Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species
Abstract
1. Introduction
1.1. Overview of Ranunculaceae Active Compounds
| Pharmacological Effects | Cell Lines | Mechanism of Action | Reference |
|---|---|---|---|
| Anti-inflammatory activity | Mouse leukemic monocyte/macrophage cell line RAW264.7 | Suppressing NF-kB and NFATc1 activation and DC-STAMP expression | [39] |
| Anti-rheumatic activities | Rheumatoid arthritis HFLS-RA fibroblast-like synoviocytes | [40] | |
| Analgesia activity | Male wildtype FVB mice and Male Mdr1a−/− FVB mice | Mdr1a deficiency | [41] |
| the rat chronic constriction injury of an infraorbital nerve model. | N-methyl-D-aspartate receptor | [42] | |
| Mice pain models caused by hot plate, acetic acid, formalin, and CFA | - | [14] | |
| Anti-cancer activity | Pancreatic cancer cell lines Miacapa-2 and PANC-1 | Suppressing cancer cell growth and increasing cell apoptosis | [43] |
| Human cervical carcinoma HeLa cells | Upregulating mRNA expression levels of eIF2α, ATF4, IRE1, XBP1, ATF6, PERK | [44] | |
| Human breast cancer cell line MDA-MB-231BO | Inhibiting cancer cell invasion by an alteration of the TGF-β/Smad signaling pathway and down-regulating of NF-κB and RANK expressions. | [45] | |
| Human OVCA A2780 cell line | Adjusting ERβ-mediated apoptosis, DNA damage and migration | [8] | |
1.2. The Choice of Natural Compounds
1.3. Molecular Targets Obtain Using Prediction Target Database
2. Results
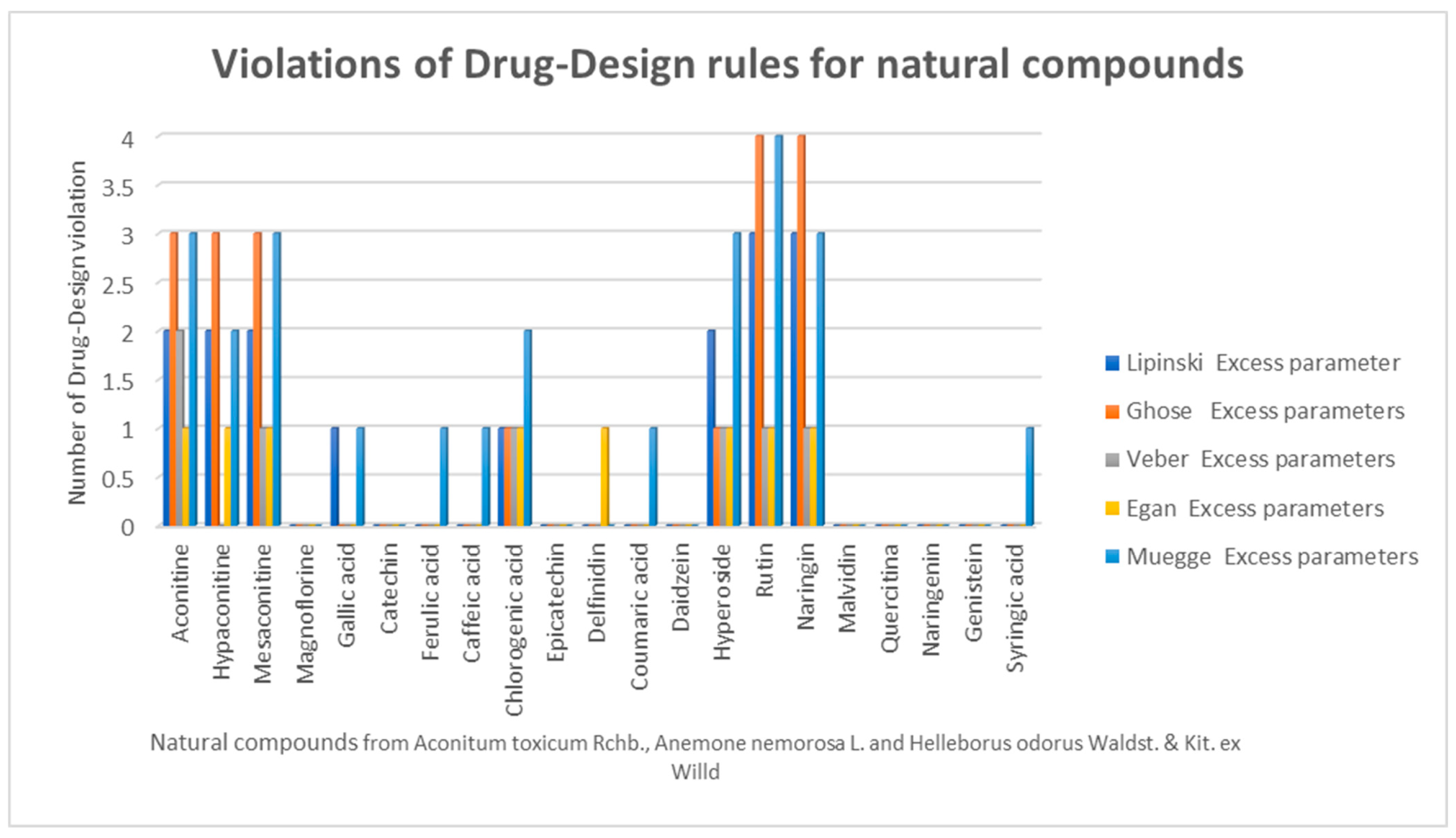
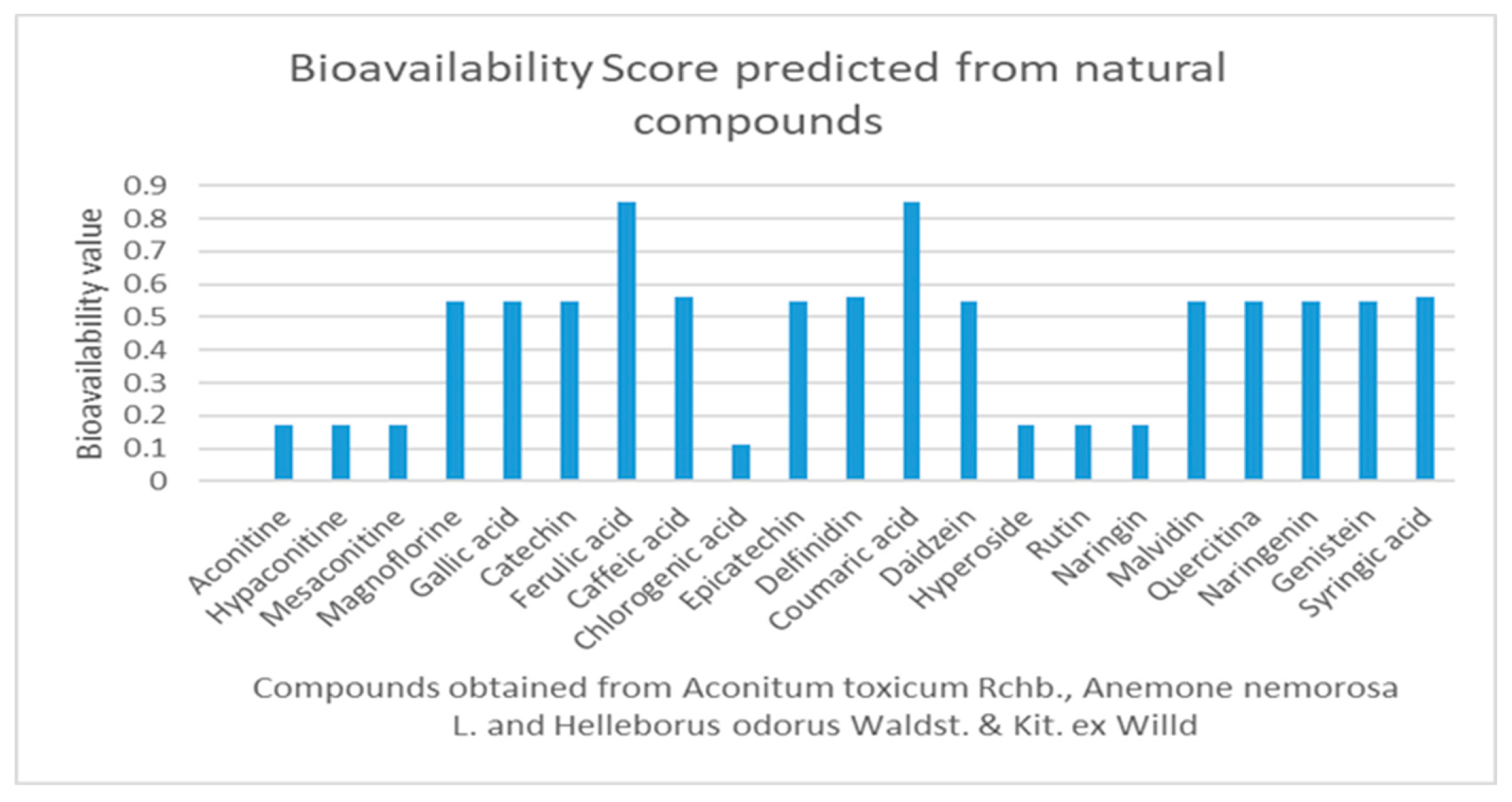
2.1. Drug-Likeness, Pharmacokinetics, and Pharmacogenomics Profiles of Compounds
2.2. Identification of Physico-Chemical Properties for the Pharmacological Profile
2.3. Pharmacokinetic, Pharmacogenomic Profile, and Toxicity of Natural Compounds (ADME-Tox)
2.4. Pharmacodynamics Profiles of Studied Compounds
2.5. Molecular Docking Results
3. Materials and Methods
3.1. Molecular Modelling of Chemical Compounds
3.2. Prediction of Compounds Drug- and Lead-Likeness Features
3.3. Identification of Important Physico-Chemical Properties for the Pharmacological Profile
3.4. Evaluation of the Pharmacokinetic and Pharmacogenomic Profile of Natural Compounds
3.5. Toxicity Profile of Natural Compounds
3.6. Pharmacodynamics Profile of Compounds
3.7. Molecular Modeling
3.8. Molecular Docking
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Yu, L.; Shi, Q.; Liu, Y.; Zhang, Y.; Wang, S.; Lai, X. An Insight into Current Advances on Pharmacology, Pharmacokinetics, Toxicity and Detoxification of Aconitine. Biomed. Pharmacother. 2022, 151, 113115. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Mai, C.-T.; Zheng, D.-C.; He, Y.-F.; Feng, S.-L.; Li, Y.-Z.; Liu, C.-X.; Zhou, H.; Liu, L. Wutou Decoction Ameliorates Experimental Rheumatoid Arthritis via Regulating NF-KB and Nrf2: Integrating Efficacy-Oriented Compatibility of Traditional Chinese Medicine. Phytomedicine 2021, 85, 153522. [Google Scholar] [CrossRef]
- Wang, J.-J.; Lou, H.-Y.; Liu, Y.; Han, H.-P.; Ma, F.-W.; Pan, W.-D.; Chen, Z. Profiling Alkaloids in Aconitum Pendulum N. Busch Collected from Different Elevations of Qinghai Province Using Widely Targeted Metabolomics. Phytochemistry 2022, 195, 113047. [Google Scholar] [CrossRef]
- Mares, C.; Udrea, A.-M.; Buiu, C.; Staicu, A.; Avram, S. Therapeutic Potentials of Aconite-like Alkaloids - Bioinformatics and Experimental Approaches. Mini Rev. Med. Chem. 2023. [Google Scholar] [CrossRef]
- Liao, Y.-P.; Shen, L.-H.; Cai, L.-H.; Chen, J.; Shao, H.-Q. Acute Myocardial Necrosis Caused by Aconitine Poisoning: A Case Report. World J. Clin. Cases 2022, 10, 12416–12421. [Google Scholar] [CrossRef]
- Chan, T.Y.K. Aconite Poisoning. Clin. Toxicol. Phila. Pa 2009, 47, 279–285. [Google Scholar] [CrossRef]
- Mi, L.; Li, Y.-C.; Sun, M.-R.; Zhang, P.-L.; Li, Y.; Yang, H. A Systematic Review of Pharmacological Activities, Toxicological Mechanisms and Pharmacokinetic Studies on Aconitum Alkaloids. Chin. J. Nat. Med. 2021, 19, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, Y.; Zheng, Y. Antitumor Effects of Aconitine in A2780 Cells via Estrogen Receptor Β-mediated Apoptosis, DNA Damage and Migration. Mol. Med. Rep. 2020, 22, 2318–2328. [Google Scholar] [CrossRef] [PubMed]
- Singhuber, J.; Zhu, M.; Prinz, S.; Kopp, B. Aconitum in Traditional Chinese Medicine—A Valuable Drug or an Unpredictable Risk? J. Ethnopharmacol. 2009, 126, 18–30. [Google Scholar] [CrossRef]
- Gao, X.; Hu, J.; Zhang, X.; Zuo, Y.; Wang, Y.; Zhu, S. Research Progress of Aconitine Toxicity and Forensic Analysis of Aconitine Poisoning. Forensic Sci. Res. 2018, 5, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wu, Q.; Li, Q.; Lv, T.; Peng, T.-F.; Yin, S.; Jin, H.-Z. Antinociceptive Diterpenoid Alkaloids from the Roots of Aconitum Austroyunnanense. J. Asian Nat. Prod. Res. 2023, 25, 132–138. [Google Scholar] [CrossRef]
- Ji, X.; Yang, M.; Or, K.H.; Yim, W.S.; Zuo, Z. Tissue Accumulations of Toxic Aconitum Alkaloids after Short-Term and Long-Term Oral Administrations of Clinically Used Radix Aconiti Lateralis Preparations in Rats. Toxins 2019, 11, 353. [Google Scholar] [CrossRef]
- Lu, H.; Mei, L.; Guo, Z.; Wu, K.; Zhang, Y.; Tang, S.; Zhu, Y.; Zhao, B. Hematological and Histopathological Effects of Subacute Aconitine Poisoning in Mouse. Front. Vet. Sci. 2022, 9, 874660. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Han, J.; Chen, J.; Zhang, Y.; Huang, Q.; Wang, Y.; Qi, X.; Liu, Z.; Leung, E.L.-H.; Wang, D.; et al. Comparison of Analgesic Activities of Aconitine in Different Mice Pain Models. PLoS ONE 2021, 16, e0249276. [Google Scholar] [CrossRef]
- Zhang, Y.; Zong, X.; Wu, J.-L.; Liu, Y.; Liu, Z.; Zhou, H.; Liu, L.; Li, N. Pharmacokinetics and Tissue Distribution of Eighteen Major Alkaloids of Aconitum Carmichaelii in Rats by UHPLC-QQQ-MS. J. Pharm. Biomed. Anal. 2020, 185, 113226. [Google Scholar] [CrossRef] [PubMed]
- PubChem Aconitine. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/245005 (accessed on 29 May 2023).
- Seiman, D.D.; Batalu, A.; Seiman, C.D.; Ciopec, M.; Udrea, A.M.; Motoc, M.; Negrea, A.; Avram, S. Pharmacological Effects of Natural Compounds Extracted from Urtica Dioica Evaluated by in Silico and Experimental Methods. Rev Chim Buchar. 2018, 69, 2377–2381. [Google Scholar] [CrossRef]
- Avram, S.; Stan, M.S.; Udrea, A.M.; Buiu, C.; Boboc, A.A.; Mernea, M. 3D-ALMOND-QSAR Models to Predict the Antidepressant Effect of Some Natural Compounds. Pharmaceutics 2021, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Iban-Arias, R.; Sebastian-Valverde, M.; Wu, H.; Lyu, W.; Wu, Q.; Simon, J.; Pasinetti, G.M. Role of Polyphenol-Derived Phenolic Acid in Mitigation of Inflammasome-Mediated Anxiety and Depression. Biomedicines 2022, 10, 1264. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, J.; Bi, W.; Ferruzzi, M.; Yemul, S.; Freire, D.; Mazzola, P.; Ho, L.; Dubner, L.; Pasinetti, G.M. Novel Application of Brain-Targeting Polyphenol Compounds in Sleep Deprivation-Induced Cognitive Dysfunction. Neurochem. Int. 2015, 89, 191–197. [Google Scholar] [CrossRef]
- de la Rosa, L.A.; Alvarez-Parrilla, E.; Shahidi, F. Phenolic Compounds and Antioxidant Activity of Kernels and Shells of Mexican Pecan (Carya Illinoinensis). J. Agric. Food Chem. 2011, 59, 152–162. [Google Scholar] [CrossRef]
- Addepalli, V.; Suryavanshi, S.V. Catechin Attenuates Diabetic Autonomic Neuropathy in Streptozotocin Induced Diabetic Rats. Biomed. Pharmacother. 2018, 108, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Agunloye, O.M.; Oboh, G.; Ademiluyi, A.O.; Ademosun, A.O.; Akindahunsi, A.A.; Oyagbemi, A.A.; Omobowale, T.O.; Ajibade, T.O.; Adedapo, A.A. Cardio-Protective and Antioxidant Properties of Caffeic Acid and Chlorogenic Acid: Mechanistic Role of Angiotensin Converting Enzyme, Cholinesterase and Arginase Activities in Cyclosporine Induced Hypertensive Rats. Biomed. Pharmacother. 2019, 109, 450–458. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, F.F.; de Paulo Farias, D.; Neri-Numa, I.A.; Pastore, G.M. Polyphenols and Their Applications: An Approach in Food Chemistry and Innovation Potential. Food Chem. 2021, 338, 127535. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, Y.; Phani Kumar, G.; Ramya, E.M.; Anilakumar, K.R. Gallic Acid Protects 6-OHDA Induced Neurotoxicity by Attenuating Oxidative Stress in Human Dopaminergic Cell Line. Neurochem. Res. 2018, 43, 1150–1160. [Google Scholar] [CrossRef]
- Sakalauskas, A.; Ziaunys, M.; Smirnovas, V. Gallic Acid Oxidation Products Alter the Formation Pathway of Insulin Amyloid Fibrils. Sci. Rep. 2020, 10, 14466. [Google Scholar] [CrossRef]
- Seo, E.-J.; Fischer, N.; Efferth, T. Phytochemicals as Inhibitors of NF-ΚB for Treatment of Alzheimer’s Disease. Pharmacol. Res. 2018, 129, 262–273. [Google Scholar] [CrossRef]
- Ogut, E.; Armagan, K.; Gül, Z. The Role of Syringic Acid as a Neuroprotective Agent for Neurodegenerative Disorders and Future Expectations. Metab. Brain Dis. 2022, 37, 859–880. [Google Scholar] [CrossRef]
- Nouri, Z.; Fakhri, S.; El-Senduny, F.F.; Sanadgol, N.; Abd-ElGhani, G.E.; Farzaei, M.H.; Chen, J.-T. On the Neuroprotective Effects of Naringenin: Pharmacological Targets, Signaling Pathways, Molecular Mechanisms, and Clinical Perspective. Biomolecules 2019, 9, 690. [Google Scholar] [CrossRef]
- Enogieru, A.B.; Haylett, W.; Hiss, D.C.; Bardien, S.; Ekpo, O.E. Rutin as a Potent Antioxidant: Implications for Neurodegenerative Disorders. Oxid. Med. Cell. Longev. 2018, 2018, 6241017. [Google Scholar] [CrossRef]
- Xie, J.; Guo, L.; Pang, G.; Wu, X.; Zhang, M. Modulation Effect of Semen Ziziphi Spinosae Extracts on IL-1β, IL-4, IL-6, IL-10, TNF-α and IFN-γ in Mouse Serum. Nat. Prod. Res. 2011, 25, 464–467. [Google Scholar] [CrossRef]
- Li, B.; Han, L.; Cao, B.; Yang, X.; Zhu, X.; Yang, B.; Zhao, H.; Qiao, W. Use of Magnoflorine-Phospholipid Complex to Permeate Blood-Brain Barrier and Treat Depression in the CUMS Animal Model. Drug Deliv. 2019, 26, 566–574. [Google Scholar] [CrossRef]
- Yi, J.H.; Moon, S.; Cho, E.; Kwon, H.; Lee, S.; Jeon, J.; Park, A.Y.; Lee, Y.H.; Kwon, K.J.; Ryu, J.H.; et al. Hyperoside Improves Learning and Memory Deficits by Amyloid Β1-42 in Mice through Regulating Synaptic Calcium-Permeable AMPA Receptors. Eur. J. Pharmacol. 2022, 931, 175188. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, Y.-P.; Liu, H.-Y.; Gu, J.-H.; Zhou, X.-F.; Yue-Qin, Z. Long-Term Oral Administration of Hyperoside Ameliorates AD-Related Neuropathology and Improves Cognitive Impairment in APP/PS1 Transgenic Mice. Neurochem. Int. 2021, 151, 105196. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Buiu, C.; Putz, M.V.; Avram, S. Learning the Relationship between the Primary Structure of HIV Envelope Glycoproteins and Neutralization Activity of Particular Antibodies by Using Artificial Neural Networks. Int. J. Mol. Sci. 2016, 17, 1710. [Google Scholar] [CrossRef] [PubMed]
- Demir, Y.; Durmaz, L.; Taslimi, P.; Gulçin, İ. Antidiabetic Properties of Dietary Phenolic Compounds: Inhibition Effects on α-Amylase, Aldose Reductase, and α-Glycosidase. Biotechnol. Appl. Biochem. 2019, 66, 781–786. [Google Scholar] [CrossRef]
- Garbiec, E.; Cielecka-Piontek, J.; Kowalówka, M.; Hołubiec, M.; Zalewski, P. Genistein-Opportunities Related to an Interesting Molecule of Natural Origin. Mol. Basel Switz. 2022, 27, 815. [Google Scholar] [CrossRef]
- Zeng, X.; He, L.; Wang, S.; Wang, K.; Zhang, Y.; Tao, L.; Li, X.; Liu, S. Aconine Inhibits RANKL-Induced Osteoclast Differentiation in RAW264.7 Cells by Suppressing NF-ΚB and NFATc1 Activation and DC-STAMP Expression. Acta Pharmacol. Sin. 2016, 37, 255–263. [Google Scholar] [CrossRef]
- Zhang, L.; Siyiti, M.; Zhang, J.; Yao, M.; Zhao, F. Anti-Inflammatory and Anti-Rheumatic Activities in Vitro of Alkaloids Separated from Aconitum Soongoricum Stapf. Exp. Ther. Med. 2021, 21, 493. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, J.; Zhao, M.; Song, W.; Qi, X.; Wang, Y.; Lu, L.; Liu, Z. Mdr1a Plays a Crucial Role in Regulating the Analgesic Effect and Toxicity of Aconitine by Altering Its Pharmacokinetic Characteristics. Toxicol. Appl. Pharmacol. 2017, 320, 32–39. [Google Scholar] [CrossRef]
- Çankal, D.; Akkol, E.K.; Kılınç, Y.; İlhan, M.; Capasso, R. An Effective Phytoconstituent Aconitine: A Realistic Approach for the Treatment of Trigeminal Neuralgia. Mediators Inflamm. 2021, 2021, 6676063. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.-L.; Xia, L.-P.; Zhou, F.-X.; Mao, G.-Z.; Xu, L.-X. Aconitine Induces Cell Apoptosis in Human Pancreatic Cancer via NF-ΚB Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4955–4964. [Google Scholar] [PubMed]
- Li, X.-M.; Liu, J.; Pan, F.-F.; Shi, D.-D.; Wen, Z.-G.; Yang, P.-L. Quercetin and Aconitine Synergistically Induces the Human Cervical Carcinoma HeLa Cell Apoptosis via Endoplasmic Reticulum (ER) Stress Pathway. PLOS ONE 2018, 13, e0191062. [Google Scholar] [CrossRef] [PubMed]
- Guo, B. Effects of Osthole, Psoralen, Aconitine on Breast Cancer MDA-MB-231BO Cell Line Inhibition in Vitro. J. Chin. Integr. Med. 2011, 9, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Udrea, A.-M.; Dinache, A.; Staicu, A.; Avram, S. Target Prediction of 5,10,15,20-Tetrakis(4′-Sulfonatophenyl)-Porphyrin Using Molecular Docking. Pharmaceutics 2022, 14, 2390. [Google Scholar] [CrossRef]
- Tozar, T.; Santos Costa, S.; Udrea, A.-M.; Nastasa, V.; Couto, I.; Viveiros, M.; Pascu, M.L.; Romanitan, M.O. Anti-Staphylococcal Activity and Mode of Action of Thioridazine Photoproducts. Sci. Rep. 2020, 10, 18043. [Google Scholar] [CrossRef]
- Liu, M.; Cao, Y.; Lv, D.; Zhang, W.; Zhu, Z.; Zhang, H.; Chai, Y. Effect of Processing on the Alkaloids in Aconitum Tubers by HPLC-TOF/MS. J. Pharm. Anal. 2017, 7, 170–175. [Google Scholar] [CrossRef]
- Avram, S.; Milac, A.-L.; Mihailescu, D. 3D-QSAR Study Indicates an Enhancing Effect of Membrane Ions on Psychiatric Drugs Targeting Serotonin Receptor 5-HT1A. Mol. Biosyst. 2012, 8, 1418–1425. [Google Scholar] [CrossRef]
- Avram, S.; Buiu, C.; Duda-Seiman, D.; Duda-Seiman, C.; Borcan, F.; Mihailescu, D. Evaluation of the Pharmacological Descriptors Related to the Induction of Antidepressant Activity and Its Prediction by QSAR/QRAR Methods. Mini Rev. Med. Chem. 2012, 12, 467–476. [Google Scholar] [CrossRef]
- Avram, S.; Mernea, M.; Limban, C.; Borcan, F.; Chifiriuc, C. Potential Therapeutic Approaches to Alzheimer’s Disease By Bioinformatics, Cheminformatics And Predicted Adme-Tox Tools. Curr. Neuropharmacol. 2020, 18, 696–719. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic Anhydrase Activators. Future Med. Chem. 2018, 10, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic Anhydrase Inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 3467–3474. [Google Scholar] [CrossRef]
- Avram, S.; Milac, A.L.; Carta, F.; Supuran, C.T. More Effective Dithiocarbamate Derivatives Inhibiting Carbonic Anhydrases, Generated by QSAR and Computational Design. J. Enzyme Inhib. Med. Chem. 2013, 28, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Mishra, C.B.; Tiwari, M.; Supuran, C.T. Progress in the Development of Human Carbonic Anhydrase Inhibitors and Their Pharmacological Applications: Where Are We Today? Med. Res. Rev. 2020, 40, 2485–2565. [Google Scholar] [CrossRef]
- Ciccone, L.; Cerri, C.; Nencetti, S.; Orlandini, E. Carbonic Anhydrase Inhibitors and Epilepsy: State of the Art and Future Perspectives. Molecules 2021, 26, 6380. [Google Scholar] [CrossRef] [PubMed]
- Kida, E.; Palminiello, S.; Golabek, A.A.; Walus, M.; Wierzba-Bobrowicz, T.; Rabe, A.; Albertini, G.; Wisniewski, K.E. Carbonic Anhydrase II in the Developing and Adult Human Brain. J. Neuropathol. Exp. Neurol. 2006, 65, 664–674. [Google Scholar] [CrossRef]
- Aspatwar, A.; Tolvanen, M.E.E.; Ortutay, C.; Parkkila, S. Carbonic Anhydrase Related Proteins: Molecular Biology and Evolution. Subcell. Biochem. 2014, 75, 135–156. [Google Scholar] [CrossRef]
- Mishra, C.B.; Kumari, S.; Angeli, A.; Bua, S.; Tiwari, M.; Supuran, C.T. Discovery of Benzenesulfonamide Derivatives as Carbonic Anhydrase Inhibitors with Effective Anticonvulsant Action: Design, Synthesis, and Pharmacological Evaluation. J. Med. Chem. 2018, 61, 3151–3165. [Google Scholar] [CrossRef]
- Balestri, F.; Moschini, R.; Mura, U.; Cappiello, M.; Corso, A.D. In Search of Differential Inhibitors of Aldose Reductase. Biomolecules 2022, 12, 485. [Google Scholar] [CrossRef]
- Veeresham, C.; Rama Rao, A.; Asres, K. Aldose Reductase Inhibitors of Plant Origin. Phytother. Res. PTR 2014, 28, 317–333. [Google Scholar] [CrossRef]
- Grewal, A.S.; Bhardwaj, S.; Pandita, D.; Lather, V.; Sekhon, B.S. Updates on Aldose Reductase Inhibitors for Management of Diabetic Complications and Non-Diabetic Diseases. Mini Rev. Med. Chem. 2016, 16, 120–162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-K.; Liu, C.-C.; Wang, S.; Cheng, H.-C.; Meadows, C.; Chang, K.-C. The Role of Aldose Reductase in Beta-Amyloid-Induced Microglia Activation. Int. J. Mol. Sci. 2022, 23, 15088. [Google Scholar] [CrossRef] [PubMed]
- SIB Swiss Institute of Bioinformatics | Expasy. Available online: https://www.expasy.org/ (accessed on 8 February 2023).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef]
- Kesarwani, K.; Gupta, R. Bioavailability Enhancers of Herbal Origin: An Overview. Asian Pac. J. Trop. Biomed. 2013, 3, 253–266. [Google Scholar] [CrossRef]
- Nistorescu, S.; Gradisteanu Pircalabioru, G.; Udrea, A.-M.; Simon, A.; Pascu, M.L.; Chifiriuc, M.-C. Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices. Coatings 2020, 10, 1230. [Google Scholar] [CrossRef]
- Udrea, A.-M.; Dinache, A.; Pagès, J.-M.; Pirvulescu, R.A. Quinazoline Derivatives Designed as Efflux Pump Inhibitors: Molecular Modeling and Spectroscopic Studies. Molecules 2021, 26, 2374. [Google Scholar] [CrossRef]
- Pervin, M.; Unno, K.; Takagaki, A.; Isemura, M.; Nakamura, Y. Function of Green Tea Catechins in the Brain: Epigallocatechin Gallate and Its Metabolites. Int. J. Mol. Sci. 2019, 20, E3630. [Google Scholar] [CrossRef]
- Supuran, C.T. Coumarin Carbonic Anhydrase Inhibitors from Natural Sources. J. Enzyme Inhib. Med. Chem. 2020, 35, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the Chemical Beauty of Drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Avram, S.; Mernea, M.; Borcan, F.; Mihailescu, D. Evaluation of the Therapeutic Properties of Mastoparan- and Sifuvirtide- Derivative Antimicrobial Peptides Using Chemical Structure-Function Relationship—in Vivo and in Silico Approaches. Curr. Drug Deliv. 2016, 13, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Moret, M.; Pachon Angona, I.; Cotos, L.; Yan, S.; Atz, K.; Brunner, C.; Baumgartner, M.; Grisoni, F.; Schneider, G. Leveraging Molecular Structure and Bioactivity with Chemical Language Models for de Novo Drug Design. Nat. Commun. 2023, 14, 114. [Google Scholar] [CrossRef]
- Dumitrascu, F.; Udrea, A.-M.; Caira, M.R.; Nuta, D.C.; Limban, C.; Chifiriuc, M.C.; Popa, M.; Bleotu, C.; Hanganu, A.; Dumitrescu, D.; et al. In Silico and Experimental Investigation of the Biological Potential of Some Recently Developed Carprofen Derivatives. Molecules 2022, 27, 2722. [Google Scholar] [CrossRef] [PubMed]
- Putz, M.V.; Duda-Seiman, C.; Duda-Seiman, D.; Putz, A.-M.; Alexandrescu, I.; Mernea, M.; Avram, S. Chemical Structure-Biological Activity Models for Pharmacophores’ 3D-Interactions. Int. J. Mol. Sci. 2016, 17, 1087. [Google Scholar] [CrossRef]
- Caron, G.; Digiesi, V.; Solaro, S.; Ermondi, G. Flexibility in Early Drug Discovery: Focus on the beyond-Rule-of-5 Chemical Space. Drug Discov. Today 2020, 25, 621–627. [Google Scholar] [CrossRef]
- Raevsky, O.A.; Grigorev, V.Y.; Polianczyk, D.E.; Raevskaja, O.E.; Dearden, J.C. Aqueous Drug Solubility: What Do We Measure, Calculate and QSPR Predict? Mini Rev. Med. Chem. 2019, 19, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Vraka, C.; Mijailovic, S.; Fröhlich, V.; Zeilinger, M.; Klebermass, E.-M.; Wadsak, W.; Wagner, K.-H.; Hacker, M.; Mitterhauser, M. Expanding LogP: Present Possibilities. Nucl. Med. Biol. 2018, 58, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Chomicki, D.; Kharchenko, O.; Skowronski, L.; Kowalonek, J.; Kozanecka-Szmigiel, A.; Szmigiel, D.; Smokal, V.; Krupka, O.; Derkowska-Zielinska, B. Physico-Chemical and Light-Induced Properties of Quinoline Azo-Dyes Polymers. Int. J. Mol. Sci. 2020, 21, 5755. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Udrea, A.M.; Gradisteanu Pircalabioru, G.; Boboc, A.A.; Mares, C.; Dinache, A.; Mernea, M.; Avram, S. Advanced Bioinformatics Tools in the Pharmacokinetic Profiles of Natural and Synthetic Compounds with Anti-Diabetic Activity. Biomolecules 2021, 11, 1692. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Udrea, A.M.; Negrea, A.; Ciopec, M.; Duteanu, N.; Postolache, C.; Duda-Seiman, C.; Duda-Seiman, D.; Shaposhnikov, S. Prevention of Deficit in Neuropsychiatric Disorders through Monitoring of Arsenic and Its Derivatives as Well as Through Bioinformatics and Cheminformatics. Int. J. Mol. Sci. 2019, 20, E1804. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Mernea, M.; Mihailescu, D.; Duda-Seiman, D.; Duda-Seiman, C. Advanced QSAR Methods Evaluated Polycyclic Aromatic Compounds Duality as Drugs and Inductors in Psychiatric Disorders. Curr. Org. Chem. 2013, 17, 2880–2890. [Google Scholar] [CrossRef]
- Hikino, H.; Konno, C.; Takata, H.; Yamada, Y.; Yamada, C.; Ohizumi, Y.; Sugio, K.; Fujimura, H. Antiinflammatory Principles of Aconitum Roots. J. Pharmacobiodyn. 1980, 3, 514–525. [Google Scholar] [CrossRef]
- Bai, L.; Li, X.; He, L.; Zheng, Y.; Lu, H.; Li, J.; Zhong, L.; Tong, R.; Jiang, Z.; Shi, J.; et al. Antidiabetic Potential of Flavonoids from Traditional Chinese Medicine: A Review. Am. J. Chin. Med. 2019, 47, 933–957. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Negrea, E.; Oancea, P.; Leonties, A.; Ana Maria, U.; Avram, S.; Raducan, A. Spectroscopic Studies on Binding of Ibuprofen and Drotaverine with Bovine Serum Albumin. J. Photochem. Photobiol. Chem. 2023, 438, 114512. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE). 2022.02 Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. 2022. Available online: https://www.chemcomp.com/Research-Citing_MOE.htm (accessed on 15 May 2023).
- Open Babel: An Open Chemical Toolbox | Journal of Cheminformatics | Full Text. Available online: https://jcheminf.biomedcentral.com/articles/10.1186/1758-2946-3-33 (accessed on 16 March 2022).
- Harrison, D.H.; Bohren, K.M.; Ringe, D.; Petsko, G.A.; Gabbay, K.H. An Anion Binding Site in Human Aldose Reductase: Mechanistic Implications for the Binding of Citrate, Cacodylate, and Glucose 6-Phosphate. Biochemistry 1994, 33, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinforma. 2008, 24, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, X.; Hu, J.; Xu, X.; Zuo, Y.; Wang, Y.; Ding, J.; Xu, H.; Zhu, S. Aconitine Induces Apoptosis in H9c2 Cardiac Cells via Mitochondria-mediated Pathway. Mol. Med. Rep. 2018, 17, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, J.; Gupta, N.; Agrawal, M.; Bala Bhaskar, A.S.; Lakshmana Rao, P.V. Modulation of ROS/MAPK Signaling Pathways by Okadaic Acid Leads to Cell Death via, Mitochondrial Mediated Caspase-Dependent Mechanism. Apoptosis Int. J. Program. Cell Death 2011, 16, 145–161. [Google Scholar] [CrossRef]
- Anjum, F.; Ali, F.; Mohammad, T.; Shafie, A.; Akhtar, O.; Abdullaev, B.; Hassan, I. Discovery of Natural Compounds as Potential Inhibitors of Human Carbonic Anhydrase II: An Integrated Virtual Screening, Docking, and Molecular Dynamics Simulation Study. Omics J. Integr. Biol. 2021, 25, 513–524. [Google Scholar] [CrossRef]
- Taslimi, P.; Caglayan, C.; Gulcin, İ. The Impact of Some Natural Phenolic Compounds on Carbonic Anhydrase, Acetylcholinesterase, Butyrylcholinesterase, and α-Glycosidase Enzymes: An Antidiabetic, Anticholinergic, and Antiepileptic Study. J. Biochem. Mol. Toxicol. 2017, 31. [Google Scholar] [CrossRef]
- Aggul, A.G.; Uzun, N.; Kuzu, M.; Taslimi, P.; Gulcin, I. Some Phenolic Natural Compounds as Carbonic Anhydrase Inhibitors: An in Vitro and in Silico Study. Arch. Pharm. (Weinheim) 2022, 355, e2100476. [Google Scholar] [CrossRef]
- Sethi, K.K.; Sahoo, S.K.; Pichikala, J.N.; Suresh, P. Carbonic Anhydrase I and II Inhibition with Natural Products: Caffeine and Piperine. J. Enzyme Inhib. Med. Chem. 2012, 27, 97–100. [Google Scholar] [CrossRef]
- BIOVIA. Dassault Systèmes, [Discovery Studio Visualizer], [V21.1.0.20298]; Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- Chahal, V.; Kakkar, R. A Combination Strategy of Structure-Based Virtual Screening, MM-GBSA, Cross Docking, Molecular Dynamics and Metadynamics Simulations Used to Investigate Natural Compounds as Potent and Specific Inhibitors of Tumor Linked Human Carbonic Anhydrase IX. J. Biomol. Struct. Dyn. 2022, 23, 1–16. [Google Scholar] [CrossRef]


| Compound | Flexibility | Refractivity | TPSA | Hydrophobicity | Solubility |
|---|---|---|---|---|---|
| Aconitine | 11 | 165.5 | 153.45 | 3.61 | −3.39 |
| Hypaconitine | 10 | 159.53 | 133.22 | 4.3 | −3.65 |
| Mesaconitine | 10 | 160.69 | 153.45 | 3.56 | −3.14 |
| Magnoflorine | 2 | 101.87 | 58.92 | −0.66 | −3.91 |
| Gallic acid | 6 | 119.15 | 64.99 | 3.97 | −6.25 |
| Catechin | 1 | 74.33 | 110.38 | 1.47 | −2.22 |
| Ferulic acid | 3 | 51.63 | 66.76 | 1.62 | −2.11 |
| Caffeic acid | 2 | 47.16 | 77.76 | 0.97 | −1.89 |
| Chlorogenic acid | 5 | 83.5 | 164.75 | 0.96 | −1.62 |
| Epicatechin | 1 | 74.33 | 110.38 | 1.47 | −2.22 |
| Delphinidin | 1 | 75.26 | 134.19 | 1.35 | −2.35 |
| Coumaric acid | 2 | 45.13 | 57.53 | 0.95 | −2.02 |
| Daidzein | 1 | 71.97 | 70.67 | 1.77 | −3.53 |
| Hyperoside | 4 | 110.16 | 210.51 | 2.11 | −3.04 |
| Rutin | 6 | 141.38 | 269.43 | 1.58 | −3.3 |
| Naringin | 6 | 134.91 | 225.06 | 2.38 | −2.98 |
| Malvidin | 3 | 87.13 | 112.52 | −1.96 | −3.6 |
| Quercetin | 1 | 78.03 | 131.36 | 1.63 | −3.16 |
| Naringenin | 1 | 71.57 | 86.99 | 1.75 | −3.49 |
| Genistein | 1 | 73.99 | 90.9 | 1.91 | −3.72 |
| Syringic acid | 3 | 48.41 | 75.99 | 1.54 | −1.84 |
| Compound | Intestinal Absorption (Human) | Bioavailability | BBB Permeability | Fraction Unbound (Human) | OCT2 Inhibitor | OCT1 Inhibitor |
|---|---|---|---|---|---|---|
| AC | yes | no | yes | 0.52 | no | no |
| Hypaconitine | yes | no | yes | 0.78 | no | no |
| Mesaconitine | yes | no | yes | 0.53 | no | no |
| Magnoflorine | no | no | yes | 0.19 | no | yes |
| Gallic acid | yes | no | yes | 0.99 | no | no |
| Catechin | yes | no | no | 1.01 | no | no |
| Ferulic acid | yes | yes | yes | 0.72 | no | no |
| Caffeic acid | yes | yes | no | 0.73 | no | no |
| Chlorogenic acid | yes | no | no | 0.63 | no | no |
| Epicatechin | yes | no | no | 1.01 | no | no |
| Delphinidin | yes | yes | no | 0.85 | no | no |
| Coumaric acid | yes | no | no | 0.50 | no | no |
| Daidzein | yes | yes | no | 0.96 | No | no |
| Hyperoside | yes | no | no | 0.79 | no | no |
| Rutin | yes | no | no | 0.96 | no | no |
| Naringin | yes | no | no | 0.73 | no | no |
| Malvidin | yes | no | yes | 0.88 | no | no |
| Quercetin | yes | no | no | 1.16 | no | no |
| Naringenin | yes | no | no | 0.93 | no | no |
| Genistein | yes | no | no | 1.09 | no | no |
| Syringic acid | yes | yes | yes | 0.55 | no | no |
| Compounds | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2C9 Substrate | CYP2D6 Inhibitor | CYP2D6 Substrate | CYP3A4 Inhibitor | CYP3A4 Substrate |
|---|---|---|---|---|---|---|---|---|
| AC | no | no | no | no | no | no | no | yes |
| Hypaconitine | no | no | no | no | no | no | no | yes |
| Mesaconitine | no | no | no | no | no | no | no | yes |
| Magnoflorine | no | no | no | no | no | yes | no | yes |
| Gallic acid | no | yes | yes | no | no | no | no | no |
| Catechin | no | no | no | no | no | yes | no | no |
| ferulic acid | no | no | no | no | no | no | no | no |
| Caffeic acid | no | no | no | no | no | no | no | no |
| Chlorogenic acid | no | no | no | yes | no | no | no | yes |
| Epicatechin | no | no | no | no | no | yes | no | no |
| Delphinidin | yes | yes | yes | no | no | no | yes | no |
| Coumaric acid | no | no | no | no | no | no | no | no |
| Daidzein | yes | yes | yes | no | no | no | no | no |
| Hyperoside | no | no | no | no | no | no | no | yes |
| Rutin | no | no | no | no | no | no | no | yes |
| Naringin | no | no | no | no | no | no | no | yes |
| Malvidin | yes | yes | yes | no | no | no | yes | no |
| Quercetin | yes | no | no | no | no | no | yes | yes |
| Naringenin | yes | yes | yes | no | no | no | yes | no |
| Genistein | yes | yes | yes | no | no | no | yes | no |
| Syringic acid | no | no | no | no | no | no | no | no |
| Compounds | Ames Mutagenesis | Avian Toxicity | Crustacea Aquatic Toxicity | Toxicities Fish | Hepatotoxicity | Mitochondrial Toxicity |
|---|---|---|---|---|---|---|
| AC | no | no | yes | yes | yes | yes |
| Hypaconitine | no | no | yes | yes | no | yes |
| Mesaconitine | no | no | no | yes | yes | yes |
| Magnoflorine | no | no | yes | yes | yes | yes |
| Gallic acid | no | no | no | yes | yes | no |
| Catechin | yes | no | yes | yes | no | yes |
| Ferulic acid | no | no | no | yes | no | no |
| Caffeic acid | no | no | no | yes | no | no |
| Chlorogenic acid | no | no | no | yes | no | yes |
| Epicatechin | yes | no | yes | yes | no | yes |
| Delphinidin | no | no | no | yes | yes | yes |
| Coumaric acid | no | no | no | yes | no | no |
| Daidzein | no | no | no | yes | yes | no |
| Hyperoside | yes | no | no | yes | yes | yes |
| Rutin | yes | no | no | yes | yes | no |
| Naringin | no | no | no | yes | yes | no |
| Malvidin | no | no | yes | yes | yes | yes |
| Quercetin | yes | no | no | yes | yes | yes |
| Naringenin | no | no | no | yes | yes | yes |
| Genistein | no | no | no | yes | yes | yes |
| Syringic acid | no | no | no | yes | no | no |
| Target Name | Target Abbreviation | Binding Probability |
|---|---|---|
| Caffeic acid | ||
| Carbonic anhydrase II | CA2 | 0.72 |
| Arachidonate 5nolipoxygenase | ALOX5 | 0.72 |
| Carbonic anhydrase VII | CA7 | 0.72 |
| Carbonic anhydrase I | CA1 | 0.7 |
| Carbonic anhydrase VI | CA6 | 0.7 |
| Chlorogenic acid | ||
| Aldose reductase | AKR1B1 | 0.87 |
| Aldonoketo reductase family 1 member B10 | AKR1B10 | 0.74 |
| Coumaric acid | ||
| Aldose reductase | AKR1B1 | 1 |
| Carbonic anhydrase II | CA2 | 1 |
| Carbonic anhydrase VII | CA7 | 1 |
| Estrogen receptor beta | ESR2 | 1 |
| Carbonic anhydrase I | CA1 | 1 |
| Carbonic anhydrase III | CA3 | 1 |
| Carbonic anhydrase VI | CA6 | 1 |
| Carbonic anhydrase XII | CA12 | 1 |
| Carbonic anhydrase XIV | CA14 | 1 |
| Carbonic anhydrase IX | CA9 | 1 |
| Carbonic anhydrase IV | CA4 | 1 |
| Carbonic anhydrase VB | CA5B | 1 |
| Carbonic anhydrase VA | CA5A | 1 |
| Ferulic acid | ||
| Carbonic anhydrase II | CA2 | 0.93 |
| Carbonic anhydrase VII | CA7 | 0.93 |
| Carbonic anhydrase I | CA1 | 0.93 |
| Carbonic anhydrase VI | CA6 | 0.93 |
| Carbonic anhydrase XII | CA12 | 0.9 |
| Carbonic anhydrase XIV | CA14 | 0.901 |
| Carbonic anhydrase IX | CA9 | 0.901 |
| Carbonic anhydrase VA | CA5A | 0.9 |
| Gallic acid without statistical significance | ||
| Syringic acid | ||
| Carbonic anhydrase II | CA2 | 1 |
| Carbonic anhydrase VII | CA7 | 1 |
| Carbonic anhydrase I | CA1 | 1 |
| Carbonic anhydrase III | CA3 | 1 |
| Carbonic anhydrase VI | CA6 | 1 |
| Carbonic anhydrase XII | CA12 | 1 |
| Carbonic anhydrase XIV | CA14 | 1 |
| Carbonic anhydrase IX | CA9 | 1 |
| Carbonic anhydrase VA | CA5A | 1 |
| AC without statistical significance | ||
| Daidzein | ||
| Aldehyde dehydrogenase | ALDH2 | 1 |
| Estrogen receptor alpha | ESR1 | 1 |
| Carbonic anhydrase VII | CA7 | 1 |
| Estrogen receptor beta | ESR2 | 1 |
| Carbonic anhydrase XII | CA12 | 1 |
| Carbonic anhydrase IV | CA4 | 1 |
| Delfinidin without statistical significance | ||
| Genistein | ||
| ThromboxanenoA synthase | TBXAS1 | 1 |
| Monoamine oxidase A | MAOA | 1 |
| Epidermal growth factor receptor erbB1 | EGFR | 1 |
| Estrogen receptor alpha | ESR1 | 1 |
| Maltasenoglucoamylase | MGAM | 1 |
| Serotonin 2a (5noHT2a) receptor | HTR2A | 1 |
| Serotonin 2c (5noHT2c) receptor | HTR2C | 1 |
| Adenosine A1 receptor (by homology) | ADORA1 | 1 |
| Estrogen receptor beta | ESR2 | 1 |
| Adenosine A2a receptor | ADORA2A | 1 |
| Estradiol 17nobetanodehydrogenase 1 | HSD17B1 | 1 |
| Estrogennorelated receptor alpha | ESRRA | 1 |
| Estrogennorelated receptor beta | ESRRB | 1 |
| ATPnobinding cassette subnofamily G member 2 | ABCG2 | 1 |
| Hypaconitine | ||
| Dopamine transporter (by homology) | SLC6A3 | 0.093576 |
| HERG | KCNH2 | 0.074565 |
| Hyperoside | ||
| Aldose reductase | AKR1B1 | 1 |
| Carbonic anhydrase II | CA2 | 1 |
| Carbonic anhydrase VII | CA7 | 1 |
| Carbonic anhydrase XII | CA12 | 1 |
| Carbonic anhydrase IV | CA4 | 1 |
| Magnoflorine | ||
| Dopamine D2 receptor | DRD2 | 0.548778 |
| Neuronal acetylcholine receptor; alpha4/beta2 | CHRNA4 CHRNB2 | 0.390156 |
| Dopamine D3 receptor | DRD3 | 0.306473 |
| Malvidin without statistical significance | ||
| Glyoxalase I | GLO1 | 0.128899 |
| Xanthine dehydrogenase | XDH | 0.112748 |
| Mesaconitine without statistical significance | ||
| Naringenin | ||
| Cytochrome P450 19A1 | CYP19A1 | 0.91 |
| Carbonic anhydrase VII | CA7 | 0.91 |
| Multidrug resistance no associated protein 1 | ABCC1 | 0.91 |
| Estradiol 17nobetanodehydrogenase 1 | HSD17B1 | 0.91 |
| Carbonic anhydrase XII | CA12 | 0.91 |
| Test is no specific androgen no binding protein | SHBG | 0.91 |
| Carbonic anhydrase IV | CA4 | 0.91 |
| Cytochrome P450 1B1 | CYP1B1 | 0.9 |
| Carbonyl reductase [NADPH] 1 | CBR1 | 0.9 |
| Naringin | ||
| Cytochrome P450 19A1 | CYP19A1 | 1 |
| Quercetin | ||
| NADPH oxidase 4 | NOX4 | 1 |
| Vasopressin V2 receptor | AVPR2 | 1 |
| Aldose reductase | AKR1B1 | 1 |
| Xanthine dehydrogenase | XDH | 1 |
| Monoamine oxidase A | MAOA | 1 |
| Insulinnolike growth factor I receptor | IGF1R | 1 |
| Rutin | ||
| NeuromedinnoU receptor 2 | NMUR2 | 1 |
| Alphano2a adrenergic receptor | ADRA2A | 1 |
| Adrenergic receptor alphano2 | ADRA2C | 1 |
| Acetylcholinesterase | ACHE | 1 |
| Aldose reductase | AKR1B1 | 0.60286 |
| Compound | LEFEB |
|---|---|
| Quercetin | −7.85 kcal/mol |
| Caffeic acid | −6.21 kcal/mol |
| Chlorogenic acid | −5.81 kcal/mol |
| Coumaric acid | −5.60 kcal/mol |
| Hyperoside | −5.59 kcal/mol |
| Rutin | −4.54 kcal/mol |
| Name | SMILES | Formula | MW (Da) |
|---|---|---|---|
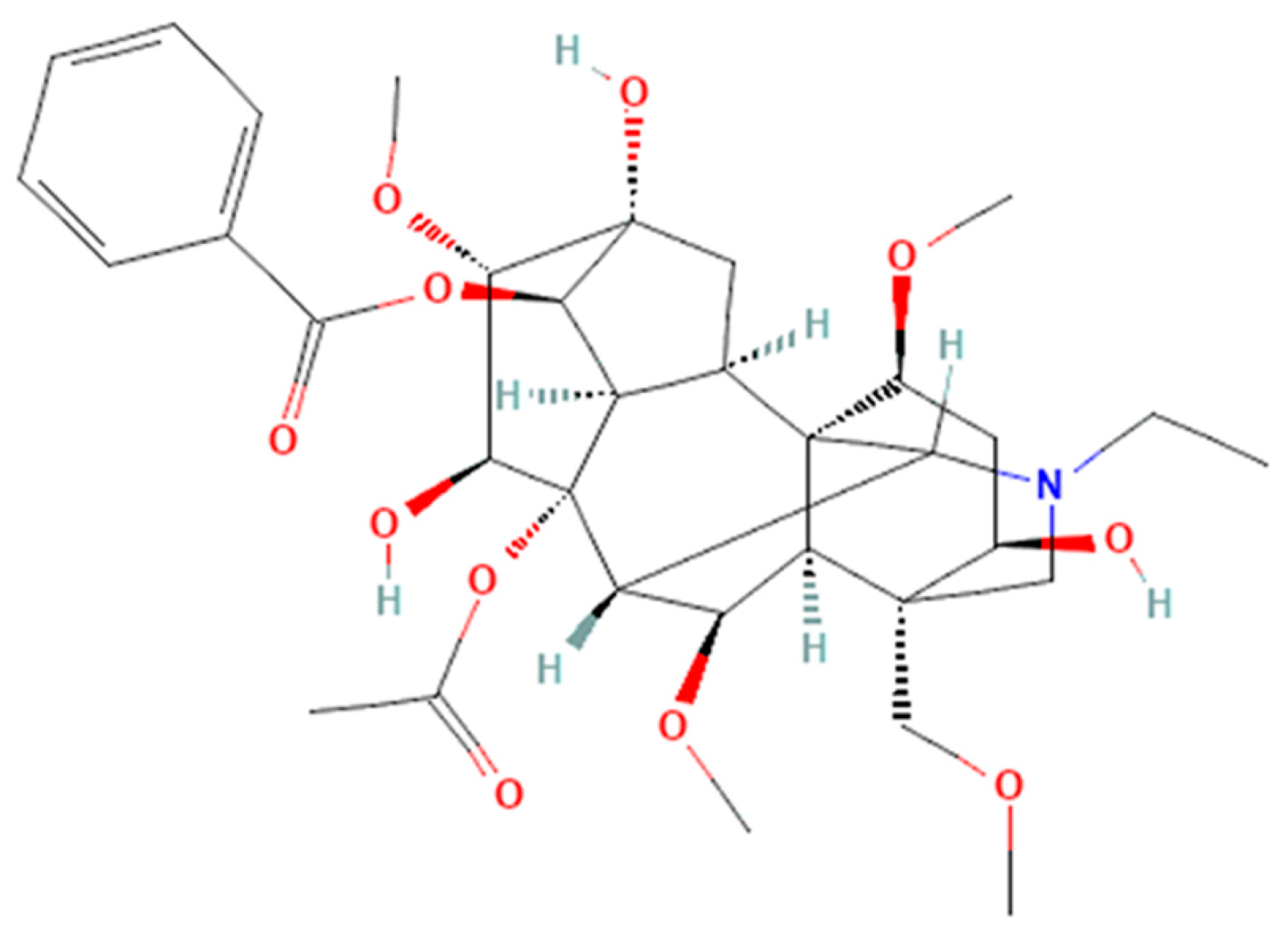
| AC | COCC12CN(CC)C3C4(C2C(OC)C3C2(C3C4CC(C3OC(=O)c3ccccc3)(C(C2O)OC)O)OC(=O)C)C(CC1O)OC | C34H47NO11 | 645.74 |
| Hypaconitine | COCC12CCC(C34C2C(OC)C(C3N(C1)C)C1(C2C4CC(C2OC(=O)c2ccccc2)(C(C1O)OC)O)OC(=O)C)OC | C33H45NO10 | 615.71 |
| Mesaconitine | COCC12CN(C)C3C4(C2C(OC)C3C2(C3C4CC(C3OC(=O)c3ccccc3)(C(C2O)OC)O)OC(=O)C)C(CC1O)OC | C33H45NO11 | 631.71 |
| Magnoflorine | COc1ccc2c(c1O)c1c(O)c(OC)cc3c1C(C2)[N+](C)(C)CC3 | C20H24NO4+ | 342.41 |
| Gallic acid | O=C(c1cc(O)c2c(c1)OC(O2)(c1ccccc1)c1ccccc1)OCc1ccccc1 | C27H20O5 | 424.44 |
| Catechin | Oc1cc2OC(c3ccc(c(c3)O)O)C(Cc2c(c1)O)O | C15H14O6 | 290.27 |
| Ferulic acid | COc1cc(C=CC(=O)O)ccc1O | C10H10O4 | 194.18 |
| Caffeic acid | OC(=O)C=Cc1ccc(c(c1)O)O | C9H8O4 | 180.16 |
| Chlorogenic acid | O=C(OC1CC(O)(CC(C1O)O)C(=O)O)C=Cc1ccc(c(c1)O)O | C16H18O9 | 354.31 |
| Epicatechin | Oc1cc2OC(c3ccc(c(c3)O)O)C(Cc2c(c1)O)O | C15H14O6 | 290.27 |
| Delfinidin | O=c1cc2oc(c3cc(O)c(c(c3)O)[O-])c(cc2c(c1)O)O | C15H9O7- | 301.23 |
| Coumaric acid | OC(=O)C=Cc1ccc(cc1)O | C9H8O3 | 164.16 |
| Daidzein | Oc1ccc(cc1)c1coc2c(c1=O)ccc(c2)O | C15H10O4 | 254.24 |
| Hyperoside | OCC1OC(Oc2c(oc3c(c2=O)c(O)cc(c3)O)c2ccc(c(c2)O)O)C(C(C1O)O)O | C21H20O12 | 464.38 |
| Rutin | Oc1cc(O)c2c(c1)oc(c(c2=O)OC1OC(COC2OC(C)C(C(C2O)O)O)C(C(C1O)O)O)c1ccc(c(c1)O)O | C27H30O16 | 610.52 |
| Naringin | OCC1OC(Oc2cc(O)c3c(c2)OC(CC3=O)c2ccc(cc2)O)C(C(C1O)O) OC1OC(C)C(C(C1O)O)O | C27H32O14 | 580.53 |
| Malvidin | COc1cc(cc(c1O)OC)c1[o+]c2cc(O)cc(c2cc1O)O | C17H15O7+ | 331.3 |
| Quercetin | Oc1cc(O)c2c(c1)oc(c(c2=O)O)c1ccc(c(c1)O)O | C15H10O7 | 302.24 |
| Naringenin | Oc1ccc(cc1)C1CC(=O)c2c(O1)cc(cc2O)O | C15H12O5 | 272.25 |
| Genistein | Oc1ccc(cc1)c1coc2c(c1=O)c(O)cc(c2)O | C15H10O5 | 270.24 |
| Syringic acid | COc1cc(cc(c1O)OC)C(=O)O | C9H10O5 | 198.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mareş, C.; Udrea, A.-M.; Şuţan, N.A.; Avram, S. Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species. Pharmaceuticals 2023, 16, 842. https://doi.org/10.3390/ph16060842
Mareş C, Udrea A-M, Şuţan NA, Avram S. Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species. Pharmaceuticals. 2023; 16(6):842. https://doi.org/10.3390/ph16060842
Chicago/Turabian StyleMareş, Cătălina, Ana-Maria Udrea, Nicoleta Anca Şuţan, and Speranţa Avram. 2023. "Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species" Pharmaceuticals 16, no. 6: 842. https://doi.org/10.3390/ph16060842
APA StyleMareş, C., Udrea, A.-M., Şuţan, N. A., & Avram, S. (2023). Bioinformatics Tools for the Analysis of Active Compounds Identified in Ranunculaceae Species. Pharmaceuticals, 16(6), 842. https://doi.org/10.3390/ph16060842








