Metabolomics and Network Analyses Reveal Phenylalanine and Tyrosine as Signatures of Anthracycline-Induced Hepatotoxicity
Abstract
1. Introduction
2. Results
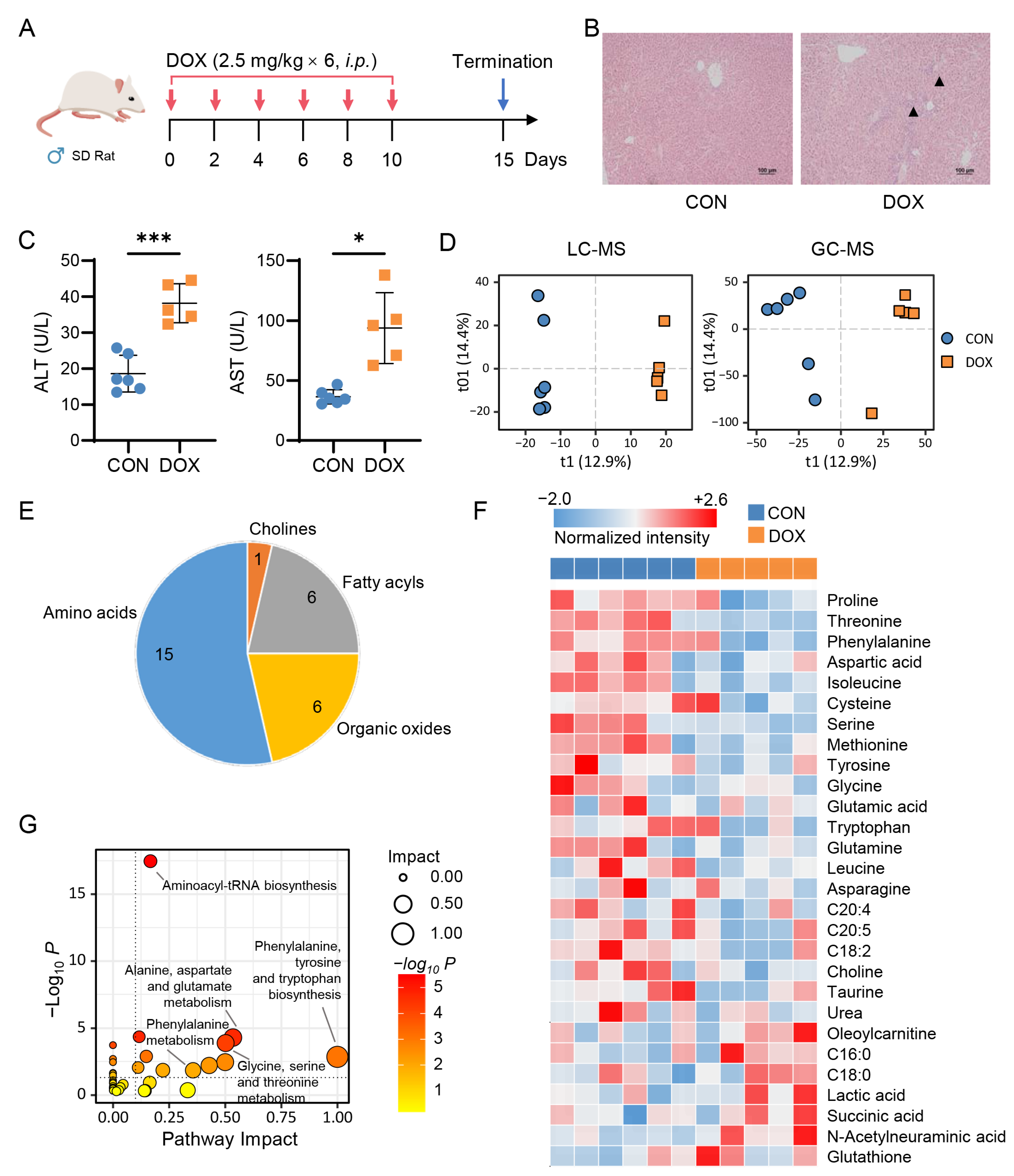
2.1. Metabolic Alteration in Mouse Liver Caused by AIH
2.2. Metabolic Alteration in Rat Liver Caused by AIH
2.3. Network-Based Discovery of AIH-Associated Metabolic Signatures
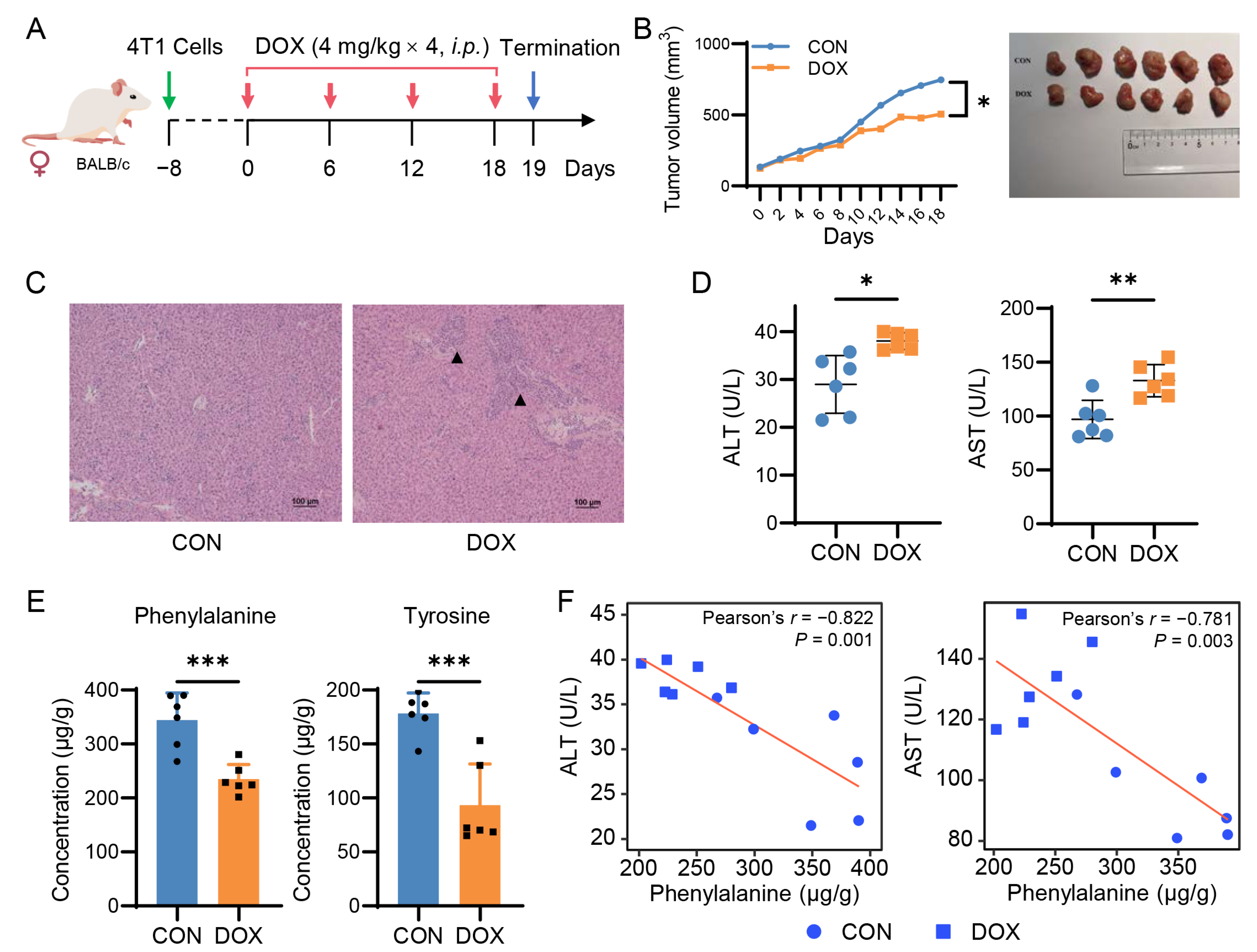
2.4. Doxorubicin Reduces Hepatic Phenylalanine and Tyrosine Levels in Breast Cancer Mice
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Animal Experiments
4.4. Histopathological and Biochemical Analysis
4.5. LC-MS and GC-MS Based Untargeted Metabolomics Analysis
4.6. Identification of Differential Metabolites
4.7. LC-MS/MS Targeted Analysis
4.8. Network Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sallustio, B.C.; Boddy, A.V. Is There Scope for Better Individualisation of Anthracycline Cancer Chemotherapy? Brit. J. Clin. Pharmaco. 2021, 87, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, M.; Guo, Z.; Ripoll, C.V.; Diab, A.; Picataggi, A.; Rawnsley, D.; Lotfinaghsh, A.; Bergom, C.; Szymanski, J.; Hwang, D.; et al. Sustained Alternate-Day Fasting Potentiates Doxorubicin Cardiotoxicity. Cell Metab. 2023. [Google Scholar] [CrossRef] [PubMed]
- Skok, Z.; Zidar, N.; Kikelj, D.; Ilaš, J. Dual Inhibitors of Human DNA Topoisomerase II and Other Cancer-Related Targets. J. Med. Chem. 2020, 63, 884–904. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J. Adverse Cardiac Effects of Cancer Therapies: Cardiotoxicity and Arrhythmia. Nat. Rev. Cardiol. 2020, 17, 474–502. [Google Scholar] [CrossRef]
- Nishiuchi, S.; Yagi, K.; Saito, H.; Zamami, Y.; Niimura, T.; Miyata, K.; Sakamoto, Y.; Fukunaga, K.; Ishida, S.; Hamano, H.; et al. Investigation of Drugs for the Prevention of Doxorubicin-Induced Cardiac Events Using Big Data Analysis. Eur. J. Pharmacol. 2022, 928, 175083. [Google Scholar] [CrossRef]
- Maayah, Z.H.; Takahara, S.; Dyck, J.R.B. The Beneficial Effects of Reducing NLRP3 Inflammasome Activation in the Cardiotoxicity and the Anti-Cancer Effects of Doxorubicin. Arch. Toxicol. 2021, 95, 1–9. [Google Scholar] [CrossRef]
- Bayles, C.E.; Hale, D.E.; Konieczny, A.; Anderson, V.D.; Richardson, C.R.; Brown, K.V.; Nguyen, J.T.; Hecht, J.; Schwartz, N.; Kharel, M.K.; et al. Upcycling the Anthracyclines: New Mechanisms of Action, Toxicology, and Pharmacology. Toxicol. Appl. Pharm. 2022, 459, 116362. [Google Scholar] [CrossRef]
- Damodar, G.; Smitha, T.; Gopinath, S.; Vijayakumar, S.; Rao, Y. An Evaluation of Hepatotoxicity in Breast Cancer Patients Receiving Injection Doxorubicin. Ann. Med. Health Sci. Res. 2014, 4, 74–79. [Google Scholar] [CrossRef]
- Sedeman, M.; Christowitz, C.; de Jager, L.; Engelbrecht, A.-M. Obese Mammary Tumour-Bearing Mice Are Highly Sensitive to Doxorubicin-Induced Hepatotoxicity. BMC Cancer 2022, 22, 1240. [Google Scholar] [CrossRef]
- Ahmed, O.M.; Elkomy, M.H.; Fahim, H.I.; Ashour, M.B.; Naguib, I.A.; Alghamdi, B.S.; Mahmoud, H.U.R.; Ahmed, N.A. Rutin and Quercetin Counter Doxorubicin-Induced Liver Toxicity in Wistar Rats via Their Modulatory Effects on Inflammation, Oxidative Stress, Apoptosis, and Nrf2. Oxid. Med. Cell Longev. 2022, 2022, 2710607. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Edison, T.N.J.I.; Velmurugan, B.K.; Jacob, J.A.; Karuppusamy, I. Toxicity of Doxorubicin (Dox) to Different Experimental Organ Systems. Life Sci. 2018, 200, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, P.L.; Renu, K.; Abilash, V.G. New Molecular and Biochemical Insights of Doxorubicin-Induced Hepatotoxicity. Life Sci. 2020, 250, 117599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, A.; Miao, J.; Sun, H.; Han, Y.; Yan, G.; Wu, F.; Wang, X. Metabolomics Biotechnology, Applications, and Future Trends: A Systematic Review. RSC Adv. 2019, 9, 37245–37257. [Google Scholar] [CrossRef]
- Maron, B.A.; Altucci, L.; Balligand, J.-L.; Baumbach, J.; Ferdinandy, P.; Filetti, S.; Parini, P.; Petrillo, E.; Silverman, E.K.; Barabási, A.-L.; et al. A Global Network for Network Medicine. NPJ Syst. Biol. Appl. 2020, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Quintás, G.; Martínez-Sena, T.; Conde, I.; Ibars, E.P.; Kleinjans, J.; Castell, J.V. Metabolomic Analysis to Discriminate Drug-Induced Liver Injury (DILI) Phenotypes. Arch. Toxicol. 2021, 95, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Timm, K.N.; Ball, V.; Miller, J.J.; Savic, D.; West, J.A.; Griffin, J.L.; Tyler, D.J. Metabolic Effects of Doxorubicin on the Rat Liver Assessed with Hyperpolarized MRI and Metabolomics. Front. Physiol. 2022, 12, 782745. [Google Scholar] [CrossRef]
- Niu, Q.-Y.; Li, Z.-Y.; Du, G.-H.; Qin, X.-M. 1H NMR Based Metabolomic Profiling Revealed Doxorubicin-Induced Systematic Alterations in a Rat Model. J. Pharm. Biomed. Anal. 2016, 118, 338–348. [Google Scholar] [CrossRef]
- Han, Z.; Guo, L.; Yu, X.; Guo, H.; Deng, X.; Yu, J.; Deng, X.; Xu, F.; Zhang, Z.; Huang, Y. Network-Driven Targeted Analysis Reveals that Astragali Radix Alleviates Doxorubicin-Induced Cardiotoxicity by Maintaining Fatty Acid Homeostasis. J. Ethnopharmacol. 2022, 287, 114967. [Google Scholar] [CrossRef]
- Amgalan, D.; Garner, T.P.; Pekson, R.; Jia, X.F.; Yanamandala, M.; Paulino, V.; Liang, F.G.; Corbalan, J.J.; Lee, J.; Chen, Y.; et al. A Small-Molecule Allosteric Inhibitor of BAX Protects against Doxorubicin-Induced Cardiomyopathy. Nat. Cancer 2020, 1, 315–328. [Google Scholar] [CrossRef]
- Oldham, S.; Fulcher, B.; Parkes, L.; Arnatkevičiūtė, A.; Suo, C.; Fornito, A. Consistency and Differences between Centrality Measures across Distinct Classes of Networks. PLoS ONE 2019, 14, e0220061. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A Three-Ring Circus: Metabolism of the Three Proteogenic Aromatic Amino Acids and Their Role in the Health of Plants and Animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Shen, D.; Luo, Y.; Che, Y.-Q. Identification of Exosomal MiRNAs Associated with the Anthracycline-Induced Liver Injury in Postoperative Breast Cancer Patients by Small RNA Sequencing. PeerJ 2020, 8, e9021. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Cui, C.; Wang, C.; Lu, S.; Zhang, M.; Chen, D.; Jiang, P. Systematic Evaluations of Doxorubicin-Induced Toxicity in Rats Based on Metabolomics. ACS Omega 2020, 6, 358–366. [Google Scholar] [CrossRef]
- Wu, L.; Wang, L.; Du, Y.; Zhang, Y.; Ren, J. Mitochondrial Quality Control Mechanisms as Therapeutic Targets in Doxorubicin-Induced Cardiotoxicity. Trends Pharmacol. Sci. 2023, 44, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Hevey, D. Network Analysis: A Brief Overview and Tutorial. Health Psychol. Behav. Med. 2018, 6, 301–328. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Sun, W.-W.; Chen, J.-C.; Zhang, H.-L.; Liu, J.; Lin, Y.; Lin, P.-C.; Wu, B.-X.; An, Y.-P.; Huang, L.; et al. Phenylalanine Impairs Insulin Signaling and Inhibits Glucose Uptake through Modification of IRβ. Nat. Commun. 2022, 13, 4291. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Gastaldelli, A.; Hyötyläinen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P.; et al. Metabolomics and Lipidomics in NAFLD: Biomarkers and Non-Invasive Diagnostic Tests. Nat. Rev. Gastroentero 2021, 18, 835–856. [Google Scholar] [CrossRef]
- Kawanaka, M.; Nishino, K.; Oka, T.; Urata, N.; Nakamura, J.; Suehiro, M.; Kawamoto, H.; Chiba, Y.; Yamada, G. Tyrosine Levels Are Associated with Insulin Resistance in Patients with Nonalcoholic Fatty Liver Disease. Hepatic Med. Evid. Res. 2015, 7, 29–35. [Google Scholar] [CrossRef]
- Sano, A.; Kakazu, E.; Hamada, S.; Inoue, J.; Ninomiya, M.; Iwata, T.; Tsuruoka, M.; Sato, K.; Masamune, A. Steatotic Hepatocytes Release Mature VLDL through Methionine and Tyrosine Metabolism in a Keap1-Nrf2–Dependent Manner. Hepatology 2021, 74, 1271–1286. [Google Scholar] [CrossRef]
- Shen, R.L.; Rathe, M.; Jiang, P.; Pontoppidan, P.E.L.; Heegaard, P.M.H.; Müller, K.; Sangild, P.T. Doxorubicin-Induced Gut Toxicity in Piglets Fed Bovine Milk and Colostrum. J. Pediatr. Gastr. Nutr. 2016, 63, 698–707. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Wu, J.; Yu, X.; Guo, L.; Zhao, L.; Ban, T.; Huang, Y. Metabolomics and Network Analyses Reveal Phenylalanine and Tyrosine as Signatures of Anthracycline-Induced Hepatotoxicity. Pharmaceuticals 2023, 16, 797. https://doi.org/10.3390/ph16060797
Liu P, Wu J, Yu X, Guo L, Zhao L, Ban T, Huang Y. Metabolomics and Network Analyses Reveal Phenylalanine and Tyrosine as Signatures of Anthracycline-Induced Hepatotoxicity. Pharmaceuticals. 2023; 16(6):797. https://doi.org/10.3390/ph16060797
Chicago/Turabian StyleLiu, Peipei, Jing Wu, Xinyue Yu, Linling Guo, Ling Zhao, Tao Ban, and Yin Huang. 2023. "Metabolomics and Network Analyses Reveal Phenylalanine and Tyrosine as Signatures of Anthracycline-Induced Hepatotoxicity" Pharmaceuticals 16, no. 6: 797. https://doi.org/10.3390/ph16060797
APA StyleLiu, P., Wu, J., Yu, X., Guo, L., Zhao, L., Ban, T., & Huang, Y. (2023). Metabolomics and Network Analyses Reveal Phenylalanine and Tyrosine as Signatures of Anthracycline-Induced Hepatotoxicity. Pharmaceuticals, 16(6), 797. https://doi.org/10.3390/ph16060797







