P.; Mariano, M.C.; Marcoulis, M.D.; Teo, C.F.; et al. Proteasome inhibitor-induced modulation reveals the spliceosome as a specific therapeutic vulnerability in multiple myeloma. Nat. Commun. 2020, 11, 1931. [Google Scholar] [CrossRef]
- Li, J.; Seupel, R.; Feineis, D.; Mudogo, V.; Kaiser, M.; Brun, R.; Brünnert, D.; Chatterjee, M.; Seo, E.J.; Efferth, T.; et al. Dioncophyllines C2, D2, and F and related naphthylisoquinoline alkaloids from the Congolese liana Ancistrocladus ileboensis with potent activities against Plasmodium falciparum and against multiple myeloma and leukemia cell lines. J. Nat. Prod. 2017, 80, 443–458. [Google Scholar] [CrossRef]
- Bringmann, G.; François, G.; Aké Assi, L.; Schlauer, J. The alkaloids of Triphyophyllum peltatum (Dioncophyllaceae). Chimia 1998, 52, 18–28. [Google Scholar] [CrossRef]
- Tajuddeen, N.; Bringmann, G. N,C-Coupled naphthylisoquinoline alkaloids: A versatile new class of axially chiral natural products. Nat. Prod. Rep. 2021, 38, 2154–2186. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Kajahn, I.; Reichert, M.; Pedersen, S.E.; Faber, J.H.; Gulder, T.; Brun, R.; Christensen, S.B.; Ponte-Sucre, A.; Moll, H.; et al. Ancistrocladinium A and B, the first N,C-coupled naphthyldihydroisoquinoline alkaloids, from a Congolese Ancistrocladus species. J. Org. Chem. 2006, 71, 9348–9356. [Google Scholar] [CrossRef] [PubMed]
- Vincenz, L.; Jäger, R.; O’Dwyer, M.; Samali, A. Endoplasmic reticulum stress and the unfolded protein response: Targeting the Achilles heel of multiple myeloma. Mol. Cancer Ther. 2013, 12, 831–843. [Google Scholar] [CrossRef]
- Chan, C.P.; Kok, K.H.; Tang, H.M.; Wong, C.M.; Jin, D.Y. Internal ribosome entry site-mediated translational regulation of ATF4 splice variant in mammalian unfolded protein response. Biochim. Biophys. Acta 2013, 1833, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.; Pries, V.; Hedberg, C.; Waldmann, H. Target identification for small bioactive molecules: Finding the needle in the haystack. Angew. Chem. Int. Ed. 2013, 52, 2744–2792. [Google Scholar] [CrossRef]
- Leslie, B.J.; Hergenrother, P.J. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem. Soc. Rev. 2008, 37, 1347–1360. [Google Scholar] [CrossRef]
- Te Raa, G.D.; Derks, I.A.; Navrkalova, V.; Skowronska, A.; Moerland, P.D.; van Laar, J.; Oldreive, C.; Monsuur, H.; Trbusek, M.; Malcikova, J.; et al. The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia 2015, 29, 1133–1142. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small nucleolar RNAs: Insight into their function in cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Jorjani, H.; Kehr, S.; Jedlinski, D.J.; Gumienny, R.; Hertel, J.; Stadler, P.F.; Zavolan, M.; Gruber, A.R. An updated human snoRNAome. Nucleic Acids Res. 2016, 44, 5068–5082. [Google Scholar] [CrossRef] [PubMed]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Rajasingh, S.; Samanta, S.; Dawn, B.; Bittel, D.C.; Rajasingh, J. Biology and clinical relevance of noncoding sno/scaRNAs. Trends Cardiovasc. Med. 2018, 28, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-I.; Wen, H. Heightened protein-translation activities in mammalian cells and the disease/treatment implications. Natl. Sci. Rev. 2020, 7, 1851–1855. [Google Scholar] [CrossRef] [PubMed]
- Muraki, M.; Ohkawara, B.; Hosoya, T.; Onogi, H.; Koizumi, J.; Koizumi, T.; Sumi, K.; Yomoda, J.; Murray, M.V.; Kimura, H.; et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 2004, 279, 24246–24254. [Google Scholar] [CrossRef]
- Sakai, Y.; Yoshida, T.; Ochiai, K.; Uosaki, Y.; Saitoh, Y.; Tanaka, F.; Akiyama, T.; Akinaga, S.; Mizukami, T. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. 2002, 55, 855–862. [Google Scholar] [CrossRef]
- Lagisetti, C.; Yermolina, M.V.; Sharma, L.K.; Palacios, G.; Prigaro, B.J.; Webb, T.R. Pre-mRNA splicing-modulatory pharmacophores: The total synthesis of herboxidiene, a pladienolide-herboxidiene hybrid analog and related derivatives. ACS Chem. Biol. 2014, 9, 643–648. [Google Scholar] [CrossRef]
- Albert, B.J.; Sivaramakrishnan, A.; Naka, T.; Czaicki, N.L.; Koide, K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J. Am. Chem. Soc. 2007, 129, 2648–2659. [Google Scholar] [CrossRef]
- Fan, L.; Lagisetti, C.; Edwards, C.C.; Webb, T.R.; Potter, P.M. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 2011, 6, 582–589. [Google Scholar] [CrossRef]
- Girardot, M.; Pecquet, C.; Chachoua, I.; Van Hees, J.; Guibert, S.; Ferrant, A.; Knoops, L.; Baxter, E.J.; Beer, P.A.; Giraudier, S.; et al. Persistent STAT5 activation in myeloid neoplasms recruits p53 into gene regulation. Oncogene 2015, 34, 1323–1332. [Google Scholar] [CrossRef]
- Bi, G.; Zhu, D.; Bian, Y.; Huang, Y.; Zhan, C.; Yang, Y.; Wang, Q. Knockdown of GTF2E2 inhibits the growth and progression of lung adenocarcinoma via RPS4X in vitro and in vivo. Cancer Cell Int. 2021, 21, 181. [Google Scholar] [CrossRef] [PubMed]
- Yevshin, I.; Sharipov, R.; Kolmykov, S.; Kondrakhin, Y.; Kolpakov, F. GTRD: A database on gene transcription regulation-2019 update. Nucleic Acids Res. 2019, 47, D100–D105. [Google Scholar] [CrossRef] [PubMed]
- Wortel, I.M.N.; van der Meer, L.T.; Kilberg, M.S.; van Leeuwen, F.N. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends Endocrinol. Metab. 2017, 28, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Quirós, P.M.; Prado, M.A.; Zamboni, N.; D’Amico, D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017, 216, 2027–2045. [Google Scholar] [CrossRef]
- Biterge, B.; Schneider, R. Histone variants: Key players of chromatin. Cell Tissue Res. 2014, 356, 457–466. [Google Scholar] [CrossRef]
- Stope, M.B. Phosphorylation of histone H2A.X as a DNA-associated biomarker (Review). World Acad. Sci. J. 2021, 3, 31. [Google Scholar] [CrossRef]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008, 36, 5678–5694. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; van Duin, M.; Sonneveld, P.; Mateos, M.V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Primers 2017, 3, 17046. [Google Scholar] [CrossRef]
- Lü, S.; Wang, J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomak. Res. 2013, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Seupel, R.; Hertlein-Amslinger, B.; Gulder, T.; Stawski, P.; Kaiser, M.; Brun, R.; Bringmann, G. Directed Synthesis of All Four Pure Stereoisomers of the N,C-coupled naphthylisoquinoline alkaloid ancistrocladinium A. Org. Lett. 2016, 18, 6508–6511. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Gulder, T.; Hertlein, B.; Hemberger, Y.; Meyer, F. Total synthesis of the N,C-coupled naphthylisoquinoline alkaloids ancistrocladinium A and B and related analogues. J. Am. Chem. Soc. 2010, 132, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.; Bauer, S.; Stühmer, T.; Mottok, A.; Scholz, C.J.; Steinbrunn, T.; Brünnert, D.; Brandl, A.; Schraud, H.; Kressmann, S.; et al. Pan-Raf co-operates with PI3K-dependent signalling and critically contributes to myeloma cell survival independently of mutated RAS. Leukemia 2017, 31, 922–933. [Google Scholar] [CrossRef]
- Bach, M.; Lehmann, A.; Brünnert, D.; Vanselow, J.T.; Hartung, A.; Bargou, R.C.; Holzgrabe, U.; Schlosser, A.; Chatterjee, M. Ugi reaction-derived alpha-Acyl aminocarboxamides bind to Phosphatidylinositol 3-Kinase-Related Kinases, inhibit HSF1-dependent heat shock response, and induce apoptosis in multiple myeloma cells. J. Med. Chem. 2017, 60, 4147–4160. [Google Scholar] [CrossRef] [PubMed]
- Stühmer, T.; Arts, J.; Chatterjee, M.; Borawski, J.; Wolff, A.; King, P.; Einsele, H.; Leo, E.; Bargou, R.C. Preclinical anti-myeloma activity of the novel HDAC-inhibitor JNJ-26481585. Br. J. Haematol. 2010, 149, 529–536. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]

| Gene Set Name [# Genes (K)] | # Genes in Overlap (k) | k/K | p-Value | fdr q-Value | Gene Regulation |
|---|---|---|---|---|---|
| GOBP_RNA_PROCESSING [1424] | 102 |  | 3.13 × 10−29 | 1.01 × 10−24 | 73 upregulated, 29 downregulated |
| GTF2A2_TARGET_GENES [522] | 50 | 6.23 × 10−20 | 1.01 × 10−15 | 46 upregulated, 4 downregulated | |
| SNAPC4_TARGET_GENES [138] | 27 |  | 3.3 × 10−19 | 3.55 × 10−15 | 26 upregulated, 1 downregulated |
| GOBP_FORMATION_OF_QUADRUPLE_SL_U4_U5_U5_U6_SNRNP [10] | 10 |  | 9.08 × 10−18 | 7.33 × 10−14 | 10 upregulated, 0 downregulated |
| GOCC_NUCLEOLUS [1386] | 76 | 2.17 × 10−15 | 1.4 × 10−11 | 54 upregulated, 22 downregulated | |
| ZHENG_CORD_BLOOD_C5_SIMILAR_TO_HSC_C6_C6_PUTATIVE_ALTERED_METABOLIC_STATE [97] | 20 |  | 4.73 × 10−15 | 2.54 × 10−11 | 20 upregulated, 0 downregulated |
| GTF2E2_TARGET_GENES [411] | 38 |  | 5.87 × 10−15 | 2.71 × 10−11 | 32 upregulated, 6 downregulated |
| GOBP_SPLICEOSOMAL_TRI_SNRNP_COMPLEX_AS_ASSEMBLY [25] | 12 |  | 1.43 × 10−14 | 5.78 × 10−11 | 12 upregulated, 0 downregulated |
| GOCC_SMALL_NUCLEAR_RIBONUCLEOPROTEIN_CN_COMPLEX [99] | 19 |  | 9.09 × 10−14 | 3.26 × 10−10 | 19 upregulated, 0 downregulated |
| GOBP_RIBONUCLEOPROTEIN_COMPLEX_BIOGENEENESIS [471] | 38 |  | 4.27 × 10−13 | 1.38 | 20 upregulated, 18 downregulated |
| Gene Set Name [# Genes (K)] | # Genes in Overlap (k) | k/K | p-Value | FDR q-Value |
|---|---|---|---|---|
| PSMB5_TARGET_GENES [307] | 68 |  | 3.31 × 10−63 | 1.07 × 10−58 |
| GTF2A2_TARGET_GENES [522] | 69 |  | 3.43 × 10−48 | 5.54 × 10−44 |
| SETD7_TARGET_GENES [991] | 83 |  | 2.09 × 10−42 | 2.25 × 10−38 |
| FISCHER_DREAM_TARGETS [968] | 81 |  | 2.43 × 10−41 | 1.96 × 10−37 |
| REACTOME_CELLULAR_RESPONSES_TO_EXTERNARNAL_STIMULI [706] | 70 |  | 1.81 × 10−40 | 1.17 × 10−36 |
| BARX2_TARGET_GENES [1723] | 101 |  | 4.94 × 10−38 | 2.66 × 10−34 |
| REACTOME_CELL_CYCLE [693] | 66 |  | 4.41 × 10−37 | 2.03 × 10−33 |
| ZNF84_TARGET_GENES [2000] | 107 |  | 7.93 × 10−37 | 3.2 × 10−33 |
| REACTOME_CELL_CYCLE_MITOTIC [561] | 60 |  | 1.39 × 10−36 | 4.98 × 10−33 |
| MARTENS_TRETINOIN_RESPONSE_DN [839] | 70 |  | 1.28 × 10−35 | 4.14 × 10−32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brünnert, D.; Seupel, R.; Goyal, P.; Bach, M.; Schraud, H.; Kirner, S.; Köster, E.; Feineis, D.; Bargou, R.C.; Schlosser, A.; et al. Ancistrocladinium A Induces Apoptosis in Proteasome Inhibitor-Resistant Multiple Myeloma Cells: A Promising Therapeutic Agent Candidate. Pharmaceuticals 2023, 16, 1181. https://doi.org/10.3390/ph16081181
Brünnert D, Seupel R, Goyal P, Bach M, Schraud H, Kirner S, Köster E, Feineis D, Bargou RC, Schlosser A, et al. Ancistrocladinium A Induces Apoptosis in Proteasome Inhibitor-Resistant Multiple Myeloma Cells: A Promising Therapeutic Agent Candidate. Pharmaceuticals. 2023; 16(8):1181. https://doi.org/10.3390/ph16081181
Chicago/Turabian StyleBrünnert, Daniela, Raina Seupel, Pankaj Goyal, Matthias Bach, Heike Schraud, Stefanie Kirner, Eva Köster, Doris Feineis, Ralf C. Bargou, Andreas Schlosser, and et al. 2023. "Ancistrocladinium A Induces Apoptosis in Proteasome Inhibitor-Resistant Multiple Myeloma Cells: A Promising Therapeutic Agent Candidate" Pharmaceuticals 16, no. 8: 1181. https://doi.org/10.3390/ph16081181
APA StyleBrünnert, D., Seupel, R., Goyal, P., Bach, M., Schraud, H., Kirner, S., Köster, E., Feineis, D., Bargou, R. C., Schlosser, A., Bringmann, G., & Chatterjee, M. (2023). Ancistrocladinium A Induces Apoptosis in Proteasome Inhibitor-Resistant Multiple Myeloma Cells: A Promising Therapeutic Agent Candidate. Pharmaceuticals, 16(8), 1181. https://doi.org/10.3390/ph16081181






