Advances in the Applications of Bioinformatics and Chemoinformatics
Abstract
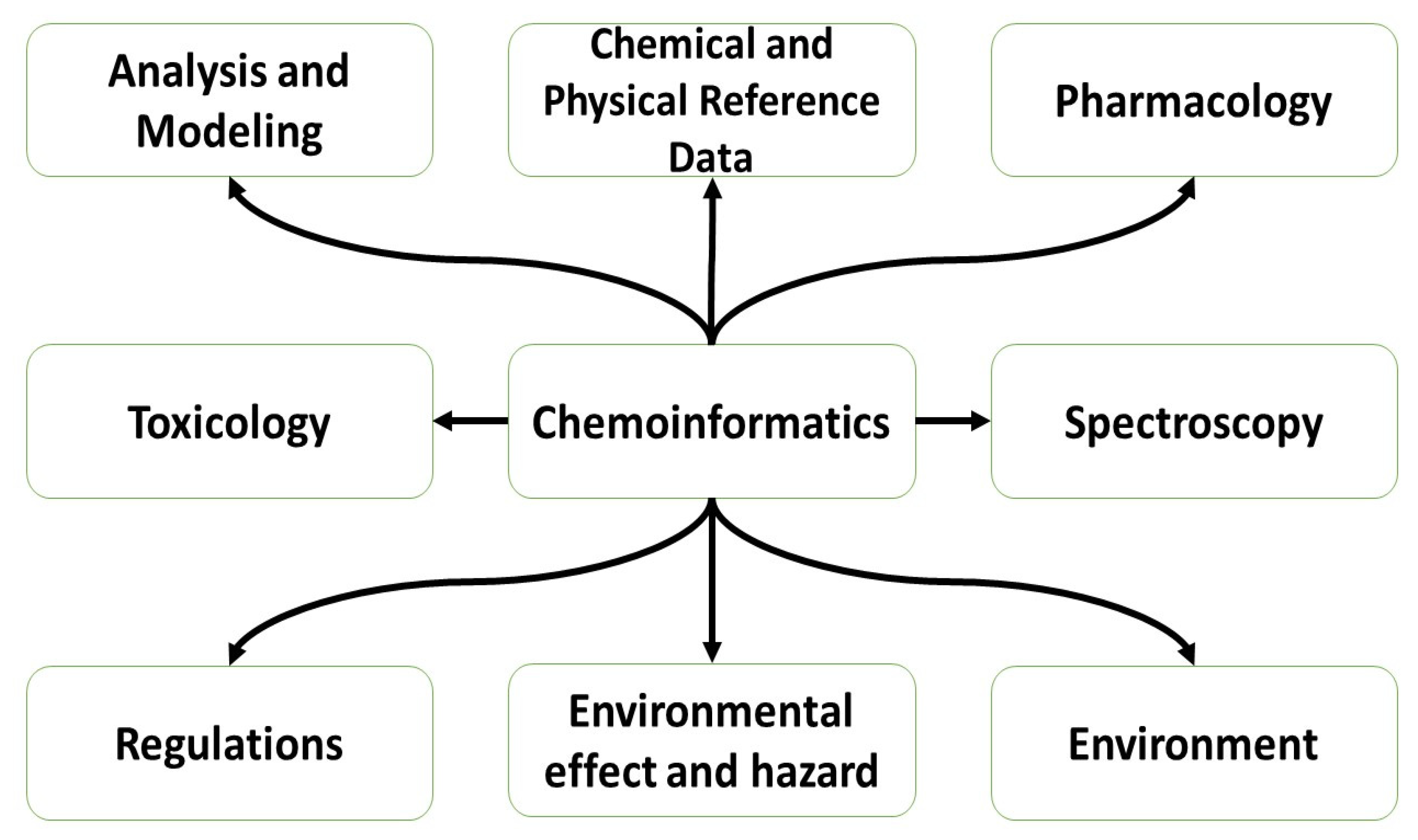
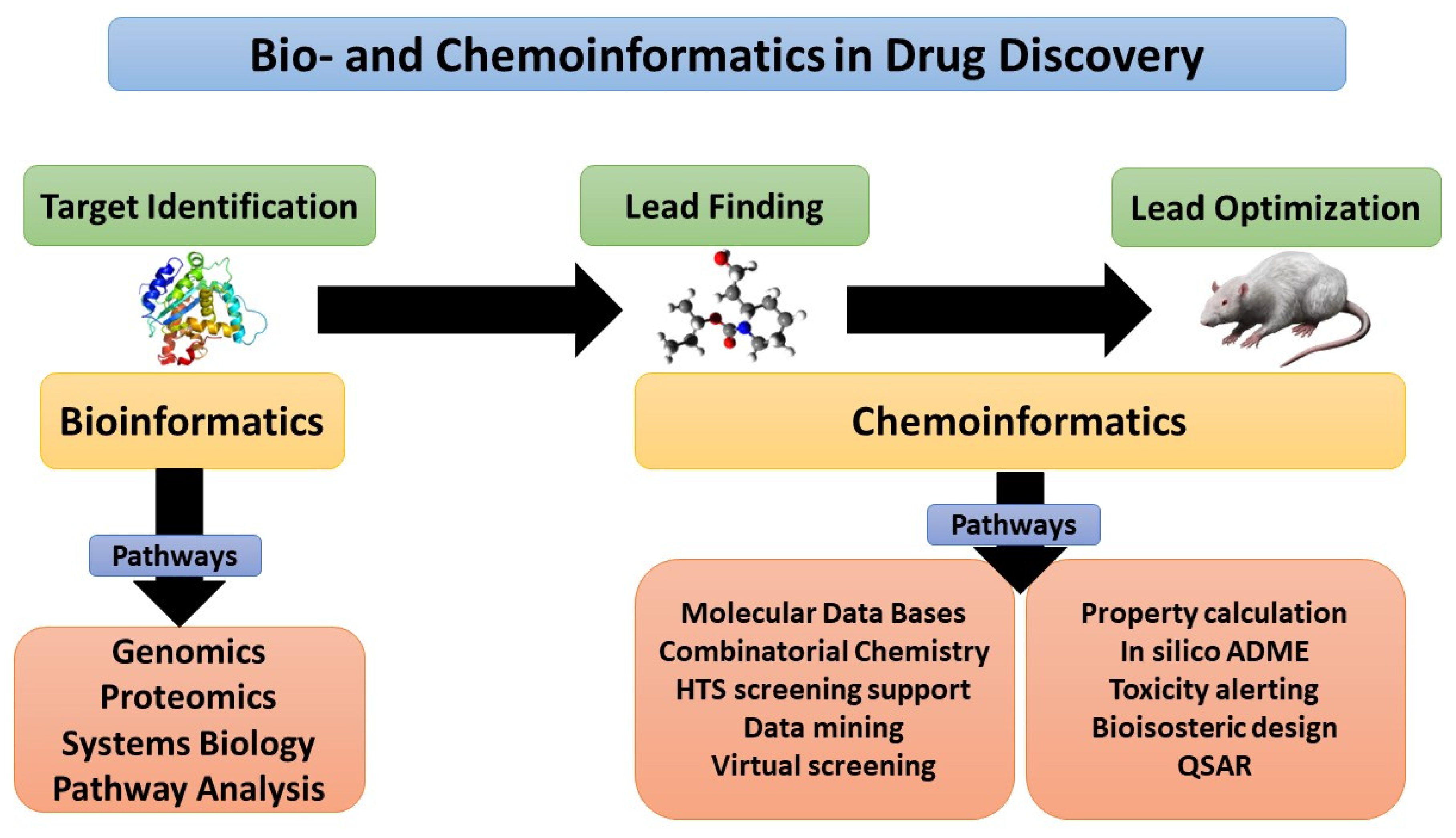
1. Introduction
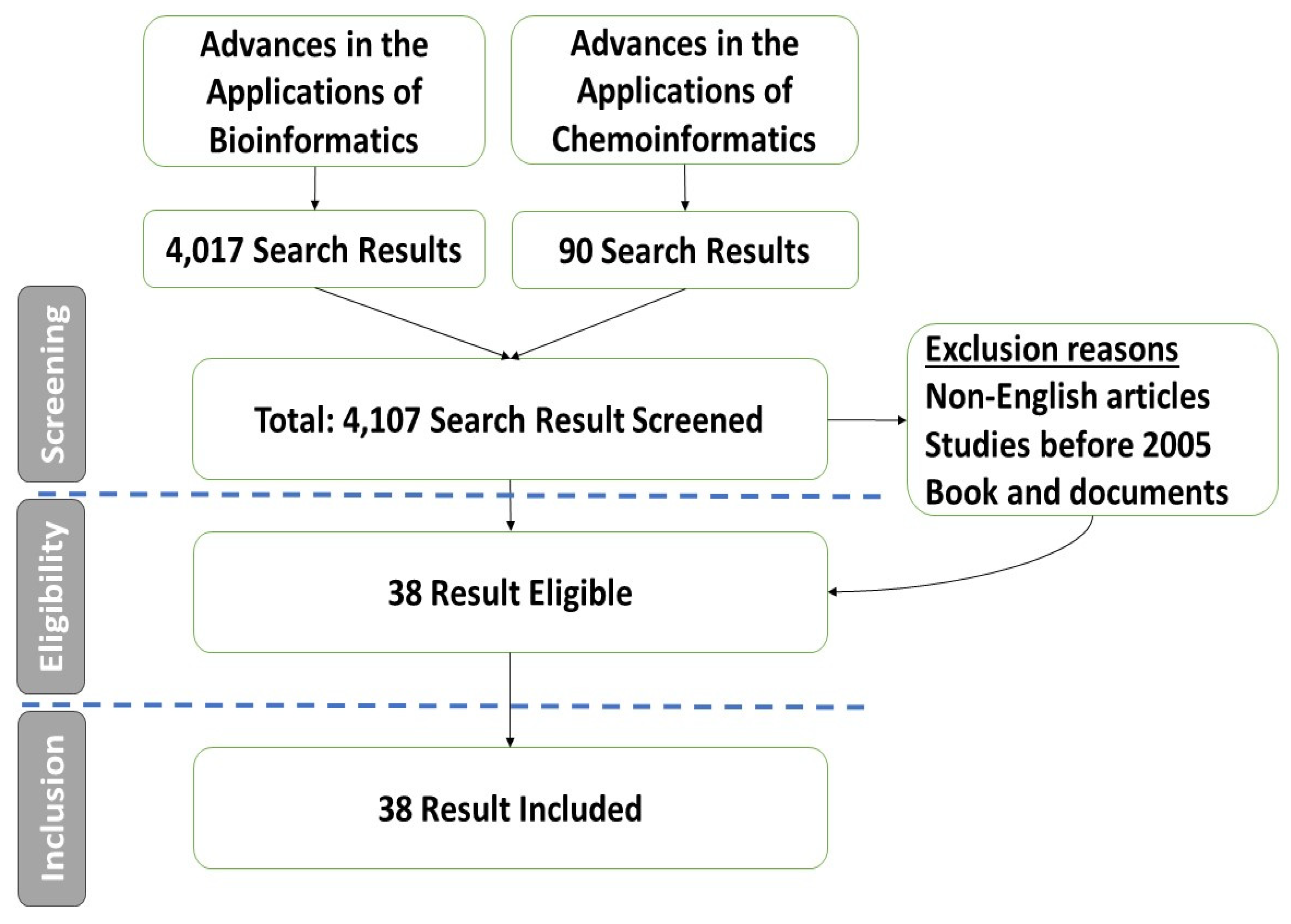
2. Materials and Methods
3. Drug Discovery and Design
3.1. Chemoinformatics and New Tetracycline Analogue
3.2. Bio- and Chemoinformatics in Identification of Novel Pyrazole and Benzimidazole Based Derivatives as Penicillin-Binding Protein 2a Inhibitors
3.3. Chemoinformatics Application in Phytochemistry
4. Clinical Applications
4.1. Bioinformatics and Heart Disease Classification
4.2. Bioinformatics and Diagnosis of Coronavirus Disease 2019
4.3. Bioinformatics and Genomic Correlation with Clinical Information and Disease State
Additionally, a research study using an Illumina short-read sequencer-based investigation of the entire genomes of nine Egyptian women showed that 12 SNPs were shared by the majority of the participants related to obesity and were concordant with their clinical diagnosis using 30x sequencing coverage. Also, the presence of the mtDNA mutation A4282G in all samples was reported.[34]
4.4. Bioinformatics and Multiple Drug Resistant Escherichia coli (E. coli) Isolation from Pediatric Cancer Patients
5. Optimization of Drug Delivery
5.1. Bio- and Chemoinformatics in Nose-To-Brain Formulation Targeting Meningitis
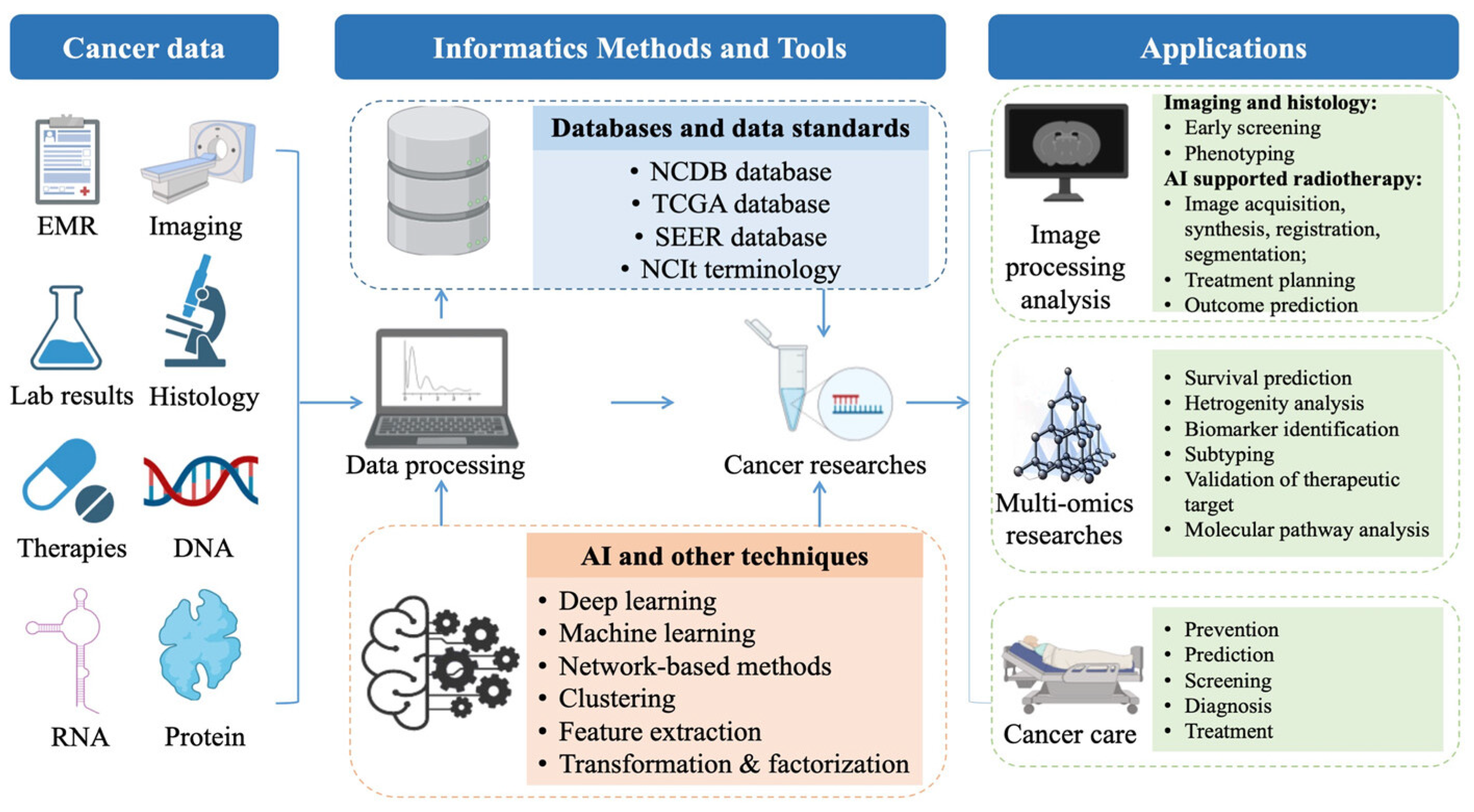
5.2. Chemoinformatics Targeting Cancer Cell Therapy
- (a)
- The incorporation of a range of informatics techniques and instruments makes it possible to scrutinize diverse cancer data and generate approaches for preventing, screening, diagnosing, and treating the disease.
- (b)
- The application of artificial intelligence (AI) algorithms holds the promise of enhancing desired therapeutic outcomes. The benefits of AI tools in interpreting medical images have been established in various environments and for a range of diseases.
- (c)
- This technology could be utilized to analyze data from multiple sources to identify patterns and early warning signs of cancer, thereby enabling prompt intervention and more effective treatment.
5.3. Bio- and Chemoinformatics in Nose-To-Brain Formulation for Treatment of Alzheimer Disease
6. Some Advances in New Algorithms and Artificial Intelligence Worldwide
6.1. Chemoinformatics and Hybrid Harris Hawks Optimization with Cuckoo Search
6.2. Chemoinformatics and Bioinformatics Integration with Artificial Intelligence (AI)
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S. Introduction to Cheminformatics. Curr. Protoc. Bioinform. 2007, 18, 14.1.1–14.1.21. [Google Scholar] [CrossRef]
- Yan, X.C.; Sanders, J.M.; Gao, Y.-D.; Tudor, M.; Haidle, A.M.; Klein, D.J.; Converso, A.; Lesburg, C.A.; Zang, Y.; Wood, H.B. Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery. J. Chem. Inf. Model. 2020, 60, 4144–4152. [Google Scholar] [CrossRef] [PubMed]
- Walters, W.P.; Patrick, W.W. Virtual Chemical Libraries. J. Med. Chem. 2019, 62, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Saldívar-González, F.I.; Huerta-García, C.S.; Medina-Franco, J.L. Chemoinformatics-based enumeration of chemical libraries: A tutorial. J. Cheminform. 2020, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.R.; Kroeplien, B. Exploring Virtual Scaffold Spaces. In Methods and Principles in Medicinal Chemistry; Brown, N., Ed.; Wiley: London, UK, 2013; pp. 83–104. [Google Scholar]
- Mak, L.; Marcus, D.; Howlett, A.; Yarova, G.; Duchateau, G.; Klaffke, W.; Bender, A.; Glen, R.C. Metrabase: A cheminformatics and bioinformatics database for small molecule transporter data analysis and (Q)SAR modeling. J. Cheminform. 2015, 7, 31. [Google Scholar] [CrossRef]
- Bhalerao, S.A.; Verma, D.R.; Rohan, L.; Teli, N.C.; Vinodkumar, S. Chemoinformatics: The Application of Informatics Methods to Solve Chemical Problems. Res. J. Pharm. Biol. Chem. Sci. 2013, 4, 475. [Google Scholar]
- Kumar, P. Clark’s Clinical Medicine; Elsevier: Amsterdam, The Netherlands, 2020; pp. 774–811. [Google Scholar]
- James, O. Clinical Pharmacology Made Ridiculously Simple; MedMaster: Metro Manila, Philippines, 2020; Volume 12, pp. 93–109. [Google Scholar]
- Levinson, W. Review of Medical Microbiology and Immunology; MC Graw Hill Press: New York, NY, USA, 2021; Volume 9, pp. 211–244. [Google Scholar]
- Oliva, B.; Gordon, G.; McNicholas, P.; Ellestad, G.; Chopra, I. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob. Agents Chemother. 2012, 36, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Metting Patricia, J. Physiology; Mc Graw Hill Education: New York, NY, USA, 2019; Volume 14, pp. 437–505. [Google Scholar]
- Aleksandrov, A.; Simonson, T. Molecular mechanics models for tetracycline analogs. J. Comput. Chem. 2009, 30, 243–255. [Google Scholar] [CrossRef]
- Kassab, M.M. Development of novel antimicrobial tetracycline analog b (iodocycline) by chemo-informatics. Ain Shams Med. J. 2022, 73, 969–981. [Google Scholar] [CrossRef]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The Basis for Resistance to β-Lactam Antibiotics by Penicillin-binding Protein 2a of Methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; de Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef]
- Shalaby, M.-A.W.; Dokla, E.M.; Serya, R.A.T.; Abouzid, K.A.M. Identification of novel pyrazole and benzimidazole based derivatives as PBP2a inhibitors: Design, synthesis, and biological evaluation. Arch. Pharm. Sci. Ain Shams Univ. 2019, 3, 228–245. [Google Scholar] [CrossRef]
- Nematallah, K.A.; Elmekkawy, S.; Abdollah, M.R.A.; Elmazar, M.M.; Abdel-Sattar, E.; Meselhy, M.R. Cheminformatics Application in the Phytochemical and Biological Study of Eucalyptus globulus L. Bark as a Potential Hepatoprotective Drug. ACS Omega 2022, 7, 7945–7956. [Google Scholar] [CrossRef] [PubMed]
- Hayat, U.; Jilani, M.I.; Rehman, R.; Nadeem, F. A Review on Eucalyptus globulus: A New Perspective in Therapeutics. Int. J. Chem. Biol. Sci. 2015, 8, 85–91. [Google Scholar]
- Mota, I.; Pinto, P.C.O.R.; Novo, C.; Sousa, G.; Guerreiro, O.; Guerra, Â.; Duarte, M.D.; Rodrigues, A.E. Eucalyptus globulus bark as A source of polyphenolic compounds with biological activity. O Papel 2013, 74, 57–64. [Google Scholar]
- Romano, B.; Pagano, E.; Montanaro, V.; Fortunato, A.L.; Milic, N.; Borrelli, F. Novel Insights into the Pharmacology of Flavonoids. Phytother. Res. 2013, 27, 1588–1596. [Google Scholar] [CrossRef]
- WHO. Cardiovascular Diseases (CDs)—Key Facts. 2017. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovasculardiseases-(cvds) (accessed on 11 July 2023).
- Srinivas, K.; Rani, B.K.; Govrdhan, A. Applications of data mining techniques in healthcare and prediction of heart attacks. Int. J. Comput. Sci. Eng. 2010, 2, 250–25562. [Google Scholar]
- Shafenoor Amin, M.; Kia Chiam, Y.; Dewi Varathan, K. Identification of significant features and data mining techniques in predicting heart disease. Telemat. Inform. 2019, 36, 82–93. [Google Scholar] [CrossRef]
- Maini, E.; Venkateswarlu, B.; Maini, B.; Marwaha, D. Machine learning–based heart disease prediction system for Indian population: An exploratory study done in South India. Med. J. Armed Forces India 2021, 77, 302–311. [Google Scholar] [CrossRef]
- El Sheikh, A.; Mahmoud, N.; Keshk, A.E. Heart Disease Classification Based on Hybrid Ensemble Stacking Technique. IJCI Int. J. Comput. Inf. 2021, 8, 1–8. [Google Scholar] [CrossRef]
- WHO. COVID-19. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 15 July 2023).
- Das, D.; Santosh, K.C.; Pal, U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys. Eng. Sci. Med. 2020, 43, 915–925. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Butt, C.; Gill, J.; Chun, D.; Babu, B.A. RETRACTED ARTICLE: Deep learning system to screen coronavirus disease 2019 pneumonia. Appl. Intell. 2023, 53, 4874. [Google Scholar] [CrossRef]
- Dawod, E.F.; Mahmoud, N.; Elsisi, A. Hybrid approach for COVID-19 detection from chest radiography. IJCI Int. J. Comput. Inf. 2021, 8, 71–76. [Google Scholar] [CrossRef]
- Hassan, N.E.; El-Masry, S.A.; Zarouk, W.; El Banna, R.A.; Mosaad, R.M.; Al-Tohamy, M.; Salamah, A.R. Obesity phenotype in relation to gene polymorphism among samples of Egyptian children and their mothers. Genes Dis. 2017, 5, 150–157. [Google Scholar] [CrossRef] [PubMed]
- ElHefnawi, M.; Hegazy, E.; Elfiky, A.; Jeon, Y.; Jeon, S.; Bhak, J.; Metwally, F.M.; Sugano, S.; Horiuchi, T.; Kazumi, A.; et al. Complete genome sequence and bioinformatics analysis of nine Egyptian females with clinical information from different geographic regions in Egypt. Gene 2020, 769, 145237. [Google Scholar] [CrossRef]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.-P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef]
- Fricke, W.F.; Welch, T.J.; McDermott, P.F.; Mammel, M.K.; LeClerc, J.E.; White, D.G.; Cebula, T.A.; Ravel, J. Comparative Genomics of the IncA/C Multidrug Resistance Plasmid Family. J. Bacteriol. 2009, 191, 4750–4757. [Google Scholar] [CrossRef]
- Sabat, A.J.; Budimir, A.; Nashev, D.; Sá-Leão, R.; van Dijl, J.M.; Laurent, F.; Grundmann, H.; Friedrich, A.W.; on behalf of the ESCMID Study Group of Epidemiological Markers (ESGEM). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance 2013, 18, 20380. [Google Scholar] [CrossRef]
- Stoesser, N.; Batty, E.M.; Eyre, D.W.; Morgan, M.; Wyllie, D.H.; Elias, C.D.O.; Johnson, J.R.; Walker, A.S.; Peto, T.E.A.; Crook, D.W. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother. 2013, 68, 2234–2244. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Tantawy, M.; Gouda, N.A.; Elzayat, M.G.; Gabra, S.; Nabih, A.; Diab, A.A.; El-Hadidi, M.; Bakry, U.; Shoeb, M.R.; et al. Genotypic characterization of multiple drug resistant Escherichia coli isolates from a pediatric cancer hospital in Egypt. Sci. Rep. 2020, 10, 4165. [Google Scholar] [CrossRef]
- Troendle, M.; Pettigrew, A. A systematic review of cases of meningitis in the absence of cerebrospinal fluid pleocytosis on lumbar puncture. BMC Infect. Dis. 2019, 19, 692. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, M.J.; McGill, F.; Solomon, T. Management of acute meningitis. Clin. Med. 2018, 18, 164–169. [Google Scholar] [CrossRef]
- Oordt-Speets, A.M.; Bolijn, R.; van Hoorn, R.C.; Bhavsar, A.; Kyaw, M.H. Global etiology of bacterial meningitis: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0198772. [Google Scholar] [CrossRef] [PubMed]
- Hathout, R.M.; Abdelhamid, S.G.; El-Housseiny, G.S.; Metwally, A.A. Comparing cefotaxime and ceftriaxone in combating meningitis through nose-to-brain delivery using bio/chemoinformatics tools. Sci. Rep. 2020, 10, 21250. [Google Scholar] [CrossRef] [PubMed]
- Gharib, A.F.; Eldeen, M.A.; Khalifa, A.S.; Elsawy, W.H.; Eed, E.M.; El Askary, A.; Eid, R.A.; Soltan, M.A.; Raafat, N. Assessment of Glutathione Peroxidase-1 (GPX1) Gene Expression as a Specific Diagnostic and Prognostic Biomarker in Malignant Pleural Mesothelioma. Diagnostics 2021, 11, 2285. [Google Scholar] [CrossRef]
- Zabady, S.; Mahran, N.; Soltan, M.A.; Eldeen, M.A.; Eid, R.A.; Albogami, S.; Fayad, E.; Matboli, M.; Habib, E.K.; Hasanin, A.H.; et al. Cyanidin-3-Glucoside Modulates hsa_circ_0001345/miRNA106b/ATG16L1 Axis Expression as a Potential Protective Mechanism against Hepatocellular Carcinoma. Curr. Issues Mol. Biol. 2022, 44, 1677–1687. [Google Scholar] [CrossRef]
- Soltan, M.A.; Eldeen, M.A.; Sajer, B.H.; Abdelhameed, R.F.A.; Al-Salmi, F.A.; Fayad, E.; Jafri, I.; Ahmed, H.E.M.; Eid, R.A.; Hassan, H.M.; et al. Integration of Chemoinformatics and Multi-Omics Analysis Defines ECT2 as a Potential Target for Cancer Drug Therapy. Biology 2023, 12, 613. [Google Scholar] [CrossRef]
- Hong, N.; Sun, G.; Zuo, X.; Chen, M.; Liu, L.; Wang, J.; Feng, X.; Shi, W.; Gong, M.; Ma, P. Application of informatics in cancer research and clinical practice: Opportunities and challenges. Cancer Innov. 2022, 1, 80–91. [Google Scholar] [CrossRef]
- Roney, C.; Kulkarni, P.; Arora, V.; Antich, P.; Bonte, F.; Wu, A.; Mallikarjuana, N.; Manohar, S.; Liang, H.-F.; Kulkarni, A.R.; et al. Targeted nanoparticles for drug delivery through the blood–brain barrier for Alzheimer’s disease. J. Control. Release 2005, 108, 193–214. [Google Scholar] [CrossRef]
- Wu, H.; Hu, K.; Jiang, X. From nose to brain: Understanding transport capacity and transport rate of drugs. Expert Opin. Drug Deliv. 2008, 5, 1159–1168. [Google Scholar] [CrossRef]
- Hathout, R.M.; El-Ahmady, S.; Metwally, A. Curcumin or bisdemethoxycurcumin for nose-to-brain treatment of Alzheimer disease? A bio/chemo-informatics case study. Nat. Prod. Res. 2018, 32, 2873–2881. [Google Scholar] [CrossRef]
- Houssein, E.H.; Hosney, M.E.; Elhoseny, M.; Oliva, D.; Mohamed, W.M.; Hassaballah, M. Hybrid Harris hawks optimization with cuckoo search for drug design and discovery in chemoinformatics. Sci. Rep. 2020, 10, 14439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K.; et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Fu, T.; Gao, W.; Zhao, Y.; Roohani, Y.; Leskovec, J.; Coley, C.W.; Xiao, C.; Sun, J.; Zitnik, M. Therapeutics Data Commons: Machine learning datasets and tasks for drug discovery and development. arXiv 2021, arXiv:2102.09548v2. [Google Scholar]
- Therapeutics Data Commons. Available online: https://tdcommons.ai/ (accessed on 22 June 2023).
- Hamzic, S.; Lewis, R.; Desrayaud, S.; Soylu, C.; Fortunato, M.; Gerebtzoff, G.; Rodríguez-Pérez, R. Predicting in vivo compound brain penetration using multi-task graph neural networks. J. Chem. Inf. Model. 2022, 62, 3180–3190. [Google Scholar] [CrossRef]
- Auslander, N.; Gussow, A.B.; Koonin, E.V. Incorporating Machine Learning into Established Bioinformatics Frameworks. Int. J. Mol. Sci. 2021, 22, 2903. [Google Scholar] [CrossRef]
- Abadi, S.; Avram, O.; Rosset, S.; Pupko, T.; Mayrose, I. ModelTeller: Model Selection for Optimal Phylogenetic Reconstruction Using Machine Learning. Mol. Biol. Evol. 2020, 37, 3338–3352. [Google Scholar] [CrossRef] [PubMed]
- Suvorov, A.; Hochuli, J.; Schrider, D.R. Accurate Inference of Tree Topologies from Multiple Sequence Alignments Using Deep Learning. Syst. Biol. 2019, 69, 221–233. [Google Scholar] [CrossRef]
- Jafari, R.; Javidi, M.M.; Rafsanjani, M.K. Using deep reinforcement learning approach for solving the multiple sequence alignment problem. SN Appl. Sci. 2019, 1, 592. [Google Scholar] [CrossRef]
- Fortin, F.A.; De Rainville, F.M.; Gardner, M.A.; Parizeau, M.; Gagńe, C. DEAP: Evolutionary Algorithms Made Easy. J. Mach. Learn. Res. 2012, 13, 2171–2175. [Google Scholar]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Oh, M.; Park, Y.; Kim, S. DeepFam: Deep learning based alignment-free method for protein family modeling and prediction. Bioinformatics 2018, 34, i254–i262. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L. Deep Recurrent Neural Network for Protein Function Prediction from Sequence. arXiv 2017, arXiv:1701.08318. [Google Scholar]
- Gussow, A.B.; Park, A.E.; Borges, A.L.; Shmakov, S.A.; Makarova, K.S.; Wolf, Y.I.; Bondy-Denomy, J.; Koonin, E.V. Ma-chine-Learning Approach Expands the Repertoire of Anti-CRISPR Protein Families. Nat. Commun. 2020, 11, 3784. [Google Scholar] [CrossRef]
- Eitzinger, S.; Asif, A.; Watters, K.E.; Iavarone, A.T.; Knott, G.J.; Doudna, J.A.; Minhas, F.U.A.A. Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 2020, 48, 4698–4708. [Google Scholar] [CrossRef]
- Gussow, A.B.; Auslander, N.; Faure, G.; Wolf, Y.I.; Zhang, F.; Koonin, E.V. Genomic Determinants of Pathogenicity in SARS-CoV-2 and Other Human Coronaviruses. Proc. Natl. Acad. Sci. USA 2020, 117, 15193–15199. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Wolf, Y.I.; Shabalina, S.A.; Koonin, E.V. A unique insert in the genomes of high-risk human papillomaviruses with a predicted dual role in conferring oncogenic risk. F1000Research 2019, 8, 1000. [Google Scholar] [CrossRef]
- Auslander, N.; Gussow, A.B.; Benler, S.; Wolf, Y.I.; Koonin, E.V. Seeker: Alignment-free identification of bacteriophage genomes by deep learning. Nucleic Acids Res. 2020, 48, e121. [Google Scholar] [CrossRef]
- Fang, Z.; Tan, J.; Wu, S.; Li, M.; Xu, C.; Xie, Z.; Zhu, H. PPR-Meta: A tool for identifying phages and plasmids from metagenomic fragments using deep learning. Gigascience 2019, 8, giz066. [Google Scholar] [CrossRef]
- Gao, J.; Thelen, J.J.; Dunker, A.K.; Xu, D. Musite, a Tool for Global Prediction of General and Kinase-specific Phosphorylation Sites. Mol. Cell. Proteom. 2010, 9, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Wang, M.; Liu, Y.; Zhao, X.-M.; Li, A. DeepPhos: Prediction of protein phosphorylation sites with deep learning. Bioinformatics 2019, 35, 2766–2773. [Google Scholar] [CrossRef]
- Caragea, C.; Sinapov, J.; Silvescu, A.; Dobbs, D.; Honavar, V. Glycosylation site prediction using ensembles of Support Vector Machine classifiers. BMC Bioinform. 2007, 8, 438. [Google Scholar] [CrossRef]
- Fukuda, H.; Tomii, K. DeepECA: An end-to-end learning framework for protein contact prediction from a multiple sequence alignment. BMC Bioinform. 2020, 21, 10–15. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Marbach, D.; Costello, J.C.; Küffner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Kellis, M.; Collins, J.J.; Stolovitzky, G.; et al. Wisdom of crowds for robust gene network inference. Nat. Methods 2012, 9, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Basu, S.; Kundu, M.; Nasipuri, M.; Plewczynski, D. PPI_SVM: Prediction of protein-protein interactions using machine learning, domain-domain affinities and frequency tables. Cell. Mol. Biol. Lett. 2011, 16, 264–278. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Q.; Ma, Q.; Yu, B. LightGBM-PPI: Predicting protein-protein interactions through LightGBM with mul-ti-information fusion. Chemom. Intell. Lab. Syst. 2019, 191, 54–64. [Google Scholar] [CrossRef]
- Aghakhani, S.; Qabaja, A.; Alhajj, R. Integration of k-means clustering algorithm with network analysis for drug-target inter-actions network prediction. Int. J. Data Min. Bioinform. 2018, 20, 185. [Google Scholar] [CrossRef]
- Shaked, I.; Oberhardt, M.A.; Atias, N.; Sharan, R.; Ruppin, E. Metabolic Network Prediction of Drug Side Effects. Cell Syst. 2016, 2, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Wildenhain, J.; Spitzer, M.; Dolma, S.; Jarvik, N.; White, R.; Roy, M.; Griffiths, E.; Bellows, D.S.; Wright, G.D.; Tyers, M. Prediction of Synergism from Chemical-Genetic Interactions by Machine Learning. Cell Syst. 2015, 1, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Rappoport, N.; Shamir, R. NEMO: Cancer subtyping by integration of partial multi-omic data. Bioinformatics 2019, 35, 3348–3356. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, Q.; Li, F.; Xu, Y.; Hu, C.; Wang, J.; Shang, D.; Zheng, X.; Yang, H.; Zhang, C.; et al. Identifying subpathway signatures for individualized anticancer drug response by integrating multi-omics data. J. Transl. Med. 2019, 17, 255. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, X.; Wang, J.; Zong, M.; Yang, H. Bioinformatics analysis of gene expression profile data to screen key genes involved in pulmonary sarcoidosis. Gene 2017, 596, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Luo, J.; Liang, C.; Cai, J.; Ding, P. A graph regularized non-negative matrix factorization method for identifying microRNA-disease associations. Bioinformatics 2018, 34, 239–248. [Google Scholar] [CrossRef]
- Xu, W.; Jiang, X.; Hu, X.; Li, G. Visualization of genetic disease-phenotype similarities by multiple maps t-SNE with Laplacian regularization. BMC Med. Genom. 2014, 7, S1. [Google Scholar] [CrossRef][Green Version]
- Abeel, T.; Helleputte, T.; Van de Peer, Y.; Dupont, P.; Saeys, Y. Robust Biomarker Identification for Cancer Diagnosis with En-semble Feature Selection Methods. Bioinformatics 2010, 26, 392–398. [Google Scholar] [CrossRef]
- Cun, Y.; Fröhlich, H. netClass: An R-package for network based, integrative biomarker signature discovery. Bioinformatics 2014, 30, 1325–1326. [Google Scholar] [CrossRef]
- Liu, L.; Chang, Y.; Yang, T.; Noren, D.P.; Long, B.; Kornblau, S.; Qutub, A.; Ye, J. Evolution-informed modeling improves outcome prediction for cancers. Evol. Appl. 2016, 10, 68–76. [Google Scholar] [CrossRef]
- Cheng, M.; Jiang, Y.; Xu, J.; Mentis, A.-F.A.; Wang, S.; Zheng, H.; Sahu, S.K.; Liu, L.; Xu, X. Spatially resolved transcriptomics: A comprehensive review of their technological advances, applications, and challenges. J. Genet. Genom. 2023; in press. [Google Scholar] [CrossRef]


| Reference | Problem Category | Goal | Bioinformatic Tools | ML Method | Bioinformatics Area |
|---|---|---|---|---|---|
| [62] | Biological sequence clustering | Protein family prediction | Clusters of Orthologous Groups (COGs) and G protein-coupled receptor (GPCR) dataset | CNN | Molecular evolution |
| [63] | Protein function prediction | BLAST and HMMER search | deep RNN | ||
| [64] | Anti-CRISPR proteins identification | MSA and PSI-BLAST | Random forest | ||
| [65] | K-mer based clustering (CD-HIT), BLAST | EXtreme Gradient Boosting | |||
| [66,67] | Viral pathogenicity feature identification | MSA, phylogenetic tree construction | SVM | ||
| [68] | Alignment free biological sequence analysis | Identification of viral genomes | BLAST, Sequence clustering, HHPRED | RNN | |
| [69] | BLAST | CNN | |||
| [70] | Post translational modifications | Phosphorylation sites prediction | Local sequence similarity | KNN | protein structure analysis |
| [71] | K-mer based clustering (CD-HIT), BLAST | CNN | |||
| [72] | Glycosylation sites prediction | curated glycosylated protein database (O-GLYCBASE) | ensemble SVM | ||
| [73] | Protein structure prediction | Protein contact prediction | MSA | CNN | |
| [74] | Prediction of distances between pairs of residues | MSA, HHPRED, PSI-BLAST | CNN | ||
| [75] | inference of biological networks | Gene regulatory network prediction | GeneNetWeaver, RegulonDB | SVM | systems biology |
| [76] | Protein-protein interaction network prediction | Domain affinity and frequency tables | SVM | ||
| [77] | Protein descriptors | Elastic-net regression | |||
| [78] | Analysis of biological networks | Drug target prediction | Network analysis tools | K-means | |
| [79] | Drug side effect prediction | Genome scale metabolic modeling | SVM | ||
| [80] | Drug Synergism prediction | A chemical-genetic interaction matrix | Random Forest Ensemble | ||
| [81] | Multi-omics integration | Cancer subtype prediction | Similarity based integration | Neighborhood based clustering | |
| [82] | Drug response prediction | Cancer hallmarks datasets, pathway data | logistic regression | ||
| [83] | Disease-associated genes investigation | Pulmonary sarcoidosis genes identification | Differential expression analysis | Hierarchical clustering | biomarker analysis for disease research |
| [84] | Identification of miRNA-disease association | Disease semantic information and miRNA functional information | NMF | ||
| [85] | Disease-phenotype visualization | OMIM database and human disease networks | t-SNE | ||
| [86] | Biomarker discovery | Cancer diagnosis | Reference gene selection | SVM | |
| [87] | Biomarker signature identification | Network-based gene selection | SVM | ||
| [88] | Cancer outcome prediction | Evolutionary conservation estimation | Random forest |
| Reference | Informatics Used | Application | Outcome |
|---|---|---|---|
| [15] | Chemoinformatics | Antibiotic discovery | Tetracycline analogue B (iodocycline). More active than tetracycline and less bacterial-resistant. |
| [27] | Bioinformatics | Disease Classification | The implementation of the ensemble model, in conjunction with brute force as a feature selection methodology, results in an exceptional accuracy rate of 97.8% for the categorization of heart disease. |
| [32] | Bioinformatics | Disease Diagnosis | Based on data from X-ray pictures and a CT scan, the findings showed a quantitative evaluation of COVID-19 using the suggested ensemble stacking technique, with percentages approaching 99%. |
| [43] | Chemo/ Bio-informatics | Special formulation for meningitis | The utilization of Ceftriaxone gelatin nanospheres or tripalmitin solid lipid nanoparticles has been proven to be a more practicable and effective nasal-to-brain formulation for the purpose of targeting meningitis in comparison to cefotaxime. |
| [19] | Chemoinformatics | Phytochemistry therapeutic discovery | The cytotoxic activity against HEPG2 and HUH-7 liver cancer cell lines attributed to the extract of Eucalyptus globulus bark was considerably high, and its absorption was found to be enhanced through the application of nanoformulation. |
| [46] | Chemoinformatics | Targeting Cancer Cells | Findings of the study demonstrate that ECT2 is capable of elevating both mRNA and protein concentrations in different types of human tumors, thereby enabling greater elimination of myeloid-derived suppressor cells (MDSC) and reducing the population of natural killer T (NKT) cells, resulting in a poor prognosis for survival. The investigation looked for medicines that could both inhibit ECT2 and function as anticancer agents. |
| [50] | Chemo/ Bio-informatics | Special formulation for Alzheimer disease | Curcumin outperformed bisdemethoxycurcumin (BDMC) in a nose-to-brain formulation for treatment of Alzheimer’s disease. |
| [18] | Chemo/ Bio-informatics | Testing Antibacterial activity against Resistant microorganisms | Three pyrazole and benzimidazole-based compounds examined showed modest bactericidal efficacy against MSSA, MRSA, and vancomycin-resistant Staphylococcus aureus (VRSA). |
| [34] | Bioinformatics | Genomic correlation with disease state | It was discovered that 12 SNPs were shared by the majority of the participants related to obesity and were concordant with their clinical diagnostics. In addition, results showed the presence of the mtDNA mutation A4282G in all samples; moreover, it is linked to chronic progressive external ophthalmoplegia |
| [39] | Bioinformatics | Multidrug-resistant organism identification | The highest represented genes among the 32 antimicrobial resistance genes discovered in pediatric cancer patients that exceeded the study threshold coverage were the aph(6)-Id gene, sul2, aph(3′)-Ia, sul1, dfrA12, aph(3″)-Ib, NDM-11, and TEM-220. Suggesting a horizontal transfer of resistance genes and plasmids between species in the context of nosocomial infections. |
| [51] | Cheminformatics | Hybrid Harris Hawks Optimization with Cuckoo Search | The experimental and statistical analyses demonstrate that the Hybrid Harris Hawks Optimization with Cuckoo Search method outperforms competitor algorithms. |
| [56,57,58,59,60,61] | Chemo/Bioinformatics | Integration with Artificial Intelligence | Different applications in molecular evolution, protein structure analysis, genomics for disease research, and system biology |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raslan, M.A.; Raslan, S.A.; Shehata, E.M.; Mahmoud, A.S.; Sabri, N.A. Advances in the Applications of Bioinformatics and Chemoinformatics. Pharmaceuticals 2023, 16, 1050. https://doi.org/10.3390/ph16071050
Raslan MA, Raslan SA, Shehata EM, Mahmoud AS, Sabri NA. Advances in the Applications of Bioinformatics and Chemoinformatics. Pharmaceuticals. 2023; 16(7):1050. https://doi.org/10.3390/ph16071050
Chicago/Turabian StyleRaslan, Mohamed A., Sara A. Raslan, Eslam M. Shehata, Amr S. Mahmoud, and Nagwa A. Sabri. 2023. "Advances in the Applications of Bioinformatics and Chemoinformatics" Pharmaceuticals 16, no. 7: 1050. https://doi.org/10.3390/ph16071050
APA StyleRaslan, M. A., Raslan, S. A., Shehata, E. M., Mahmoud, A. S., & Sabri, N. A. (2023). Advances in the Applications of Bioinformatics and Chemoinformatics. Pharmaceuticals, 16(7), 1050. https://doi.org/10.3390/ph16071050






