Application of the Analytical Procedure Lifecycle Concept to a Quantitative 1H NMR Method for Total Dammarane-Type Saponins
Abstract
1. Introduction
2. Results and Discussion
2.1. Risk Identification and Assessment Results Based on the Ishikawa Diagram and FMECA
2.2. DoE-Based Analytical Process Design (APD)
2.2.1. CAPPs Screening Results Based on PBD
PBD Screening Experiments
CAPPs Screening Results
2.2.2. The Results of CCD-based CAPP Optimization
CCD Experimental Design Results
MODR Calculation Results Based on the Optimization Model
2.3. Procedure Performance Qualification (APPQ)
2.3.1. Determination of Acquisition Parameters
2.3.2. APPQ Index Examination
MODR Robustness Examination
Linearity, Accuracy Investigation, and Calculation of LOD and LOQ
Precision and Sample Stability Investigation
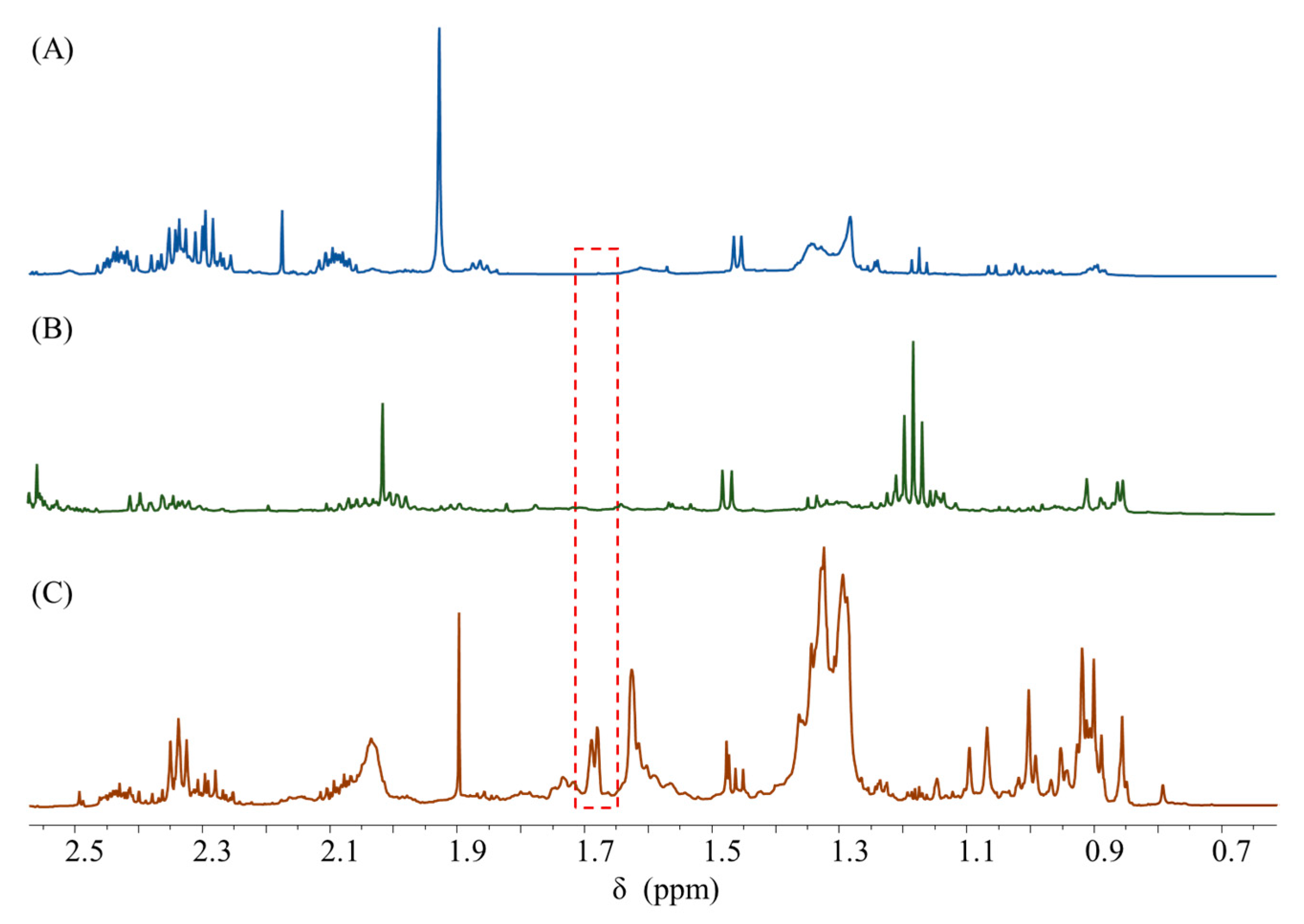
Signal Specificity Investigation
Measurement Uncertainty
2.4. CPPV Example: Method Transfer and Application
2.4.1. 1H qNMR Analysis of Total Dammarane-Type Ginsenosides in the Shenmai Injection
2.4.2. 1H qNMR Analysis of Total Notoginsenosides in the Xuesaitong Injection
2.4.3. 1H qNMR Analysis of Total Gypenosides in the Gynostemma Process Intermediates
3. Materials and Methods
3.1. Reagents and Materials
3.2. Primary Analysis Conditions
3.2.1. Selection of Deuterated Reagents
3.2.2. Selection of Pulse Sequences
3.2.3. Selection of Internal Standard
3.2.4. Selection of Signals for Quantification
3.2.5. Data Processing
3.3. Determination of ATPs
3.4. Risk Assessment Methodology
3.5. DoE Methods
3.6. Indexes of APPQ
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ruan, J.; Zheng, C.; Qu, L.; Liu, Y.; Han, L.; Yu, H.; Zhang, Y.; Wang, T. Plant resources, 13C-NMR spectral characteristic and pharmacological activities of dammarane-type triterpenoids. Molecules 2016, 21, E1047. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, X.; Qu, F.; Guo, Z.; Zhao, Y. Dammarane triterpenoids for pharmaceutical use: A patent review (2005–2014). Expert Opin. Ther. Pat. 2015, 25, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, J.; Zhao, F.; Pan, J.; Qu, H. 1H-qNMR quantification of total ginsenosides in Shenmai Injection. Chin. J. Chin. Mater. Med. 2022, 47, 587–592. [Google Scholar] [CrossRef]
- Ynag, L.; Chen, J.; Xu, X.; Gu, L.; Zhang, S.; You, Z.; Zheng, G.; Chen, J.; Xin, Y. Simultaneous determination of eleven components in Shenmai injection by UHPLC. Chin. J. Pharm. Anal. 2019, 39, 1660–1665. [Google Scholar] [CrossRef]
- Stavrianidi, A.; Stekolshchikova, E.; Porotova, A.; Rodin, I.; Shpigun, O. Combination of HPLC–MS and QAMS as a new analytical approach for determination of saponins in ginseng containing products. J. Pharm. Biomed. Anal. 2017, 132, 87–92. [Google Scholar] [CrossRef]
- Abashev, M.; Stekolshchikova, E.; Stavrianidi, A. Quantitative aspects of the hydrolysis of ginseng saponins: Application in HPLC-MS analysis of herbal products. J. Ginseng Res. 2021, 45, 246–253. [Google Scholar] [CrossRef]
- Wang, R.; Wang, H.; Fan, X.; Duan, M.; Li, H.; Wang, S. Analysis of chemical constituents in Shenmai Injection by LC-Q-TOF-MS and LC-IT-MS. Chin. J. Chin. Mater. Med. 2020, 45, 105–114. [Google Scholar] [CrossRef]
- Verch, T.; Campa, C.; Chéry, C.; Frenkel, R.; Graul, T.; Jaya, N.; Nakhle, B.; Springall, J.; Starkey, J.; Wypych, J.; et al. Analytical Quality by Design, Life Cycle Management, and Method Control. AAPS J. 2022, 24, 34. [Google Scholar] [CrossRef]
- Tiwari, R.; Kumar, A.; Solanki, P.; Dhobi, M.; Sundaresan, V.; Kalaiselvan, V.; Raghuvanshi, R.S. Analytical quality-by-design (AQbD) guided development of a robust HPLC method for the quantification of plumbagin from Plumbago species. J. Liq. Chromatogr. Relat. Technol. 2021, 44, 529–537. [Google Scholar] [CrossRef]
- Baglow, R.L. The reliability parameter and its importance for life cycle management. Microelectron. Reliab. 1975, 14, 91–104. [Google Scholar] [CrossRef]
- Parr, M.K.; Schmidt, A.H. Life cycle management of analytical methods. J. Pharm. Biomed. Anal. 2018, 147, 506–517. [Google Scholar] [CrossRef]
- Park, G.; Kim, M.K.; Go, S.H.; Choi, M.; Jang, Y.P. Analytical quality by design (AQbD) approach to the development of analytical procedures for medicinal plants. Plants 2022, 11, 2960. [Google Scholar] [CrossRef]
- Kim, M.K.; Park, S.C.; Park, G.; Choi, E.; Ji, Y.; Jang, Y.P. Analytical quality by design methodology for botanical raw material analysis: A case study of flavonoids in Genkwa Flos. Sci. Rep. 2021, 11, 11936. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Chen, Y.; Shen, X.; Jiang, H.; Gong, X.; Yan, J. Establishing the chromatographic fingerprint of traditional Chinese medicine standard decoction based on quality by design approach: A case study of Licorice. J. Sep. Sci. 2019, 42, 1144–1154. [Google Scholar] [CrossRef]
- Dai, S.; Xu, B.; Zhang, Y.; Sun, F.; Li, J.; Shi, X.; Qiao, Y. Robust design space development for HPLC analysis of five chemical components in Panax notoginseng saponins. J. Liq. Chromatogr. Relat. Technol. 2016, 39, 504–512. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, Y.; Chen, H.; Chen, T.; Pan, J.; Wang, X.; Qu, H. Development of an analytical method by defining a design space: A case study of saponin determination for Panax notoginseng extracts. Anal. Methods 2016, 8, 2282–2289. [Google Scholar] [CrossRef]
- Kim, M.K.; Park, G.; Hong, S.P.; Jang, Y.P. Analytical quality by design methodology approach for simultaneous quantitation of paeoniflorin and decursin in herbal medicine by RP-HPLC analysis. Nat. Prod. Sci. 2021, 27, 264–273. [Google Scholar] [CrossRef]
- Parab Gaonkar, V.; Hullatti, K. Quality assessment and RP-HPLC method development for estimation of curcuminoids in Curcuma longa: A quality by design approach. J. Liq. Chromatogr. Relat. Technol. 2021, 44, 95–102. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, L.; Wang, X.; Gong, X.; Qu, H. Development of an HPLC-MS method for the determination of four terpene trilactones in Ginkgo biloba leaf extract via quality by design. Biomed. Chromatogr. 2021, 35, e5170. [Google Scholar] [CrossRef]
- Ancillotti, C.; Orlandini, S.; Ciofi, L.; Pasquini, B.; Caprini, C.; Droandi, C.; Furlanetto, S.; Del Bubba, M. Quality by design compliant strategy for the development of a liquid chromatography–tandem mass spectrometry method for the determination of selected polyphenols in Diospyros kaki. J. Chromatogr. A 2018, 1569, 79–90. [Google Scholar] [CrossRef]
- Silva, P.; Silva, C.L.; Perestrelo, R.; Nunes, F.M.; Câmara, J.S. Application of quality-by-design approach in the analytical method development for quantification of sugars in sugarcane honey by reversed-phase liquid chromatography. Food Anal. Methods 2020, 13, 1634–1649. [Google Scholar] [CrossRef]
- Van Tricht, E.; Geurink, L.; Backus, H.; Germano, M.; Somsen, G.W.; Sänger-van de Griend, C.E. One single, fast and robust capillary electrophoresis method for the direct quantification of intact adenovirus particles in upstream and downstream processing samples. Talanta 2017, 166, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Perovani, I.S.; Serpellone, C.O.; de Oliveira, A.R.M. An appraisal of experimental designs: Application to enantioselective capillary electromigration techniques. Electrophoresis 2021, 42, 1726–1743. [Google Scholar] [CrossRef] [PubMed]
- Aboushady, D.; Hanafi, R.S.; Parr, M.K. Quality by design approach for enantioseparation of terbutaline and its sulfate conjugate metabolite for bioanalytical application using supercritical fluid chromatography. J. Chromatogr. A 2022, 1676, 463285. [Google Scholar] [CrossRef] [PubMed]
- Muteki, K.; Morgado, J.E.; Reid, G.L.; Wang, J.; Xue, G.; Riley, F.W.; Harwood, J.W.; Fortin, D.T.; Miller, I.J. Quantitative Structure retention relationship models in an analytical quality by design framework: Simultaneously accounting for compound properties, mobile-phase conditions, and stationary-phase properties. Ind. Eng. Chem. Res. 2013, 52, 12269–12284. [Google Scholar] [CrossRef]
- Freitas, J.; Silva, P.; Vaz-Pires, P.; Câmara, J.S. A systematic AQbD approach for optimization of the most influential experimental parameters on analysis of fish spoilage-related volatile amines. Foods 2020, 9, E1321. [Google Scholar] [CrossRef]
- Robu, S.; Romila, A.; Buzia, O.D.; Spac, A.F.; Diaconu, C.; Tutunaru, D.; Lisa, E.; Nechita, A. Contribution to the optimization of a gas chromatographic method by QbD approach used for analysis of essential oils from Salvia officinalis. Rev. Chim. 2019, 70, 2015–2020. [Google Scholar] [CrossRef]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Gödecke, T.; Napolitano, J.G.; Rodríguez-Brasco, M.F.; Chen, S.-N.; Jaki, B.U.; Lankin, D.C.; Pauli, G.F. Validation of a generic quantitative 1H NMR method for natural products analysis. Phytochem. Anal. 2013, 24, 581–597. [Google Scholar] [CrossRef]
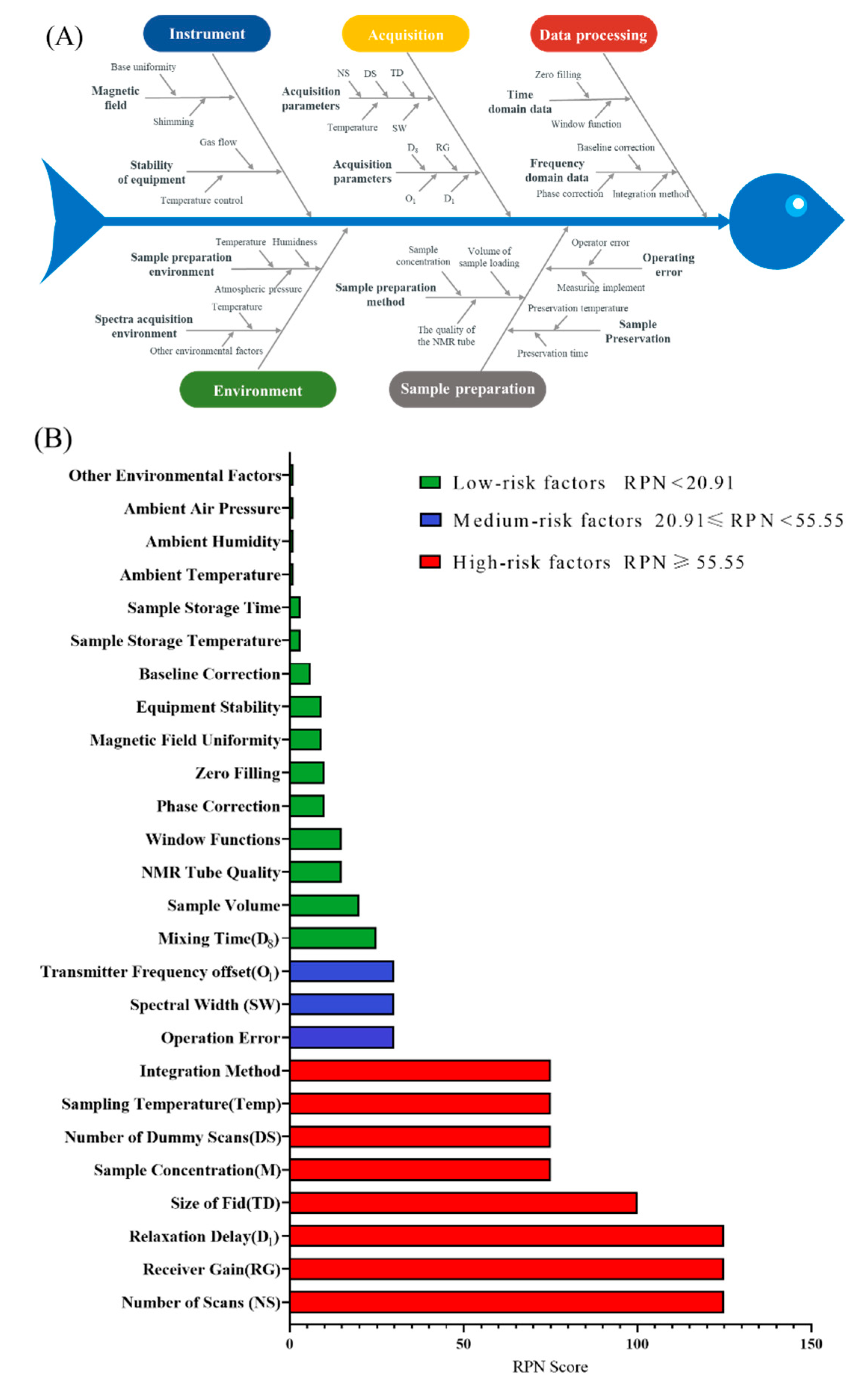
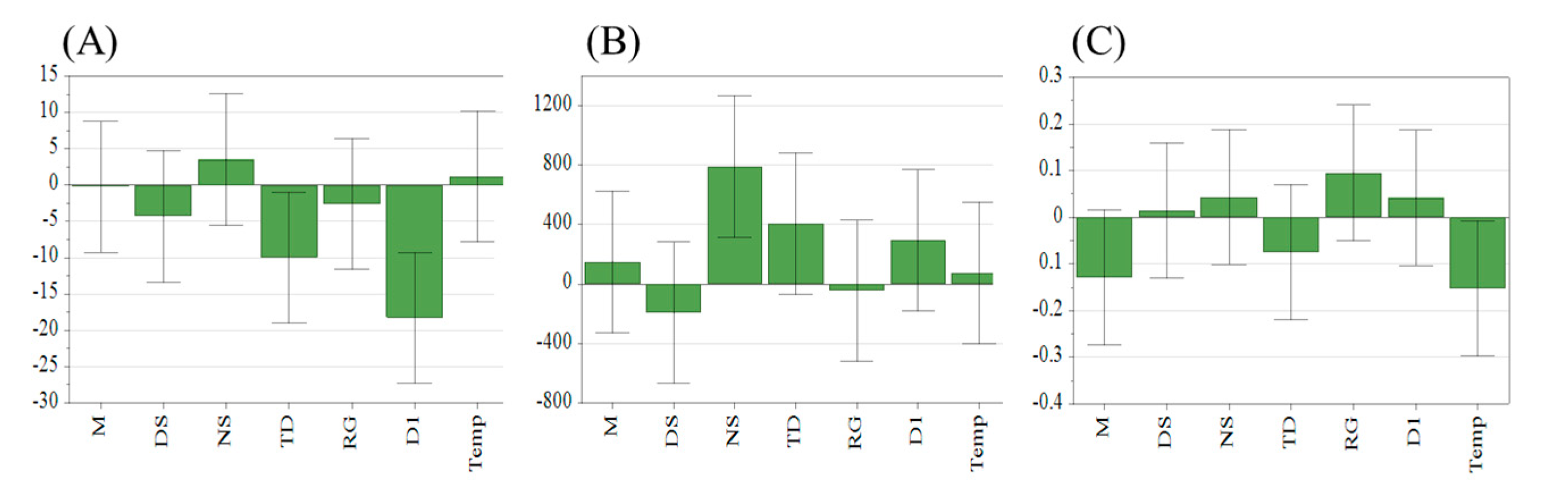
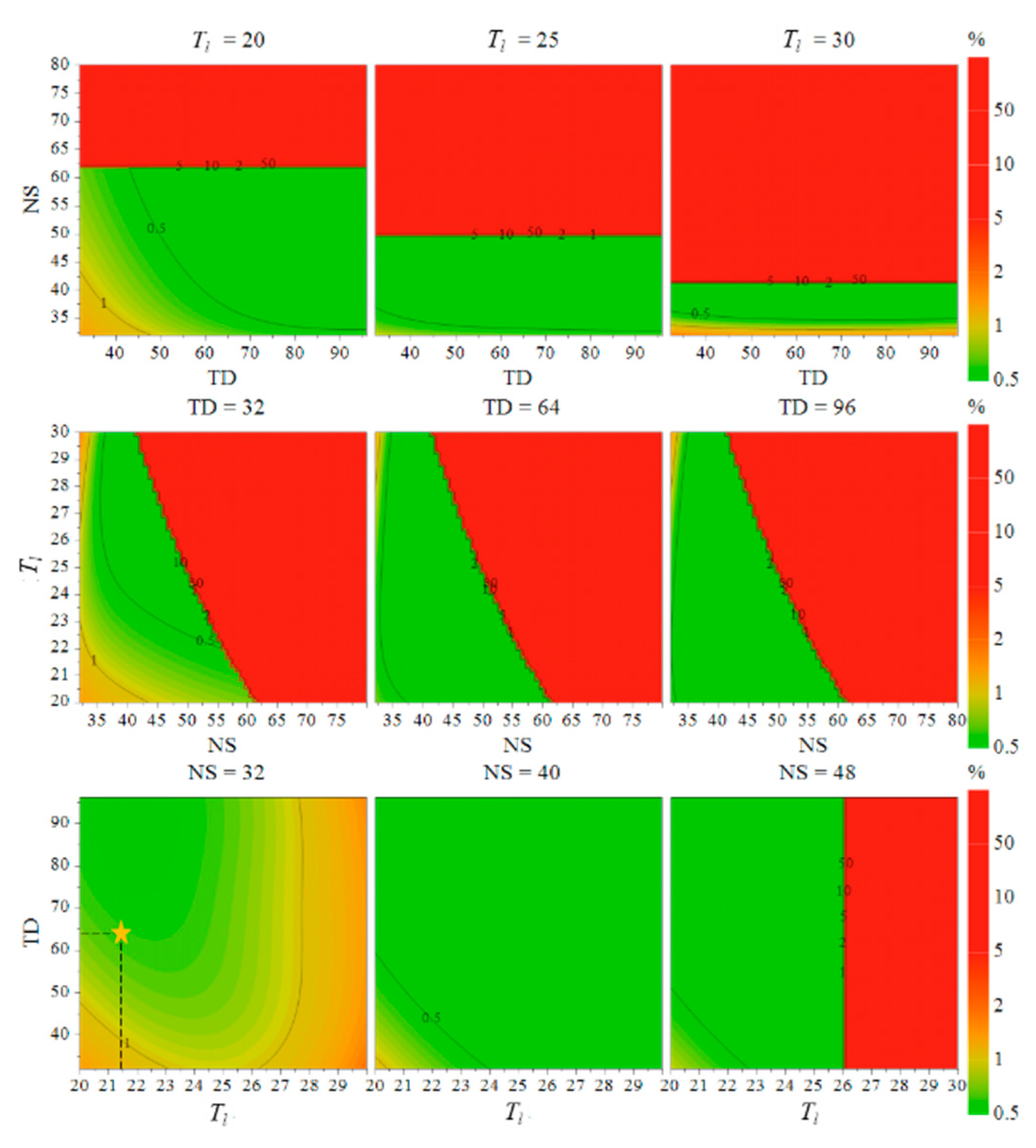

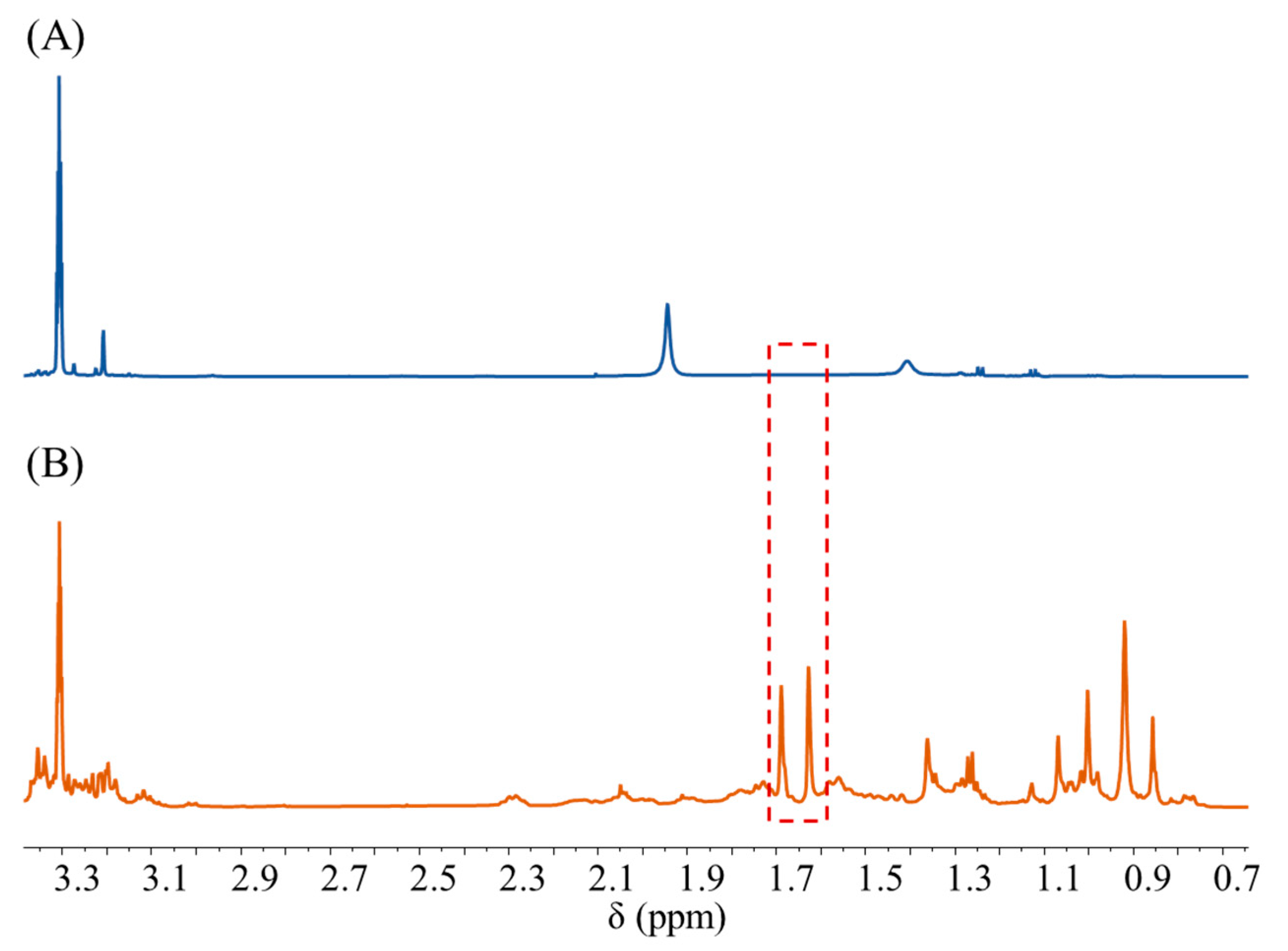
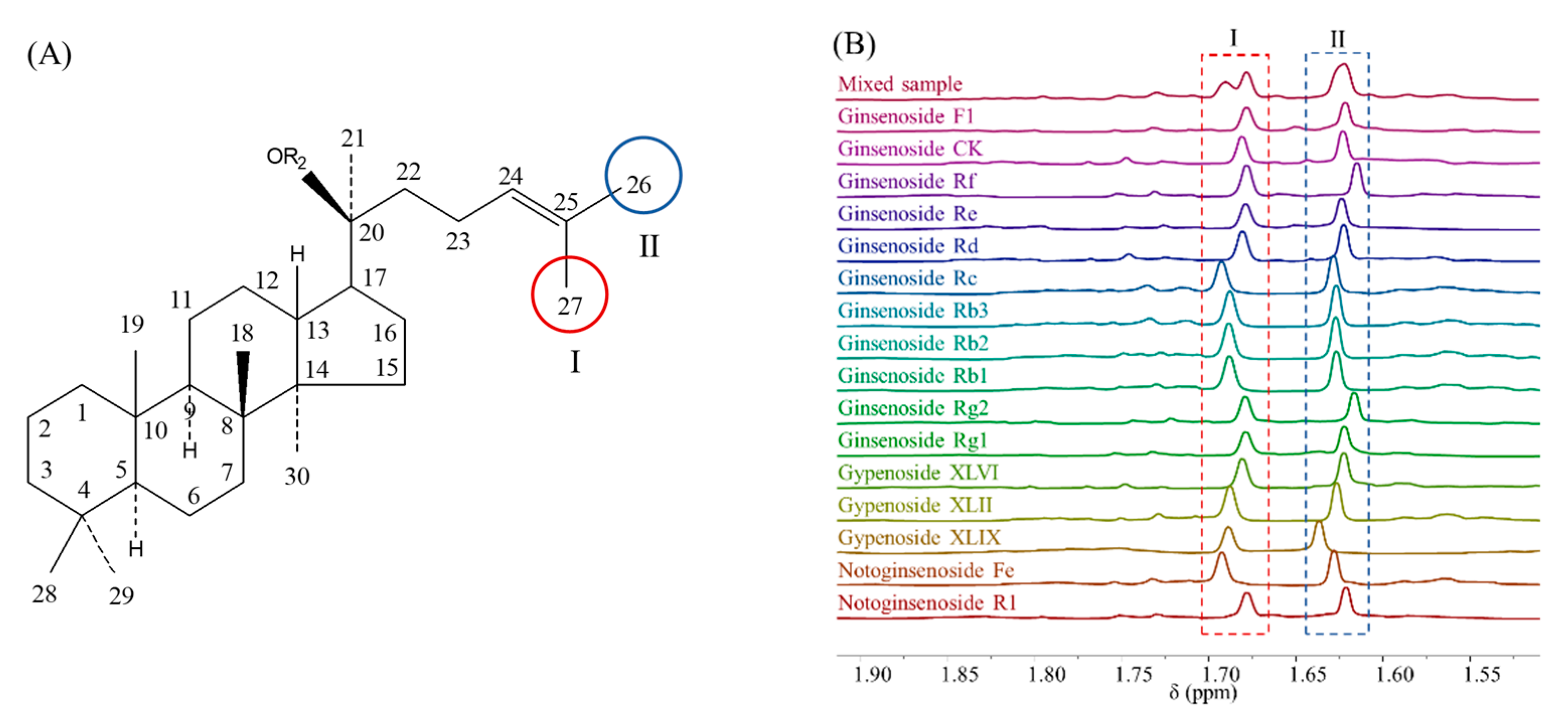
| NO. | x1 (mmol/L) | x2 (Times) | x3 (Times) | x4 (pcs) | x5 | x6 (Seconds) | x7 (K) |
|---|---|---|---|---|---|---|---|
| 1 | 5 (+1) | 0 (−1) | 128 (+1) | 16,384 (−1) | 40.3 (−1) | 2 (−1) | 300 (+1) |
| 2 | 5 | 8 (+1) | 16 (−1) | 65,536 (+1) | 40.3 | 2 | 296 (−1) |
| 3 | 0.5 (−1) | 8 | 128 | 16,384 | 161 (+1) | 2 | 296 |
| 4 | 5 | 0 | 128 | 65,536 | 40.3 | 30 (+1) | 296 |
| 5 | 5 | 8 | 16 | 65,536 | 161 | 2 | 300 |
| 6 | 5 | 8 | 128 | 16,384 | 161 | 30 | 296 |
| 7 | 0.5 | 8 | 128 | 65,536 | 40.3 | 30 | 300 |
| 8 | 0.5 | 0 | 128 | 65,536 | 161 | 2 | 300 |
| 9 | 0.5 | 0 | 16 | 65,536 | 161 | 30 | 296 |
| 10 | 5 | 0 | 16 | 16,384 | 161 | 30 | 300 |
| 11 | 0.5 | 8 | 16 | 16,384 | 40.3 | 30 | 300 |
| 12 | 0.5 | 0 | 16 | 16,384 | 40.3 | 2 | 296 |
| 13 | 2 (0) | 4 (0) | 64 (0) | 32,768 (0) | 90.5 (0) | 10 (0) | 298 (0) |
| 14 | 2 | 4 | 64 | 32,768 | 90.5 | 10 | 298 |
| 15 | 2 | 4 | 64 | 32,768 | 90.5 | 10 | 298 |
| NO. | Accuracy (%) | SNR | Resolution |
|---|---|---|---|
| 1 | 77.07 | 2446.8 | 0.71 |
| 2 | 17.65 | 1316.0 | 0.62 |
| 3 | 55.75 | 1601.0 | 1.35 |
| 4 | 0.291 | 4174.4 | 0.68 |
| 5 | 27.69 | 1343.4 | 0.61 |
| 6 | 0.709 | 2771.8 | 1.29 |
| 7 | 0.411 | 3407.8 | 0.96 |
| 8 | 23.07 | 3404.6 | 0.75 |
| 9 | 0.747 | 1660.5 | 0.72 |
| 10 | 9.572 | 1589.5 | 0.72 |
| 11 | 4.133 | 965.6 | 1.36 |
| 12 | 51.68 | 658.6 | 1.15 |
| 13 | 6.202 | 3288.7 | 0.63 |
| 14 | 6.055 | 3451.9 | 0.60 |
| 15 | 6.263 | 3318.1 | 0.60 |
| NO. | Parameter Setting | Evaluation Indexes | ||||
|---|---|---|---|---|---|---|
| X1 (Times) | X2 (pcs) | X3 (Seconds) | Accuracy (%) | SNR | Tq (Minutes) | |
| 1 | 32 (−1) | 32,768 (−1) | 20 (−1) | 1.742 | 1314.49 | 13.68 |
| 2 | 32 | 98,304 (+1) | 20.00 | 1.327 | 1270.59 | 13.68 |
| 3 | 80 (+1) | 32,768 | 20.00 | 1.567 | 1904.43 | 31.93 |
| 4 | 80 | 98,304 | 20.00 | 0.947 | 1908.12 | 31.93 |
| 5 | 32 | 32,768 | 30.00 (+1) | 0.785 | 1225.88 | 19.68 |
| 6 | 32 | 98,304 | 30.00 | 0.396 | 1217.23 | 19.68 |
| 7 | 80 | 32,768 | 30.00 | 0.186 | 2006.74 | 45.93 |
| 8 | 80 | 98,304 | 30.00 | 0.037 | 2010.88 | 45.93 |
| 9 | 56 | 16,384 (−α) | 25.00 | 2.161 | 1621.74 | 27.80 |
| 10 | 56 | 131,072 (+α) | 25.00 | 0.066 | 1678.88 | 27.80 |
| 11 | 16 (−α) | 65,536 | 25.00 | 1.238 | 843.20 | 9.27 |
| 12 | 96 (+α) | 65,536 | 25.00 | 0.130 | 2215.89 | 46.35 |
| 13 | 56 | 65,536 | 16.59 (−α) | 1.830 | 1689.57 | 19.40 |
| 14 | 56 | 65,536 | 33.41 (+α) | 0.049 | 1609.00 | 36.22 |
| 15 | 56 (0) | 65,536 (0) | 25.00 (0) | 0.723 | 1577.34 | 27.80 |
| 16 | 56 | 65,536 | 25.00 | 0.623 | 1637.77 | 27.80 |
| 17 | 56 | 65,536 | 25.00 | 0.718 | 1652.33 | 27.80 |
| Parameter Items | Regression Coefficient | ||
|---|---|---|---|
| Accuracy (%) | SNR | Tq (Minutes) | |
| b0 | 0.855 | 1633.050 | 27.802 |
| X1 | −0.228 | 345.696 | 10.240 |
| X2 | −0.338 | - | - |
| X3 | −0.485 | −4.90011 | 4.619 |
| X12 | - | −29.765 | 0.002 |
| X32 | - | - | 0.002 |
| X X13 | - | 36.901 | 1.701 |
| r2 | 0.853 | 0.988 | 1.000 |
| r2adj | 0.819 | 0.983 | 1.000 |
| No. | UV (mmol/L) | 1H qNMR (mmol/L) | Relative Deviation (%) |
|---|---|---|---|
| 1 | 36.45 | 36.53 | −0.21 |
| 2 | 39.56 | 41.12 | −3.78 |
| 3 | 43.07 | 42.26 | 1.92 |
| 4 | 40.86 | 41.80 | −2.25 |
| 5 | 38.94 | 40.80 | −4.55 |
| 6 | 36.45 | 36.68 | −0.61 |
| Mean values | 39.22 | 39.86 | −1.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Yang, J.; Zhao, F.; Xie, X.; Pan, J.; Qu, H. Application of the Analytical Procedure Lifecycle Concept to a Quantitative 1H NMR Method for Total Dammarane-Type Saponins. Pharmaceuticals 2023, 16, 947. https://doi.org/10.3390/ph16070947
Li W, Yang J, Zhao F, Xie X, Pan J, Qu H. Application of the Analytical Procedure Lifecycle Concept to a Quantitative 1H NMR Method for Total Dammarane-Type Saponins. Pharmaceuticals. 2023; 16(7):947. https://doi.org/10.3390/ph16070947
Chicago/Turabian StyleLi, Wenzhu, Jiayu Yang, Fang Zhao, Xinyuan Xie, Jianyang Pan, and Haibin Qu. 2023. "Application of the Analytical Procedure Lifecycle Concept to a Quantitative 1H NMR Method for Total Dammarane-Type Saponins" Pharmaceuticals 16, no. 7: 947. https://doi.org/10.3390/ph16070947
APA StyleLi, W., Yang, J., Zhao, F., Xie, X., Pan, J., & Qu, H. (2023). Application of the Analytical Procedure Lifecycle Concept to a Quantitative 1H NMR Method for Total Dammarane-Type Saponins. Pharmaceuticals, 16(7), 947. https://doi.org/10.3390/ph16070947





